Published online Aug 15, 2001. doi: 10.3748/wjg.v7.i4.583
Revised: March 5, 2001
Accepted: March 12, 2001
Published online: August 15, 2001
AIM: To identify hepatitis C virus (HCV) core protein epitopes recognized by HLA-A2 restricted cytotoxic T lymphocyte (CTL).
METHODS: Utilizing the method of computer prediction followed by a 4 h 51Cr release assay confirmation.
RESULTS: The results showed that peripheral blood mononuclear cells (PBMC) obtained from two HLA-A2 positive donors who were infected with HCV could lyse autologous target cells labeled with peptide “ALAHGVRAL (core 150-158)”. The rates of specific lysis of the cells from the two donors were 37.5% and 15.8%, respectively. Blocking of the CTL response with anti-CD4 mAb caused no significant decrease of the specific lysis. But blocking of CTL response with anti-CD8 mAb could abolish the lysis.
CONCLUSION: The peptide (core 150-158) is the candidate epitope recognized by HLA-A2 restricted CTL.
-
Citation: Zhou HC, Xu DZ, Wang XP, Zhang JX, Ying-Huang, Yan YP, Zhu Y, Jin BQ. Identification of the epitopes on HCV core protein recognized by HLA-A
2 restricted cytotoxic T lymphocytes. World J Gastroenterol 2001; 7(4): 583-586 - URL: https://www.wjgnet.com/1007-9327/full/v7/i4/583.htm
- DOI: https://dx.doi.org/10.3748/wjg.v7.i4.583
Hepatitis C virus (HCV) is a single-stranded RNA virus responsible for the majority non-A non-B hepatitis[1,2]. More than 50%-60% of acute infection lead to chronic disease, and once chronicity is established, spontaneous recovery is exceptional. The related mechanism is still unknown[3-5]. Recent studies demonstrate that major histocompatibility complex (MHC) class I-restricted cytotoxic T lymphocytes (CTL) of patients with chronic hepatitis C recognize epitopes from different regions of both structural and nonstructural HCV proteins[6-12]. Some scholars speculate that CTL-mediated cellular immune response probably plays an important role in viral clearance[13,14].
CD8+ CTL interact through their polymorphic T cell receptor with HLA class I molecules containing endogenously synthesized peptides of 9-11 on the surface of infected cells. The presence of allele-specific amino acid motifs has been demonstrated by sequencing of peptides eluted from MHC molecules. Among the best studied motifs is that of HLA-A2, which is prevalent in a high percentage of population. Several reports[7,8,15-20] described the method of using HCV derived synthetic peptides containing the HLA-A2.1 binding motif to identify and characterize the HLA-A2 restricted CTL in the peripheral blood of patients with chronic HCV infection. We[21] have designed a computer programme to score the reported HCV peptides. Our results revealed that all the reported peptides were with a relative high score of 144 points or higher. Based on the previous study, we attempted to identify the epitopes recognized by the HLA-A2 restricted CTL on HCV core protein utilizing the method of computer prediction followed by 4 h 51Cr release assay.
Subjects Six patients with chronic hepatitis C and 2 healthy controls were selected from among those monitored at Xijing blood center. Table 1 summarizes patient characteristics and history of treatment. All subjects had not received any antiviral treatment for at least one year.
| Subjects | HLA-A | Anti-HCV | HCV-RNA |
| Experiment | |||
| Li | A2A31 | + | + |
| Zhang | A2A11 | + | + |
| Tang | A2A33 | + | + |
| Zhang | A2A11 | + | + |
| Patient control | |||
| Li | A11 | + | + |
| + | A3A33 | + | |
| Health control | |||
| Zheng | A2A11 | - | - |
| Wang | A2A24 | - | - |
HLA typing HLA typing of PBMC from patients and from normal donors was determined by microcytotoxicity, using trays (One lamda, Canoga Park, CA). The HLA haplotypes of subjects participating in this study are shown in Table 1.
Prediction of candidate HLA-A2 restricted CTL epitopes Based on previous study, we use our computer programme to predict HLA-A2 restricted CTL on HCV C protein. In brief, a computer programme with the function of finding peptides containing HLA-A2 allele-specific peptide motif was written in C language. The HCV cDNA is translated into HCV amino acid sequence from which the peptides was chosen, and the selected peptides include those with a length of 9-11 amino acids, a leucine (L), isoleucine (I) or methionine (M) at position 2 and a leucine (L) or Valine (V) at the last position. According to Nijiman’s score system, we scored six points for an anchor residue, four points for a strong and two points for a weak residue. The score for a given peptide is obtained by multiplication of the scores for each amino acid position. Predicted candidate CTL epitopes with scores of 144 or higher.
Synthetic peptides Peptides YLLPRRGPRL (core 35-44), NLGKVIDTL (core 118-126), DLMGYIPLV (core 132-140) and ALAHGVRAL (core 150-158) were selected from the predicted results and synthesized in automated multiple peptide synthesizer (American Research Genetics, Inc). All peptides were > 90% pure and diluted to 1g·L-1 with RPMI1640 medium before use (Gibco, Grand Ialand, N.Y.).
CTL generation PBMC from donors were separated on Ficoll-Hypaque density gradients (Shanghai Huajing, Inc), washed three times in phosphate-buffered saline (PBS), resuspended in RPMI1640 medium (Gibco, BRL.) supplemented with L-glutamine (10 g·L-1 ), penicillin (5 × 104 U·L-1), streptomycin (50 mg·L-1) and Hepes (5 mol·L-1) containing 100 mL·L-1 fetal calf serum (FCS) and plated in 24-well plates at 4 × 106 cells per well. PBMC were stimulated with concanavalin A (ConA, 20 μg per well) during the first week. On d3, 1 mL of complete medium supplemented with rIL-2 at 2 × 103 U·L-1 final concentration was added into each well. On d7, the cultures were restimulated with the peptides plus rIL-2 and irradiated (30Gy) autologous PBMC feeder cells, and the cultured PBMC were restimulated five days later with the original peptides plus rIL-2. On d16, the stimulated cells were used as effectors in CTL assay.
Preparation of autoguous B lymphoblastoid cell line After Ficoll-Hypaque separation, PBMC were suspended in the RPMI1640 medium containing 200 mL·L-1 FCS and then plated in 24-well culture plate at a concentration of 2 × 106 cells per well. EBV-transfected B cell lines were established by culturing 2 × 106 PBMC with 100 g·L-1 of cyclosporin A and 1 mL B95-8 EBV culture supernatant (provided by Dr. Jin, the Fourth Military Medical University, Xi’an). After transformation, the lymphoblastoid cell lines (B-LCL) were maintained in RPMI1640 medium with 200 mL·L-1 FCS, with media change twice each week. The cell lines were maintained at 37 °C in a humidified chamber with 50 mL·L-1 CO2 and used as targets.
CTL assay Target cells were incubated overnight with synthetic peptides at 200 mg·L-1, and then were labeled with 3.7 MBq 1-Cr for 1h and washed three times with PBS. Cytotoxity activity was determined in a standard 4 h Cr release assay using U -bottom 96 welL plates containing 5000 autogenous targets per well. All assays were performed in triplicate with effector target cell (E/T) ratios of 100:1, 50: 1, and 1:1. Maximum release was determined on the basis of lysis of labeled target cells with 50 g·L-1 Triton X-100. We examined spontaneous release by incubating target cells in the absence of effector cells. It was less than 25% of the maximum release. Percent cytotoixity was determined by the formula: 100 × [(experiment release-spontaneous release)/(maximum release-spontaneous release)].
Blocking of CTL response by antibodies CTL responses were tested in the presence of anti-CD8 or anti-CD4 monoclonal antibody added to the 96-well plates at the indicated concentrations used for the CTL assay.
Seven high-scoring peptides (≥ 144 points) were selected from HCV C protein using our computer programme. Among them, peptide ① and peptide ②, namely peptide YLLPRRGPRL and peptide DLMGYIPLV, have been reported to be epitopes recognized by HLA-A2.1 restricted CTL. Predicted peptide ⑦, namely FLLALLSCL (core 177-185) was almost the same as the reported peptide LLALLSCLTV (core 178-187). The rest predicted peptides have not been proved to be epitopes recognized by HLA-A2 restricted CTL. Four peptides (peptide ①, ②, ④, ⑤), were selected randomly from the seven predicted peptides to be used in CTL assay (Table 2).
| No | Peptide sequence | Peptide site | Score |
| ① | YLLPRRGPRL | 35-44 | 144 |
| ② | NLGKVIDTL | 118-126 | 576 |
| ③ | TLTCGFADL | 125-133 | 144 |
| ④ | DLMGYIPLV | 132-140 | 576 |
| ⑤ | ALAHGVRAL | 150-158 | 576 |
| ⑥ | NLPGCSFSIFL | 168-176 | 288 |
| ⑦ | FLLALLSCL | 177-185 | 144 |
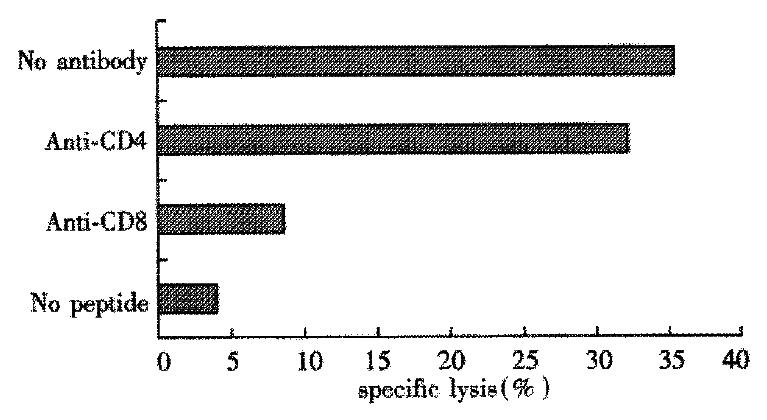
In this experiment PBMC from 8 subjects were stimulated individually with the four peptides from HCV C protein, and cultures were tested after 16d of expansion for peptide-specific CTL activity. Two of the four donors of HLA-A2 and HCV RNA positive responded to peptide ⑤. After induced by peptide ⑤, the two donors’ PBMC can lyse autologous target cells pulsed with peptide ⑤ and the specific lysis was 37.5% and 15.8%, respectively (Table 3). Treatment of the CTL specific for peptide ⑤ with anti-CD8 mAb, but not anti-CD4 mAb, plue complement markedly reduced cytotoxic activity on target cells (Figure 1).
| Subject | HLA type | 51Cr-release (%lysis) | ||||||
| A | B | ① | ② | ④ | ⑤ | |||
| Experiment | ||||||||
| Li | 2 | 31 | 51 | 16 | -2.6 | 9.6 | 0.5 | NT |
| Zhang | 2 | 11 | 8 | 27 | 0.4 | NT | 2.4 | 6.9 |
| Tang | 2 | 33 | 44 | 55 | -3.8 | 5.1 | 3.3 | 37.5 |
| Zhang | 2 | 11 | 62 | 35 | -1.1 | 9.8 | 8.4 | 12.2 |
| Healthy control | ||||||||
| Zheng | 2 | 11 | 62 | 39 | -2.9 | 8.0 | 6.8 | -0.2 |
| Wang | 2 | 24 | 61 | 46 | 0.2 | NT | 3.7 | 10.0 |
| Patient control | ||||||||
| Li | 11 | 62 | 35 | -2.6 | 2.6 | 2.3 | -1.8 | |
| Zhu | 3 | 33 | 17 | 35 | -2.5 | 1.6 | 0 | 2.5 |
CTL mediated cellular immune response probably plays an important role in anti HCV infection. Many researchers reported[22-30] that CTL specific for HCV were discovered in PBMC and liver infiltrated lymphocytes of patients infected with HCV, and that the epitopes recognized by CTL were identified. Owing to the fact that HLA-A2 exhibits a high gene frequency in populations, Cerny et al[7] focused their study on the epitopes recognized by HLA-A3 restricted CTL and have determined several epitopes on every protein region recognized by CTL. However, because of the great work and high cost, it is quite difficult to manipulate in general laboratory. In this study, we attempted to identify the HCV peptides containing HLA-A2 binding motifs, and to confirm the prediction via 4 h 51Cr release assay.
According to the reference[31-32], determining epitopes recognized by CTL included two steps: synthesis of many peptides with multi-peptide overlapping method, and identification of the peptides with experimental means. Up till now, peptides YLLPRRGPRLL (core 35-44), DLMGYIPLV (core 132-140), and LLALLSCLTV (core 178-187) were determined by using this method. The method is direct and reliable, but difficult to manipulate. We analyzed HCV core protein using computer programme. The results demonstrated that there were only 7 peptides with scores of 144 or higher. Of those[21], two peptides, YLLPRRGPRL (core 35-44), DLMGYIPLV (core 132-140) were ever reported in other studies, which could be recognized by HLA-A2 restricted CTL. Another peptide, FLLALLSCL (core 177-185) was in agreement with the reported peptide LLALLSCLTV (core 178-187). Purposeful study of the 7 peptides would simplify experimental processes and save cost. Of those, 4 peptides (Nos ①, ②, ④, ⑤), were synthesized and applied in CTL assay.
Four synthesized peptides of the HCV core protein were tested using CTL assay. Four donors were positive for HLA-A2. Among donors positive for HCV RNA, 2 donors’ PBMC were found to have lysed autologous target cell-labeled with peptide ⑤. The specific lysis rate was 37.5% and 15.8% respectively. The other 3 peptides didn’t show obvious CTL induction action. CTL response was very weak in two healthy and HLA-A2 positive donors, and also in two HCV RNA + HLA-A2- donors.
According to the reference[33], the lysis might be considered specific with the lysis rate ≥ 15%. The specific lysis rate was up to 37.5% in Tang with effector/target cell (E/T) ratio of 50:1, and 15.8% in Zhang with E/T ratio of 100:1. Blocking of the CTL response with anti-CD4 mAb did not decrease the specific lysis significantly. But blocking of the CTL response with anti-CD8 mAb could abolish the lysis. It indicated that[34-40] the lysis was mediated by CD8+ T cells rather than CD4+ T cells, and that the epitope ⑤ was probably the candidate epitope recognized by HLA-A2 restricted CTL.
Although 3 peptides, including 2 reported in other studies, YLLPRRGPRL (core 35-44), DLMGYIPLV (core 132-140), didn’t demonstrate obvious CTL induced activity, we could not draw a conclusion that they were not HLA-A2 restricted CTL recognized epitopes. The two reported epitopes were recognized by HLA-A2.1 restricted CTL, but in this study, we did not determine the HLA-A2 subtypes. Various subtypes of HLA-A2 restricted CTL probably recognized different epitopes[41-50]. Another pssible reason is that HCV protein sequence of HCV-infected patients might not be in complete accordance with the synthesized peptides. To clarify the reasons, further study is still necessary.
Edited by Lu HM
| 1. | Farci P. Choo QL, Kuo G, Weiner AJ, Overby LR, Bradley DW, Houghton M. Isolation of a cDNA clone derived from a blood-borne non-A, non-B viral hepatitis genome [Science 1989; 244: 359-362]. J Hepatol. 2002;36:582-585. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 4996] [Cited by in F6Publishing: 4592] [Article Influence: 131.2] [Reference Citation Analysis (0)] |
| 2. | Major ME, Feinstone SM. The molecular virology of hepatitis C. Hepatology. 1997;25:1527-1538. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 226] [Cited by in F6Publishing: 232] [Article Influence: 8.6] [Reference Citation Analysis (0)] |
| 3. | Meier V, Mihm S, Braun Wietzke P, Ramadori G. HCV-RNA positivity in peripheral blood mononuclear cells of patients with chronic HCV infection: does it really mean viral replication? World J Gastroenterol. 2001;7:228-234. [PubMed] [DOI] [Cited in This Article: ] [Cited by in CrossRef: 20] [Cited by in F6Publishing: 23] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 4. | Allander T, Beyene A, Jacobson SH, Grillner L, Persson MA. Patients infected with the same hepatitis C virus strain display different kinetics of the isolate-specific antibody response. J Infect Dis. 1997;175:26-31. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 28] [Cited by in F6Publishing: 29] [Article Influence: 1.1] [Reference Citation Analysis (0)] |
| 5. | Iwarson S, Norkrans G, Wejstål R. Hepatitis C: natural history of a unique infection. Clin Infect Dis. 1995;20:1361-1370. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 30] [Cited by in F6Publishing: 31] [Article Influence: 1.1] [Reference Citation Analysis (0)] |
| 6. | Schirren CA, Jung MC, Gerlach JT, Worzfeld T, Baretton G, Mamin M, Hubert Gruener N, Houghton M, Pape GR. Liver-derived hepatitis C virus (HCV)-specific CD4(+) T cells recognize multiple HCV epitopes and produce interferon gamma. Hepatology. 2000;32:597-603. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 78] [Cited by in F6Publishing: 80] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 7. | Cerny A, McHutchison JG, Pasquinelli C, Brown ME, Brothers MA, Grabscheid B, Fowler P, Houghton M, Chisari FV. Cytotoxic T lymphocyte response to hepatitis C virus-derived peptides containing the HLA A2.1 binding motif. J Clin Invest. 1995;95:521-530. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 231] [Cited by in F6Publishing: 255] [Article Influence: 8.8] [Reference Citation Analysis (0)] |
| 8. | Battegay M, Fikes J, Di Bisceglie AM, Wentworth PA, Sette A, Celis E, Ching WM, Grakoui A, Rice CM, Kurokohchi K. Patients with chronic hepatitis C have circulating cytotoxic T cells which recognize hepatitis C virus-encoded peptides binding to HLA-A2.1 molecules. J Virol. 1995;69:2462-2470. [PubMed] [Cited in This Article: ] |
| 9. | Kaneko T, Nakamura I, Kita H, Hiroishi K, Moriyama T, Imawari M. Three new cytotoxic T cell epitopes identified within the hepatitis C virus nucleoprotein. J Gen Virol. 1996;77:1305-1309. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 26] [Cited by in F6Publishing: 26] [Article Influence: 0.9] [Reference Citation Analysis (0)] |
| 10. | Pereboeva LA, Pereboev AV, Wang LF, Morris GE. Hepatitis C epitopes from phage-displayed cDNA libraries and improved diagnosis with a chimeric antigen. J Med Virol. 2000;60:144-151. [PubMed] [DOI] [Cited in This Article: ] [Cited by in F6Publishing: 1] [Reference Citation Analysis (0)] |
| 11. | Pereboeva LA, Pereboev AV, Morris GE. Identification of antigenic sites on three hepatitis C virus proteins using phage-displayed peptide libraries. J Med Virol. 1998;56:105-111. [PubMed] [DOI] [Cited in This Article: ] [Cited by in F6Publishing: 1] [Reference Citation Analysis (0)] |
| 12. | Kurokohchi K, Akatsuka T, Pendleton CD, Takamizawa A, Nishioka M, Battegay M, Feinstone SM, Berzofsky JA. Use of recombinant protein to identify a motif-negative human cytotoxic T-cell epitope presented by HLA-A2 in the hepatitis C virus NS3 region. J Virol. 1996;70:232-240. [PubMed] [Cited in This Article: ] |
| 13. | Hu GJ, Wang RY, Han DS, Alter HJ, Shih JW. Characterization of the humoral and cellular immune responses against hepatitis C virus core induced by DNA-based immunization. Vaccine. 1999;17:3160-3170. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 35] [Cited by in F6Publishing: 41] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 14. | Nelson DR, Marousis CG, Ohno T, Davis GL, Lau JY. Intrahepatic hepatitis C virus-specific cytotoxic T lymphocyte activity and response to interferon alfa therapy in chronic hepatitis C. Hepatology. 1998;28:225-230. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 103] [Cited by in F6Publishing: 110] [Article Influence: 4.2] [Reference Citation Analysis (0)] |
| 15. | Caselmann WH, Serwe M, Lehmann T, Ludwig J, Sproat BS, Engels JW. Design, delivery and efficacy testing of therapeutic nucleic acidsused to inhibit hepatitis C virus gene expression in vitro and in vivo. World J Gastroenterol. 2000;6:626-629. [PubMed] [Cited in This Article: ] |
| 16. | Penna A, Chisari FV, Bertoletti A, Missale G, Fowler P, Giuberti T, Fiaccadori F, Ferrari C. Cytotoxic T lymphocytes recognize an HLA-A2-restricted epitope within the hepatitis B virus nucleocapsid antigen. J Exp Med. 1991;174:1565-1570. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 268] [Cited by in F6Publishing: 286] [Article Influence: 8.7] [Reference Citation Analysis (0)] |
| 17. | Nijman HW, Houbiers JG, Vierboom MP, van der Burg SH, Drijfhout JW, D'Amaro J, Kenemans P, Melief CJ, Kast WM. Identification of peptide sequences that potentially trigger HLA-A2.1-restricted cytotoxic T lymphocytes. Eur J Immunol. 1993;23:1215-1219. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 151] [Cited by in F6Publishing: 159] [Article Influence: 5.1] [Reference Citation Analysis (0)] |
| 18. | Shirai M, Arichi T, Nishioka M, Nomura T, Ikeda K, Kawanishi K, Engelhard VH, Feinstone SM, Berzofsky JA. CTL responses of HLA-A2.1-transgenic mice specific for hepatitis C viral peptides predict epitopes for CTL of humans carrying HLA-A2.1. J Immunol. 1995;154:2733-2742. [PubMed] [Cited in This Article: ] |
| 19. | Parker KC, Bednarek MA, Hull LK, Utz U, Cunningham B, Zweerink HJ, Biddison WE, Coligan JE. Sequence motifs important for peptide binding to the human MHC class I molecule, HLA-A2. J Immunol. 1992;149:3580-3587. [PubMed] [Cited in This Article: ] |
| 20. | Botarelli P, Brunetto MR, Minutello MA, Calvo P, Unutmaz D, Weiner AJ, Choo QL, Shuster JR, Kuo G, Bonino F. T-lymphocyte response to hepatitis C virus in different clinical courses of infection. Gastroenterology. 1993;104:580-587. [PubMed] [Cited in This Article: ] |
| 21. | Zhou HC, Dong MQ, Xu DZ, Yan YP, Zhang JX, Song HB, Li YG. Prediction of hepatitis C virus peptides binding to HLA-A2 molecule using computer. Xibao Yu Fenzi Mianyixue Zazhi. 1996;12:41-45. [Cited in This Article: ] |
| 22. | Cohard M, Liu Q, Perkus M, Gordon E, Brotman B, Prince AM. Hepatitis C virus-specific CTL responses in PBMC from chimpanzees with chronic hepatitis C: determination of CTL and CTL precursor frequencies using a recombinant canarypox virus (ALVAC). J Immunol Methods. 1998;214:121-129. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 9] [Cited by in F6Publishing: 9] [Article Influence: 0.3] [Reference Citation Analysis (0)] |
| 23. | Diepolder HM, Gerlach JT, Zachoval R, Hoffmann RM, Jung MC, Wierenga EA, Scholz S, Santantonio T, Houghton M, Southwood S. Immunodominant CD4+ T-cell epitope within nonstructural protein 3 in acute hepatitis C virus infection. J Virol. 1997;71:6011-6019. [PubMed] [Cited in This Article: ] |
| 24. | Hoffmann RM, Diepolder HM, Zachoval R, Zwiebel FM, Jung MC, Scholz S, Nitschko H, Riethmüller G, Pape GR. Mapping of immunodominant CD4+ T lymphocyte epitopes of hepatitis C virus antigens and their relevance during the course of chronic infection. Hepatology. 1995;21:632-638. [PubMed] [DOI] [Cited in This Article: ] |
| 25. | Martins CL, Lawman MJ, Scholl T, Mebus CA, Lunney JK. African swine fever virus specific porcine cytotoxic T cell activity. Arch Virol. 1993;129:211-225. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 51] [Cited by in F6Publishing: 55] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 26. | Erickson AL, Houghton M, Choo QL, Weiner AJ, Ralston R, Muchmore E, Walker CM. Hepatitis C virus-specific CTL responses in the liver of chimpanzees with acute and chronic hepatitis C. J Immunol. 1993;151:4189-4199. [PubMed] [Cited in This Article: ] |
| 27. | Bjorkman PJ, Saper MA, Samraoui B, Bennett WS, Strominger JL, Wiley DC. Structure of the human class I histocompatibility antigen, HLA-A2. Nature. 1987;329:506-512. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 2545] [Cited by in F6Publishing: 2451] [Article Influence: 66.2] [Reference Citation Analysis (0)] |
| 28. | Vidalin O, Tanaka E, Spengler U, Trépo C, Inchauspé G. Targeting of hepatitis C virus core protein for MHC I or MHC II presentation does not enhance induction of immune responses to DNA vaccination. DNA Cell Biol. 1999;18:611-621. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 45] [Cited by in F6Publishing: 48] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 29. | Koziel MJ, Dudley D, Afdhal N, Grakoui A, Rice CM, Choo QL, Houghton M, Walker BD. HLA class I-restricted cytotoxic T lymphocytes specific for hepatitis C virus. Identification of multiple epitopes and characterization of patterns of cytokine release. J Clin Invest. 1995;96:2311-2321. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 221] [Cited by in F6Publishing: 232] [Article Influence: 8.0] [Reference Citation Analysis (0)] |
| 30. | Yan FM, Chen AS, Hao F, Zhao XP, Gu CH, Zhao LB, Yang DL, Hao LJ. Hepatitis C virus may infect extrahepatic tissues in patients with hepatitis C. World J Gastroenterol. 2000;6:805-811. [PubMed] [Cited in This Article: ] |
| 31. | Kita H, Moriyama T, Kaneko T, Harase I, Nomura M, Miura H, Nakamura I, Yazaki Y, Imawari M. HLA B44-restricted cytotoxic T lymphocytes recognizing an epitope on hepatitis C virus nucleocapsid protein. Hepatology. 1993;18:1039-1044. [PubMed] [DOI] [Cited in This Article: ] |
| 32. | Schupper H, Hayashi P, Scheffel J, Aceituno S, Paglieroni T, Holland PV, Zeldis JB. Peripheral-blood mononuclear cell responses to recombinant hepatitis C virus antigens in patients with chronic hepatitis C. Hepatology. 1993;18:1055-1060. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 32] [Cited by in F6Publishing: 29] [Article Influence: 0.9] [Reference Citation Analysis (0)] |
| 33. | Koziel MJ, Dudley D, Afdhal N, Choo QL, Houghton M, Ralston R, Walker BD. Hepatitis C virus (HCV)-specific cytotoxic T lymphocytes recognize epitopes in the core and envelope proteins of HCV. J Virol. 1993;67:7522-7532. [PubMed] [Cited in This Article: ] |
| 34. | Kakimi K, Kuribayashi K, Iwashiro M, Masuda T, Sakai M, Ling W, Kubo Y, Kobayashi H, Higo K, Seki M. Hepatitis C virus core region: helper T cell epitopes recognized by BALB/c and C57BL/6 mice. J Gen Virol. 1995;76:1205-1214. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 11] [Cited by in F6Publishing: 11] [Article Influence: 0.4] [Reference Citation Analysis (0)] |
| 35. | Song MK, Lee SW, Suh YS, Lee KJ, Sung YC. Enhancement of immunoglobulin G2a and cytotoxic T-lymphocyte responses by a booster immunization with recombinant hepatitis C virus E2 protein in E2 DNA-primed mice. J Virol. 2000;74:2920-2925. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 43] [Cited by in F6Publishing: 46] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 36. | Shirai M, Pendleton CD, Ahlers J, Takeshita T, Newman M, Berzofsky JA. Helper-cytotoxic T lymphocyte (CTL) determinant linkage required for priming of anti-HIV CD8+ CTL in vivo with peptide vaccine constructs. J Immunol. 1994;152:549-556. [PubMed] [Cited in This Article: ] |
| 37. | Calin-Laurens V, Trescol-Biémont MC, Gerlier D, Rabourdin-Combe C. Can one predict antigenic peptides for MHC class I-restricted cytotoxic T lymphocytes useful for vaccination? Vaccine. 1993;11:974-978. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 12] [Cited by in F6Publishing: 12] [Article Influence: 0.4] [Reference Citation Analysis (0)] |
| 38. | Koziel MJ, Dudley D, Wong JT, Dienstag J, Houghton M, Ralston R, Walker BD. Intrahepatic cytotoxic T lymphocytes specific for hepatitis C virus in persons with chronic hepatitis. J Immunol. 1992;149:3339-3344. [PubMed] [Cited in This Article: ] |
| 39. | Berzofsky JA, Ahlers JD, Derby MA, Pendleton CD, Arichi T, Belyakov IM. Approaches to improve engineered vaccines for human immunodeficiency virus and other viruses that cause chronic infections. Immunol Rev. 1999;170:151-172. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 83] [Cited by in F6Publishing: 83] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 40. | Chien DY, Arcangel P, Medina-Selby A, Coit D, Baumeister M, Nguyen S, George-Nascimento C, Gyenes A, Kuo G, Valenzuela P. Use of a novel hepatitis C virus (HCV) major-epitope chimeric polypeptide for diagnosis of HCV infection. J Clin Microbiol. 1999;37:1393-1397. [PubMed] [Cited in This Article: ] |
| 41. | Wang T, Guo H, Wang G. [Long-term evolution of C-terminal of HCV NS3 protein containing the predicted CTL epitopes in two HCV patients]. Zhonghua Ganzangbing Zazhi. 1999;7:207-210. [PubMed] [Cited in This Article: ] |
| 42. | Kamei A, Tamaki S, Taniyama H, Takamura S, Nishimura Y, Kagawa Y, Uno-Furuta S, Kaito M, Kim G, Toda M. Induction of hepatitis C virus-specific cytotoxic T lymphocytes in mice by an intrahepatic inoculation with an expression plasmid. Virology. 2000;273:120-126. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 7] [Cited by in F6Publishing: 8] [Article Influence: 0.3] [Reference Citation Analysis (0)] |
| 43. | Chang KM, Rehermann B, McHutchison JG, Pasquinelli C, Southwood S, Sette A, Chisari FV. Immunological significance of cytotoxic T lymphocyte epitope variants in patients chronically infected by the hepatitis C virus. J Clin Invest. 1997;100:2376-2385. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 253] [Cited by in F6Publishing: 260] [Article Influence: 9.6] [Reference Citation Analysis (0)] |
| 44. | Lee AY, Polakos NK, Otten GR, Ulmer JB, Houghton M, Paliard X. Quantification of the number of cytotoxic T cells specific for an immunodominant HCV-specific CTL epitope primed by DNA immunization. Vaccine. 2000;18:1962-1968. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 14] [Cited by in F6Publishing: 16] [Article Influence: 0.7] [Reference Citation Analysis (0)] |
| 45. | Oseroff C, Sette A, Wentworth P, Celis E, Maewal A, Dahlberg C, Fikes J, Kubo RT, Chesnut RW, Grey HM. Pools of lipidated HTL-CTL constructs prime for multiple HBV and HCV CTL epitope responses. Vaccine. 1998;16:823-833. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 33] [Cited by in F6Publishing: 35] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 46. | Tsai SL, Chen YM, Chen MH, Huang CY, Sheen IS, Yeh CT, Huang JH, Kuo GC, Liaw YF. Hepatitis C virus variants circumventing cytotoxic T lymphocyte activity as a mechanism of chronicity. Gastroenterology. 1998;115:954-965. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 105] [Cited by in F6Publishing: 106] [Article Influence: 4.1] [Reference Citation Analysis (0)] |
| 47. | Alexander J, Guercio MFD, Fikes JD, Chesnut RW, Chisari FV, Chang KM, Appella E, Sette A. Recognition of a novel natu-rally processed, A2 restricted, HCV-NS4 epitope triggers IFN-gamma release in absence of detectable cytopathicity. Human Immunol. 1998;59:776-782. [DOI] [Cited in This Article: ] [Cited by in Crossref: 21] [Cited by in F6Publishing: 21] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 48. | Mondelli MU, Cerino A, Lisa A, Brambilla S, Segagni L, Cividini A, Bissolati M, Missale G, Bellati G, Meola A. Antibody responses to hepatitis C virus hypervariable region 1: evidence for cross-reactivity and immune-mediated sequence variation. Hepatology. 1999;30:537-545. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 51] [Cited by in F6Publishing: 50] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 49. | Nakano I, Fukuda Y, Katano Y, Hayakawa T. Conformational epitopes detected by cross-reactive antibodies to envelope 2 glycoprotein of the hepatitis C virus. J Infect Dis. 1999;180:1328-1333. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 9] [Cited by in F6Publishing: 8] [Article Influence: 0.3] [Reference Citation Analysis (0)] |
| 50. | Zhao XP, Shen HX, Tian DY, Zhang DS, Peng ZH, Yang DL, Hao LJ. Expression and significance of HCV RNA and HCV NS5 antigen in liver tissues of patients with hepatitis C. Shijie Huaren Xiaohua Zazhi. 1999;7:516-518. [Cited in This Article: ] |