Copyright
©The Author(s) 2025.
World J Gastroenterol. Aug 7, 2025; 31(29): 109090
Published online Aug 7, 2025. doi: 10.3748/wjg.v31.i29.109090
Published online Aug 7, 2025. doi: 10.3748/wjg.v31.i29.109090
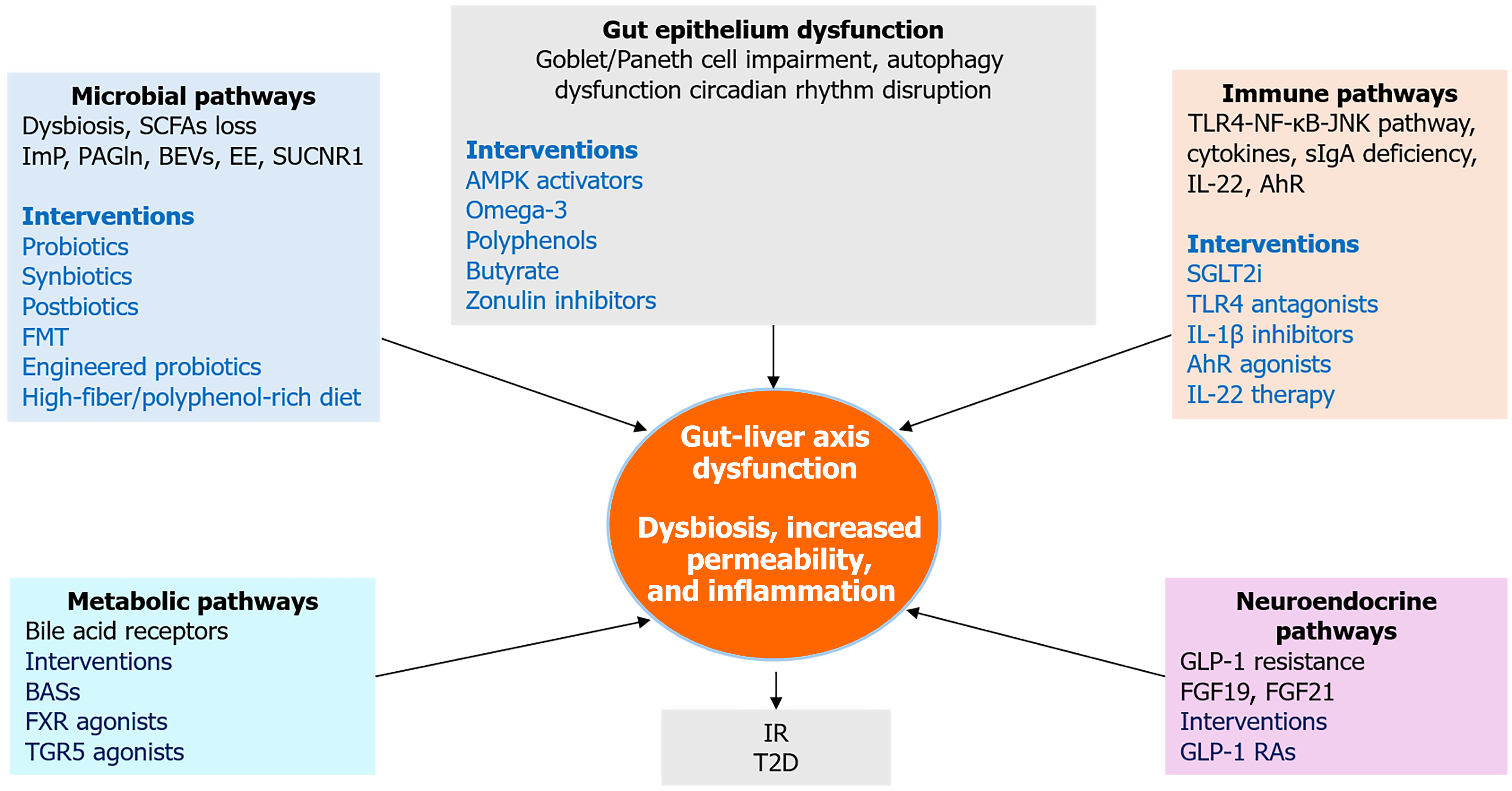
Figure 5 Integrative therapeutic and mechanistic pathways linking gut and liver in type 2 diabetes.
This integrative framework illustrates how five key mechanistic domains, microbial dysbiosis, gut epithelial dysfunction, immune activation, neuroendocrine disruption, and metabolic signaling which converge to drive gut-liver axis dysfunction in type 2 diabetes. Each domain includes representative molecular mediators and therapeutic interventions. The central node highlights dysbiosis, increased permeability, and inflammation as core features linking upstream disruptions to downstream insulin resistance and hepatic steatosis. SCFAs: Short-chain fatty acids; ImP: Imidazole propionate; PAGln: Phenylacetylglutamine; BEVs: Bacterial extracellular vesicles; EE: Endogenous ethanol; FMT: Fecal microbiota transplantation; BASs: Bile acid sequestrants; FXR: Farnesoid X receptor; TGR5: Takeda G protein-coupled receptor 5; AMPK: Adenosine 5’-monophosphate-activated protein kinase; IR: Insulin resistance; T2D: Type 2 diabetes; TLR4: Toll-like receptor 4; JNK: C-Jun N-terminal kinase; NF-κB: Nuclear factor kappa B; sIgA: Secretory immunoglobulin A; IL-1β: Interleukin-1 beta; IL-22: Interleukin-22; AhR: Aryl hydrocarbon receptor; SGLT2i: Sodium-glucose cotransporter-2 inhibitors; FGF19: Fibroblast growth factor 19; FGF21: Fibroblast growth factor 21; GLP-1: Glucagon-like peptide-1; GLP-1 RAs: Glucagon-like peptide-1 receptor agonists.
- Citation: Abdalla MMI. Gut-liver axis in diabetes: Mechanisms and therapeutic opportunities. World J Gastroenterol 2025; 31(29): 109090
- URL: https://www.wjgnet.com/1007-9327/full/v31/i29/109090.htm
- DOI: https://dx.doi.org/10.3748/wjg.v31.i29.109090