Copyright
©The Author(s) 2025.
World J Gastroenterol. Jul 28, 2025; 31(28): 108990
Published online Jul 28, 2025. doi: 10.3748/wjg.v31.i28.108990
Published online Jul 28, 2025. doi: 10.3748/wjg.v31.i28.108990
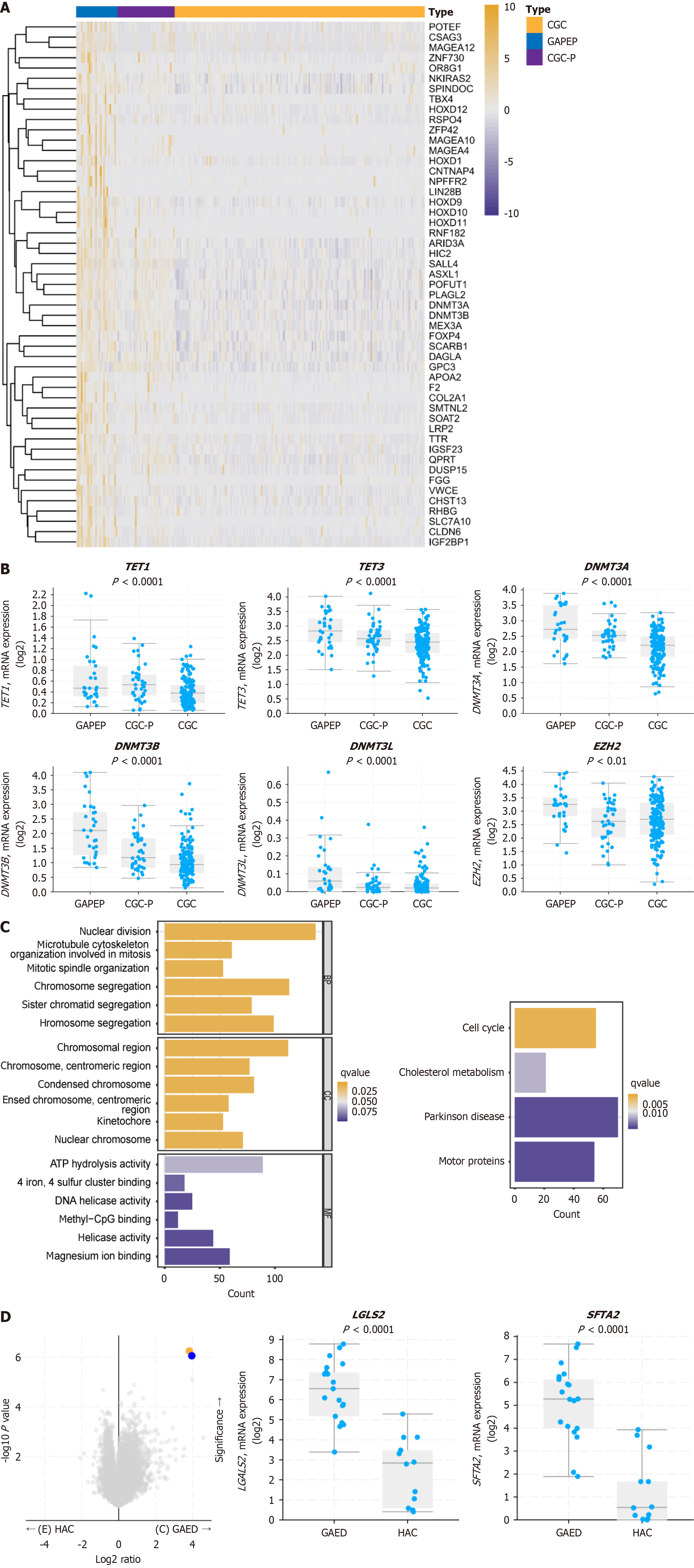
Figure 5 Multi-omics profiling reveals methylation-driven molecular features of gastric adenocarcinoma with primitive enterocyte phe
- Citation: Li HQ, Zheng LQ, Huang WT, Yu XB, Zhang X, Lin L, Lv SS, Yan XY, Chen XY. Clinicopathological significance of histological diversity in gastric adenocarcinoma with primitive enterocyte phenotype: A methylation-driven aggressive entity. World J Gastroenterol 2025; 31(28): 108990
- URL: https://www.wjgnet.com/1007-9327/full/v31/i28/108990.htm
- DOI: https://dx.doi.org/10.3748/wjg.v31.i28.108990