Copyright
©The Author(s) 2025.
World J Gastroenterol. Jul 28, 2025; 31(28): 107361
Published online Jul 28, 2025. doi: 10.3748/wjg.v31.i28.107361
Published online Jul 28, 2025. doi: 10.3748/wjg.v31.i28.107361
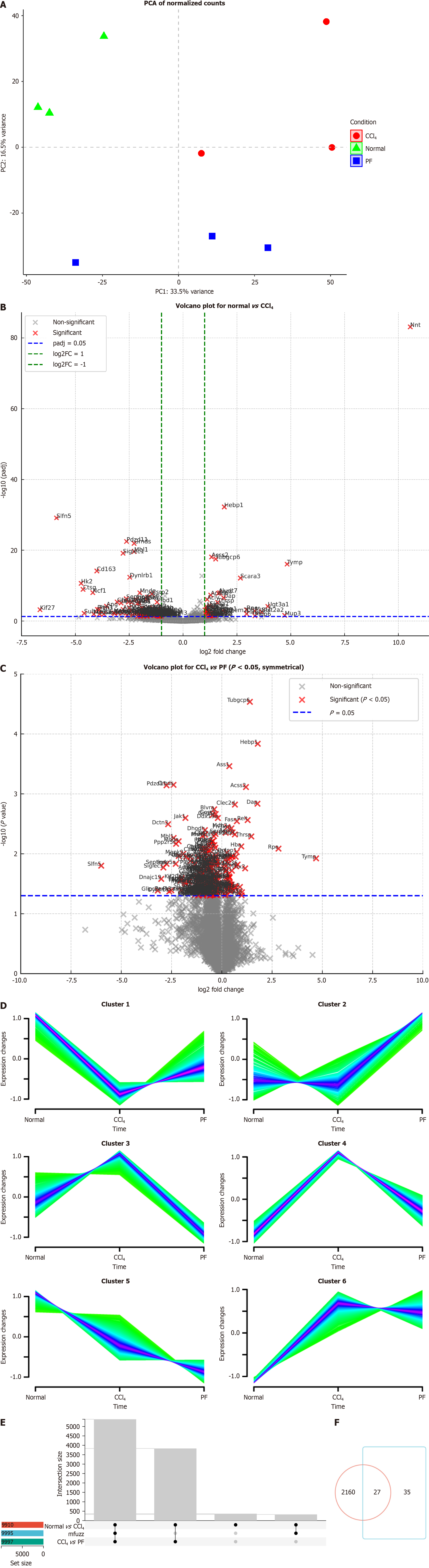
Figure 2 Proteomics analysis of differential protein expression.
A: Principal component analysis (PCA) of normalized protein counts PCA of protein expression across normal, carbon tetrachloride (CCl₄) and PF562271 (PF) conditions. The first principal component (PC1) explains 39.3% of variance, while PC2 explains 9.5%. The three conditions show clear separation; B: Volcano plot showing differentially expressed proteins between the normal and CCl₄ groups. Significant proteins (P < 0.05, log2 fold change > 1 or < -1) are highlighted; C: Volcano plot comparing the CCl₄ and PF groups. Significant proteins are marked, indicating differential expression between these two conditions (P < 0.05); D: Mfuzz clustering of protein expression profiles. Clusters 1, 3, and 4 are shown, highlighting distinct expression trends in normal, CCl₄, and PF conditions; E: UpSet plot showing overlaps of differentially expressed proteins between comparisons (normal vs CCl₄, CCl₄ vs PF) and clusters from Mfuzz analysis; F: Venn diagram showing overlap of differentially expressed proteins between the normal vs CCl₄ and CCl₄ vs PF groups. The diagram indicates unique and shared proteins across the conditions.
- Citation: Wu HY, Han L, Ran T, Sun Y, Zhang QX, Huang T, Zou GL, Zhang Y, Zhou YM, Lin GY, Chen SJ, Wang JL, Pan C, Lu F, Pu HF, Zhao XK. FBP1 as a key regulator of focal adhesion kinase-mediated hepatic stellate cell activation: Multi-omics and experimental validation. World J Gastroenterol 2025; 31(28): 107361
- URL: https://www.wjgnet.com/1007-9327/full/v31/i28/107361.htm
- DOI: https://dx.doi.org/10.3748/wjg.v31.i28.107361