Copyright
©The Author(s) 2023.
World J Gastroenterol. Dec 14, 2023; 29(46): 6060-6075
Published online Dec 14, 2023. doi: 10.3748/wjg.v29.i46.6060
Published online Dec 14, 2023. doi: 10.3748/wjg.v29.i46.6060
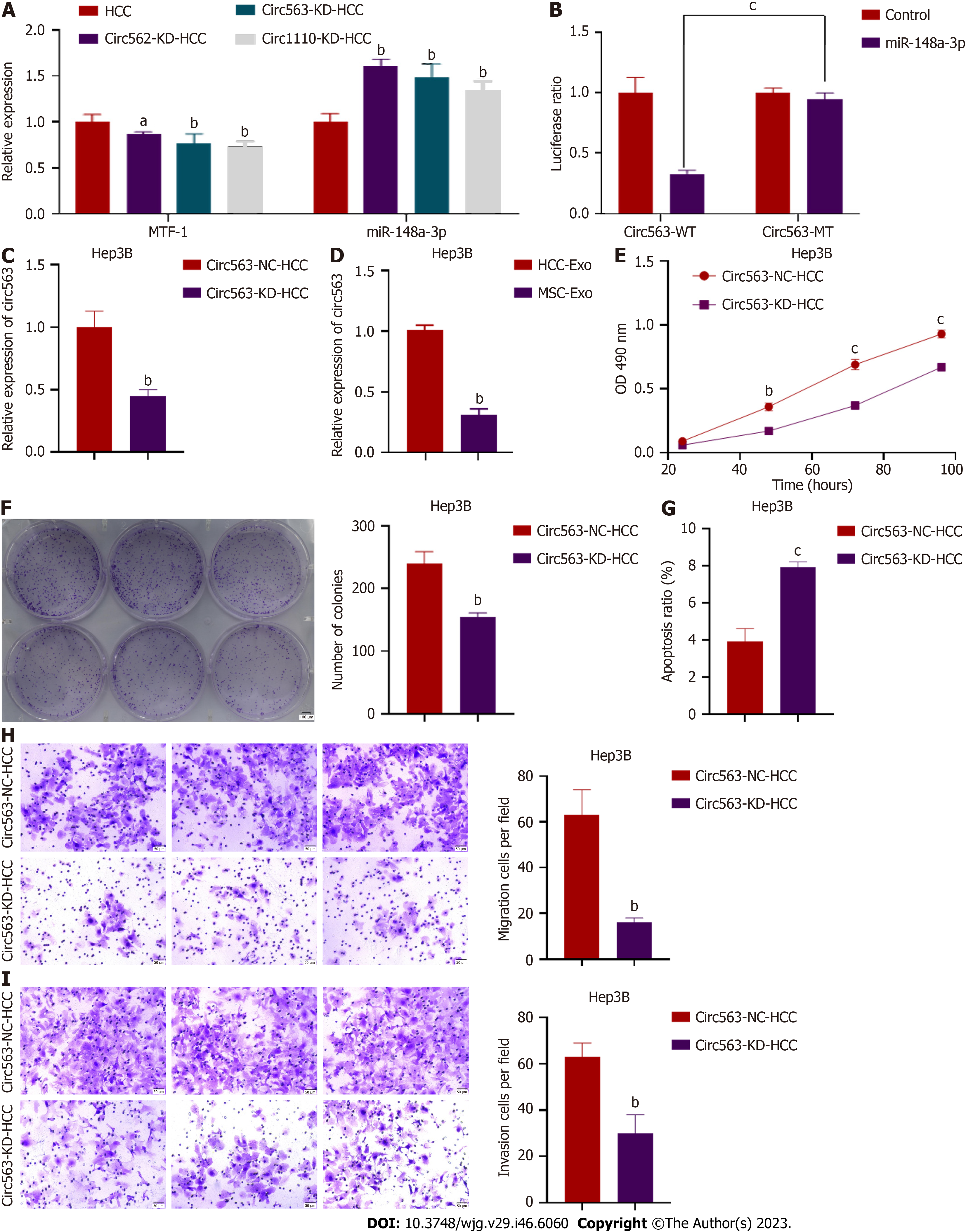
Figure 2 Identification of hsa_circRNA_0000563.
A: Three circular RNAs (circRNAs) (circ562, circ563, and circ1110) were predicted to most likely bind to miR-148a-3p, and the miR-148a-3p and metal-regulatory transcription factor-1 (MTF-1) levels in hepatocellular carcinoma (HCC) cells transfected with the predicted circRNA were detected by quantitative real-time polymerase chain reaction (RT-PCR); B: A dual-luciferase reporter assay was performed to confirm the direct binding between circ563 and miR-148a-3p based on their complementary sequences; C: The knockdown efficiency was verified by RT-PCR; D: The expression levels of circ563 in exosomes were assessed by RT-PCR; E and F: 3-(4,5-Dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide and colony-forming assay results showed that circ563 silencing suppressed cell proliferation and reduced the number of colonies; G: Knockdown of circ563 enhanced Hep3B cell apoptosis as determined by flow cytometry; H and I: Transwell assays revealed that reducing the levels of circ563 impaired the migratory potential and invasiveness of HCC cells. aP < 0.05, bP < 0.01, cP < 0.001. circRNA: Circular RNA; HCC: Hepatocellular carcinoma; Exo: Exosome.
- Citation: Lyu ZZ, Li M, Yang MY, Han MH, Yang Z. Exosome-mediated transfer of circRNA563 promoting hepatocellular carcinoma by targeting the microRNA148a-3p/metal-regulatory transcription factor-1 pathway. World J Gastroenterol 2023; 29(46): 6060-6075
- URL: https://www.wjgnet.com/1007-9327/full/v29/i46/6060.htm
- DOI: https://dx.doi.org/10.3748/wjg.v29.i46.6060