Copyright
©The Author(s) 2022.
World J Gastroenterol. Oct 7, 2022; 28(37): 5420-5443
Published online Oct 7, 2022. doi: 10.3748/wjg.v28.i37.5420
Published online Oct 7, 2022. doi: 10.3748/wjg.v28.i37.5420
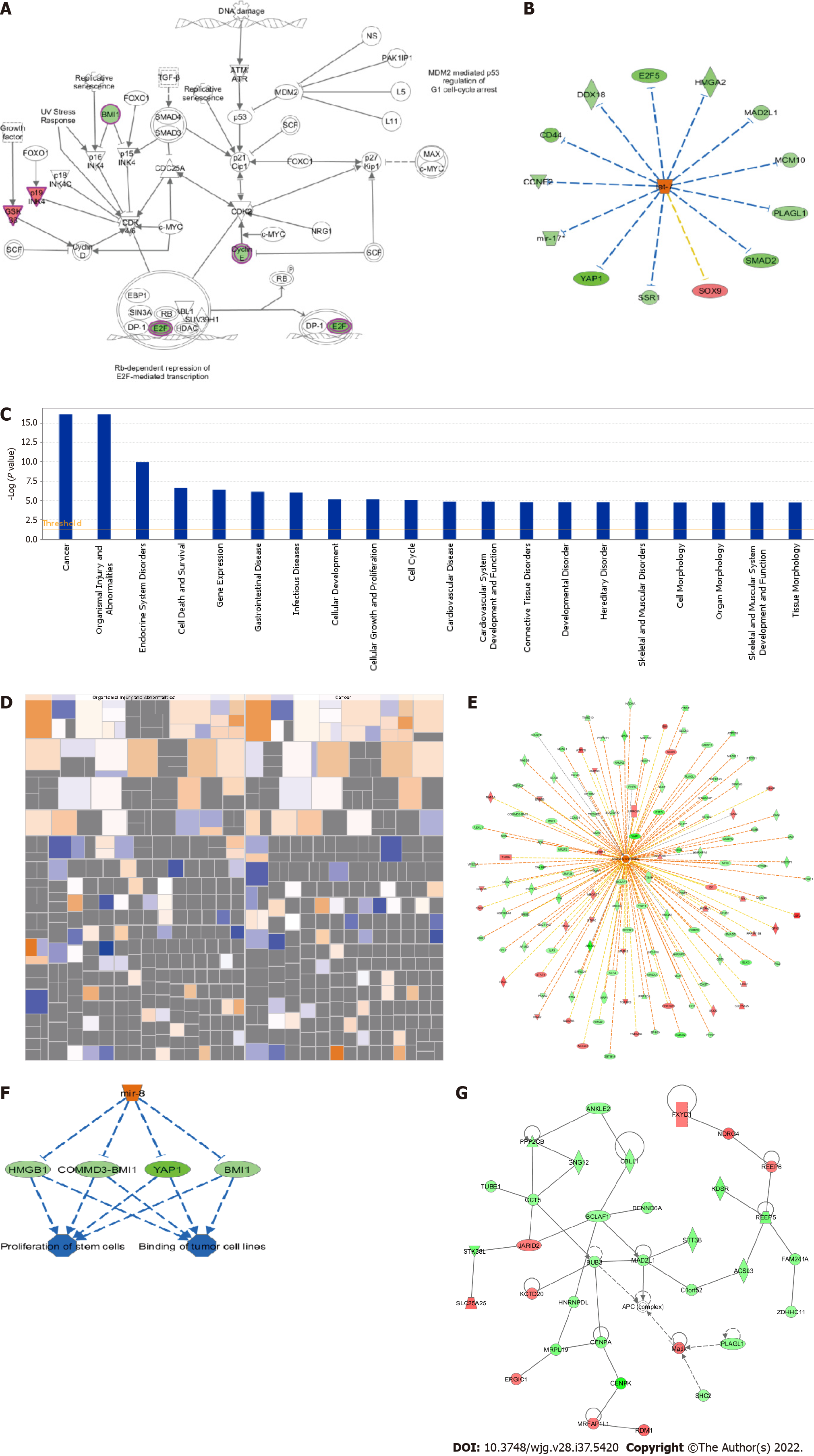
Figure 3 Centromere protein K functions in colon cancer by regulating the expression of related genes.
A: Trend of molecules in the experimental results in the classical pathway; B: Upstream regulator network diagram (the orange line indicates consistent activation of upstream regulators and genes, the blue line indicates consistent upstream regulators and gene inhibition, the yellow line indicates inconsistent expression trends between upstream regulators and genes, and the gray line indicates that there is no prediction information related to the expression state in the dataset); C: Analysis and statistics of disease and functional enrichment (the transverse coordinate is the path name and the longitudinal coordinate is significance level of enrichment (negative logarithmic transformation in the bottom 10); D: Disease and functional heatmap of the effect of disease and functional changes at gene expression levels; E: Activation and inhibition relationship between genes and disease or function; F: Interplay between genes and regulators and function in the dataset; G: Network of interaction relationships between molecules of a disease or function-related relationship. TGF-β: Transforming growth factor beta; FOXO: Forkhead box O; SMAD: Suppressor of mothers against decapentaplegic; MDM: Mouse double minute; ATM: Ataxia telangiectasia mutated; ATR: Ataxia telangiectasia and Rad3 related; PAK: p21 activated kinases; NS: Non-structural; MAX: MYC associated factor X; c-MYC: Cellular myelocytomatosis oncogene; SCF: Stem cell factor; INK: CDKN2A, cyclin dependent kinase inhibitor 2A; BMI 1: BMI1 proto-oncogene; GSK: Guanosine kinase; CDK: Cyclin-dependent kinase; NRG: Neuregulin; EBP1: ErbB3-binding protein 1; SIN3A: Suppressor interacting 3a; SUV39H1: SUV39H1 histone lysine methyltransferase; DP-1: Dodeca-satellite-binding protein 1; E2F: Early region 2 binding factor; RB: Retinoblastoma; HDAC: Histone deacetylase; CD44: Cluster of differentiation-44; DDX18: DEAD-box helicase 18; HMGA: High mobility group A; MAD2L1: Mitotic arrest deficient 2 like 1; MCM: Mei-mini-chromosome maintenance; PLAGL: Pleiomorphic adenoma gene-like; SOX: SRY-box transcription factor; SSR: Signal sequence receptor subunit; YAP1: Yes1 associated transcriptional regulator; CCNE2: Cyclin E2; COMMD3: COMM domain containing 3.
- Citation: Li X, Han YR, Xuefeng X, Ma YX, Xing GS, Yang ZW, Zhang Z, Shi L, Wu XL. Lentivirus-mediated short hairpin RNA interference of CENPK inhibits growth of colorectal cancer cells with overexpression of Cullin 4A. World J Gastroenterol 2022; 28(37): 5420-5443
- URL: https://www.wjgnet.com/1007-9327/full/v28/i37/5420.htm
- DOI: https://dx.doi.org/10.3748/wjg.v28.i37.5420