Published online Nov 14, 2021. doi: 10.3748/wjg.v27.i42.7311
Peer-review started: April 11, 2021
First decision: May 24, 2021
Revised: May 31, 2021
Accepted: September 14, 2021
Article in press: September 14, 2021
Published online: November 14, 2021
An increased amount of Fusobacterium nucleatum (F. nucleatum) is frequently detected in the gastric cancer-associated microbiota of the Taiwanese population. F. nucleatum is known to exert cytotoxic effects and play a role in the progression of colorectal cancer, though the impact of F. nucleatum colonization on gastric cancer cells and patient prognosis has not yet been examined.
To identify F. nucleatum-dependent molecular pathways in gastric cancer cells and to determine the impact of F. nucleatum on survival in gastric cancer.
Coculture of F. nucleatum with a gastric cancer cell line was performed, and changes in gene expression were investigated. Genes with significant changes in expression were identified by RNA sequencing. Pathway analysis was carried out to determine deregulated cellular functions. A cohort of gastric cancer patients undergoing gastrectomy was recruited, and nested polymerase chain reaction was performed to detect the presence of F. nucleatum in resected cancer tissues. Statistical analysis was performed to determine whether F. nucleatum colonization affects patient survival.
RNA sequencing and subsequent pathway analysis revealed a drastic interferon response induced by a high colonization load. This response peaked within 24 h and subsided after 72 h of incubation. In contrast, deregulation of actin and its regulators was observed during prolonged incubation under a low colonization load, likely altering the mobility of gastric cancer cells. According to the clinical specimen analysis, approximately one-third of the gastric cancer patients were positive for F. nucleatum, and statistical analysis indicated that the risk for colo
F. nucleatum colonization leads to deregulation of actin dynamics and likely changes cancer cell mobility. Cohort analysis demonstrated that F. nucleatum colonization leads to poorer prognosis in H. pylori-positive patients with late-stage gastric cancer. Hence, combined colonization of F. nucleatum and H. pylori is a predictive biomarker for poorer survival in late-stage gastric cancer patients treated with gastrectomy.
Core Tip: Fusobacterium nucleatum (F. nucleatum) is frequently enriched in the gastric cancer-associated microbiota. Here, we showed that F. nucleatum solicits a rapid but transient interferon response from gastric cancer cells. In addition, F. nucleatum leads to deregulation of the genes functioning in regulation of actin filament dynamics, likely changing cell mobility. In a patient cohort receiving gastrectomy, combined infection of F. nucleatum and Helicobacter pylori infection incurs poorer survival, indicating these two pathogens act synergistically to promote invasiveness of gastric cancer. Our finding suggests that F. nucleatum is a biomarker as treatment outcome predicator.
- Citation: Hsieh YY, Tung SY, Pan HY, Chang TS, Wei KL, Chen WM, Deng YF, Lu CK, Lai YH, Wu CS, Li C. Fusobacterium nucleatum colonization is associated with decreased survival of helicobacter pylori-positive gastric cancer patients. World J Gastroenterol 2021; 27(42): 7311-7323
- URL: https://www.wjgnet.com/1007-9327/full/v27/i42/7311.htm
- DOI: https://dx.doi.org/10.3748/wjg.v27.i42.7311
Disruption of the normal gastrointestinal flora is often associated with pathogenic conditions. In general, the extreme acidity and thick protective mucosa of the gastric environment limit the complexity and abundance of the microbiota and prevent direct gastric epithelium colonization by pathogenic microbes[1]. However, Helicobacter pylori (H. pylori), which is well recognized as a risk factor for gastric cancer, is able to penetrate the mucosa layer and establish long-term colonization[2]. Indeed, virulence factors produced by this bacterium facilitate the transformation of gastric mucosal cells and lead to a drastic increase in the risk of gastric cancer[2,3].
Although it is well established that H. pylori is more frequently found in gastric cancer patients than in noncancer controls[4], recent microbiota profiling studies have revealed that the abundance of H. pylori in the gastric microbiota is frequently decreased in gastric cancer patients compared with that in noncancer patients[5,6]. Our hypothesis to account for the decrease in H. pylori abundance in gastric cancer is microbial succession: Once colonization occurs, H. pylori creates a niche microenvironment on the gastric epithelium that facilitates the colonization of secondary settler bacteria. Accordingly, the predominance of H. pylori in the microenvironment can be replaced by other bacteria after a prolonged colonization period. It is possible that the secondary gastric microbes also participate in promoting the development of gastric cancer[7].
Advanced sequencing technology has enabled profiling of the microbiota without the need to isolate pure cultures, and in a previous study employing this experimental approach, we identified Fusobacterium nucleatum (F. nucleatum) as being enriched in the gastric cancer-associated microbiota. An increased presence of F. nucleatum in the colorectal cancer-associated microbiota has also been reported[8,9]. F. nucleatum colonization correlates with high microsatellite instability, disruption of the mismatch repair mechanism, and poor prognosis[10]. The genomic instability that is observed is likely mediated by the metabolites produced by F. nucleatum. One such metabolite is hydrogen sulfide, which has been shown to generate reactive oxygen species, induce DNA damage, and cause single-nucleotide mutations[11]. Hence, it is possible that F. nucleatum promotes oncogenesis by acting as a DNA-damaging agent. In fact, F. nucleatum has been shown to promote the growth and metastasis of colorectal cancer[12,13], and the level of F. nucleatum in the colorectal cancer-associated microbiota correlates with poor patient prognosis[14,15]. Therefore, F. nucleatum may be used as a prognostic biomarker for colorectal cancer.
Detection of Fusobacterium DNA using polymerase chain reaction (PCR) facilitates screening of colorectal cancer by increasing the sensitivity of the standard fecal immunochemical test[15]. Although the role of F. nucleatum in colorectal cancer has been intensively studied, it remains unclear whether this bacterium exerts a similar oncogenic effect on the gastric epithelium. In this study, a nested PCR-based method was developed to detect the presence of F. nucleatum in resected gastric cancer tissue specimens. Based on statistical analysis, we found that the risk of F. nucleatum colonization is greatly increased in patients with late-stage gastric cancer. Moreover, F. nucleatum colonization was associated with a poor prognosis in H. pylori-positive patients. Our findings suggest that invasion of F. nucleatum into the gastric cancer-associated microenvironment promotes gastric cancer aggressiveness and subsequently leads to poorer prognosis.
F. nucleatum strain ATCC25586[16] was obtained from Bioresource Collection and Research Center (Hsinchu, Taiwan). The bacteria were cultured on EG culture medium containing 2.5 g Lab-Lemco powder, 10 g proticasepeptone, 5 g yeast extract, 4 g glucose, 0.5 g starch, 0.2 g L-cystine, 0.5 g L-cysteine HCl, 4 g Na2HPO4, 15 g Bacto-Agar, and 50 mL defibrinated horse blood per liter under anaerobic conditions using a BD GasPak system (Thermo Fisher Scientific, Waltham, MA, United States). The bacteria were scrapped from a plate and resuspended in Dulbecco's Modified Eagle’s Medium. The number of cells per millimeter in the resuspended medium was determined using light microscopy. The correlation between the observed cell number and colony forming units was determined by reculturing the bacteria after serial dilutions. The gastric cancer cell line 008L-C2 used in this study was originally isolated from resected gastric cancer tissue. The cells were cultured in Dulbecco's Modified Eagle’s Medium supplemented with 10% fetal bovine serum. F. nucleatum strain ATCC25586 was cultured with 008L-C2 gastric cancer cells under anaerobic conditions at 37 °C and 5% CO2 and harvested when the coculture experiment was performed. In coculture experiments, the initial multiplicity of infection (MOI) was 10 and 100. After 0, 24, and 72 hours of coculture with F. nucleatum, the cells were collected and washed twice with phosphate-buffered saline. Total RNA was extracted from the washed cells using TRIzol reagent (Thermo Fisher Scientific) according to the manufacturer’s protocol, and RNA expression analysis was conducted using next-generation sequencing.
The integrity and concentration of purified RNA samples were determined using capillary electrophoresis with a TapeStation 2200 instrument (Agilent, Santa Clara, CA, United States) and fluorometric analysis (Qubit fluorometer; Invitrogen, Waltham, MA, United States). Libraries for RNA sequencing analysis were prepared using a SureSelect strand-specific mRNA library preparation kit (Agilent); the manufacturer's protocol was closely followed. The libraries were pooled and sequenced using a NextSeq 550 sequencer. Quality filtering, mapping, annotation, and calculation of gene expression levels were performed using CLC Genomic Workbench v.12.0.3 (Qiagen, Redwood City, CA, United States). The RNA level is expressed as transcripts per million. The sequencing data were deposited in the Sequence Read Archive, National Center for Biotechnology Information, United States. The BioProject ID is PRJNA
Resected cancer tissues from patients undergoing gastrectomy were obtained from Chiayi Chang Gung Memorial Hospital Tissue Bank. The acquisition and use of clinical specimens in this study were carried out in accordance with the Declaration of Helsinki. This study was approved by the Institutional Review Board of Chiayi Chang Gung Memorial Hospital (Institutional Review Board approval No. 201700169A3, No. 201700172A3, and No. 201700173A3). Frozen biopsies were briefly rinsed in phos
The tissue specimens were examined for the presence of H. pylori using a standard rapid urease test. The presence of F. nucleatum in the specimens was determined by nested PCR detection of the NusG gene. The forward and reverse primer sequences used for the first-stage PCR were 5’-TGTTAGAGGAAAGCCCAAGAAG and 5’-CTTCTTCCATAGGAATAGGGTCAG, respectively. The initial amplification cycle (denaturation) was as follows: 94 °C for 5 min; followed by 36 cycles at 94 °C for 30 s, 55 °C for 30 s, and 72 °C for 30 s and a final extension step at 72 °C for 5 min. The product of the first-stage amplification was directly used in the second-stage PCR. The forward and reverse primer sequences of the second-stage PCR were 5’-GCTTGAAATGGAAGCTACAAGAG and 5’-GGTCAGAACCAACTCCTACAAA, respectively. The second-stage PCR amplification cycle parameter was identical to that of the first-stage PCR. The sequence of the PCR product was determined by dideoxynucleotide sequencing to confirm that the amplified product is the F. nucleatum NusG target sequence. The PCR product was examined by nondenaturing 6% polyacrylamide gel electrophoresis and documented. Statistical analysis was carried out using SAS/STAT statistical analysis software v. 9.4 (College Station, TX, United States). Survival probability was calculated with Kaplan-Meier analysis.
In our previous metagenomic analysis, we discovered that F. nucleatum colonizes and becomes enriched in the gastric cancer-associated microbiota[17]. Among the 11 cancer biopsies collected in that study, four showed F. nucleatum colonization, suggesting that F. nucleatum is a frequent cocolonizer among gastric cancer patients in the southwestern region of Taiwan. Moreover, clinical data confirmed that F. nucleatum colonization is frequently observed in gastric cancer patients. Nevertheless, the pathogenic effects of F. nucleatum have not been investigated in gastric cancer cells. Thus, to explore the role of F. nucleatum in gastric cancer, we examined gastric cancer cell growth using an in vitro coculture system using the gastric cancer cell line 008L-C2, derived from resected gastric cancer tissue. During the incubation period, an increase in F. nucleatum abundance was observed under a microscope, though we did not determine the precise doubling time of F. nucleatum.
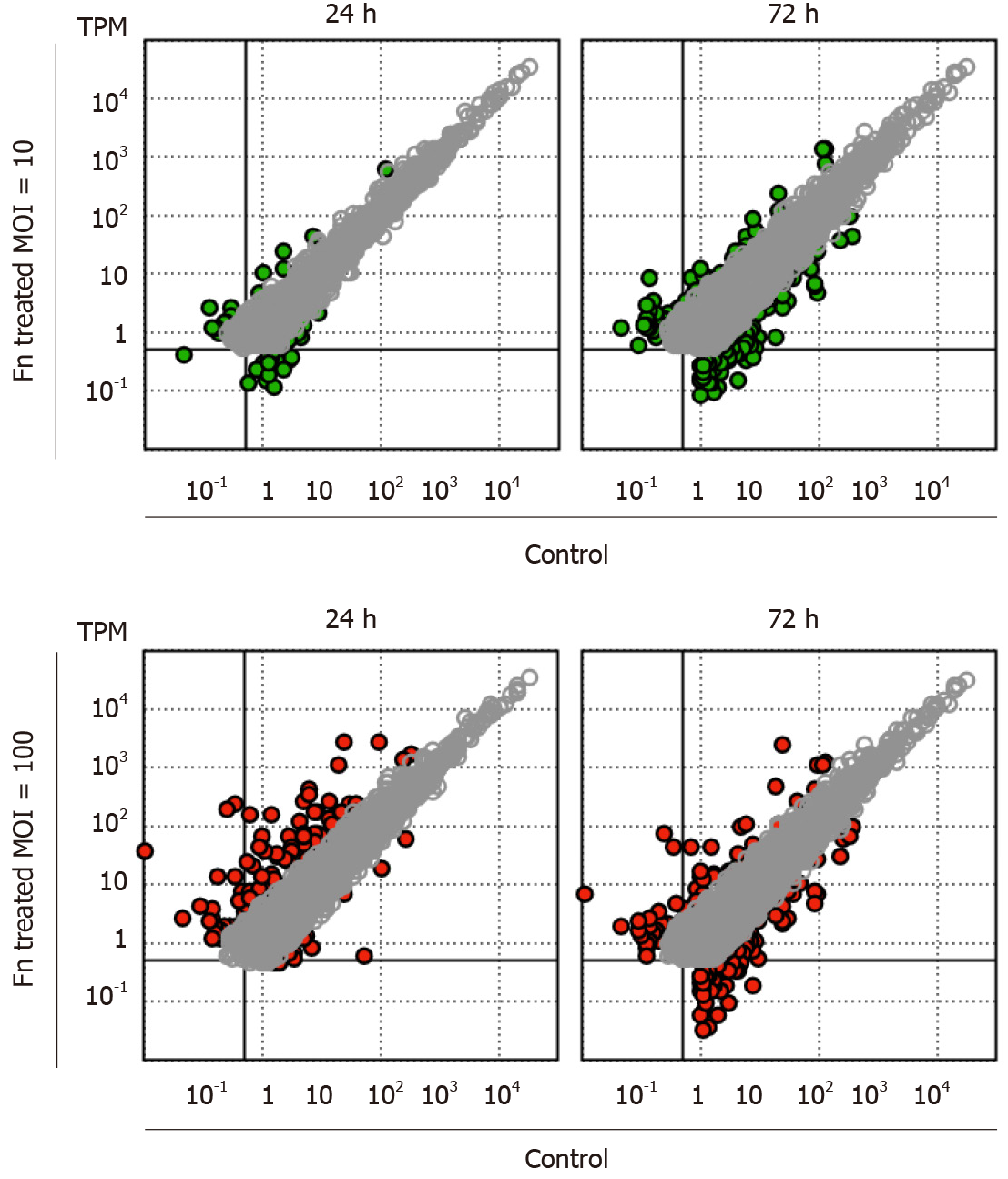
RNA sequencing analysis revealed that the presence of F. nucleatum is associated with dose- and time-dependent changes in the gene expression profile of cancer cells. After 24 h of coculture with F. nucleatum at a low MOI, only a limited number of marginally expressed genes (transcripts per million < 1) exhibited more than four-fold changes (Figure 1). In contrast, the expression level of a specific set of genes was strongly upregulated at a high MOI (100) (Figure 1). After 72 h of coculture, low-MOI treatment led to a significantly higher number of genes with more than four-fold increases in expression (Figure 1). However, the number of strongly upregulated genes in high MOI-treated cells decreased after longer incubation, and there was an increased number of genes with more than four-fold decreased expression (Figure 1). Our results indicate a rapid and strong cellular response to a large amount of F. nucleatum. Additionally, F. nucleatum caused a prolonged change in gastric cancer cells.
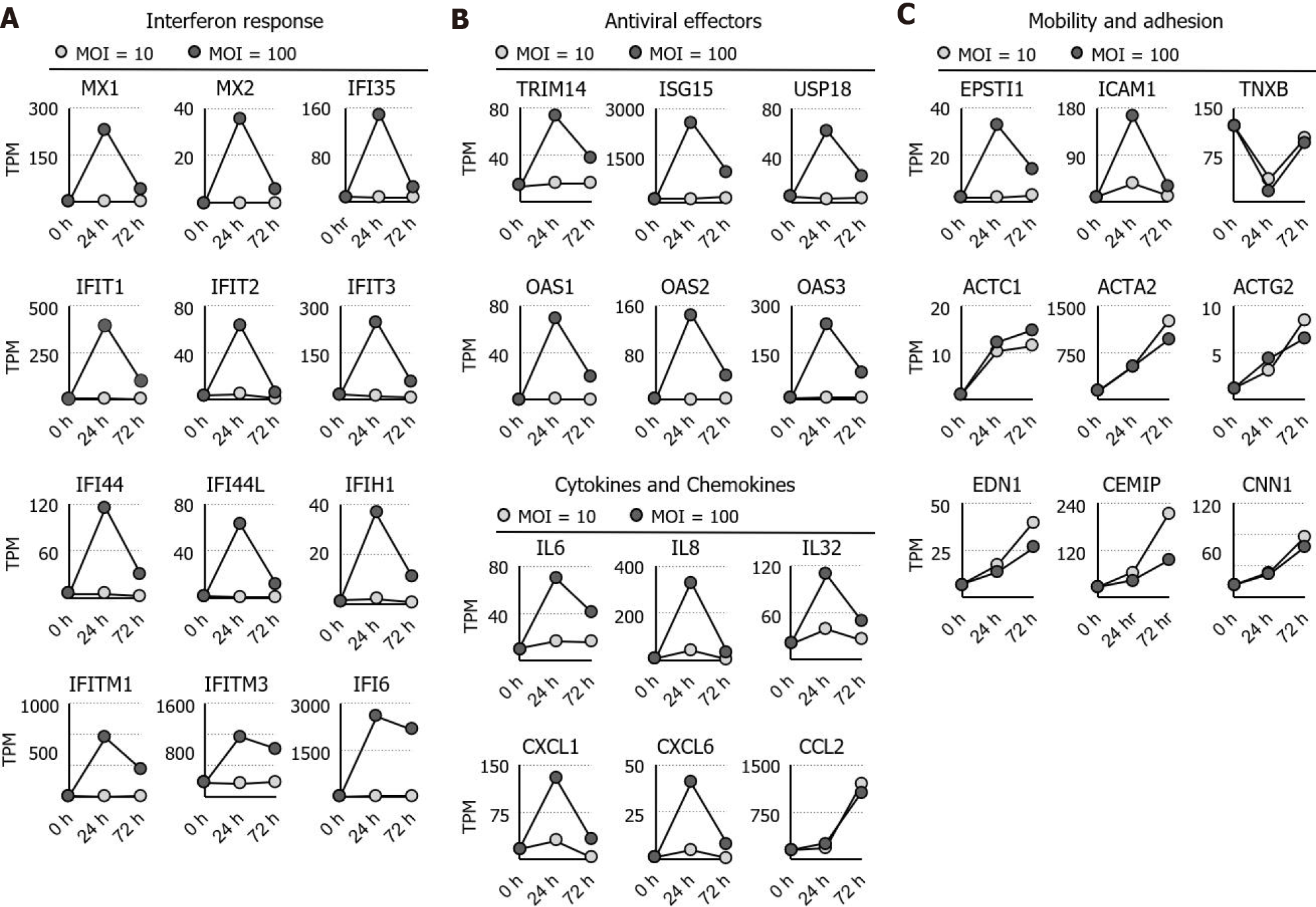
Ontological analysis indicated that interferon and its response genes as well as inflammatory cytokines are immediately activated by high MOI treatment with F. nucleatum. The activated genes, including MX1, MX2, interferon induced protein (IFI)35, IFIT1, IFIT2, IFIT3, IFI44, IFI44L, IFITM1, IFITM3, IFIH1, and IFI6[18], are well-known interferon response genes. Furthermore, tripartite motif-containing 14[19,20], interferon -stimulated gene 15[21], ubiquitin specific peptidase 18[22], 2'-5'-oligoadenylate synthetase (OAS)1, OAS2, and OAS3[23,24] were found to be involved in interferon-dependent antiviral activity (Figure 2A and B). In addition to interferon response genes, simultaneous increases in interleukin (IL)6, IL8, IL32, C-X-C motif ligand (CXCL)1, CXCL2, and CXCL6 expression were observed in the high-MOI-treated cells after 24 h of exposure (Figure 2C). In contrast to the short-term activation profile observed for interleukins and chemokines, C-C ligand 2 remained highly expressed even after 72 h of exposure (Figure 2A). Not only genes involved in the regulation of immunological functions but also those related to cell mobility and adhesion were dysregulated by F. nucleatum colonization. For instance, epithelial-stromal interaction 1[25,26] and intercellular adhesion molecule 1[27], both of which are involved in cell-matrix interaction, were upregulated at 24 h and returned to basal levels after 72 h of incubation (Figure 2B). Other cytoskeleton and cell adhesion genes were activated in a distinct pattern, and actin alpha 2; actin, alpha cardiac muscle precursor 1; actin gamma 2, smooth muscle; calponin 1, endothelin 1, and cell migration inducing hyaluronan binding protein were continuously stimulated by F. nucleatum (Figure 2C). Increased expression of these genes was time dependent but not dose (MOI) dependent, indicating that chronic colonization with a small number of F. nucleatum bacteria may lead to long-term strong effects on cytoskeletal dynamics and cell mobility.
Overall, our data show that coculturing gastric cancer cells with F. nucleatum results in a short-term immune response and continuous dysregulation of actin-related signaling in vitro. The change in actin cytoskeleton dynamics was associated with an increase in cell mobility, thereby promoting the invasiveness and metastasis of gastric cancer. Therefore, we investigated whether F. nucleatum colonization influences the survival of gastric cancer patients after gastrectomy. Gastric cancer progression is marked by tumor spread and distant metastasis, which are associated with increased cancer cell mobility. To test our hypothesis, we examined resected gastric cancer tissues from patients who underwent gastrectomy. The survival rate of these patients was 67%, which is higher than the 37% survival rate reported by Cancer Registry Annual Report, 2018, Taiwan. The clinical and pathological characteristics of the patient cohort are presented in Table 1.
| Sex | Age | Stage | TNM classification | Tumor size | H. pylori | F. nucleatum |
| M | 70 | IA | pT1aN0 | 1.2 cm × 0.5 cm | Positive | Positive |
| F | 78 | IA | pT1bN0 | 7 cm × 5 cm | Positive | Negative |
| F | 84 | IA | pT1aN0Mx | 2 cm × 2 cm | Positive | Negative |
| M | 73 | IA | pT1aN0Mx | 0.5 cm × 0.3 cm | Positive | Negative |
| M | 73 | IA | pT1aN0 | 1.5 cm × 0.3 cm | Positive | Negative |
| M | 73 | IA | pT1aN0MX | 6 cm × 6 cm | Negative | Negative |
| M | 64 | IA | pT1bN0MX | 9.5 cm × 1.5 cm | Negative | Negative |
| M | 67 | IA | pT1bN0 | 2.5 cm × 2 cm | Positive | Negative |
| M | 66 | IA | pT1bN0 | 0.8 cm × 0.5 cm | Positive | Negative |
| M | 65 | IA | pT1bN0 | 3 cm × 2.7 cm | Positive | Negative |
| F | 68 | IB | pT2N0Mx | 6 cm × 4 cm | Negative | Positive |
| M | 73 | IB | pT2N0Mx | 1.5 cm × 1.5 cm | Positive | Positive |
| M | 60 | IB | pT2N0 | 4.5 cm × 4 cm | Positive | Positive |
| F | 68 | IB | pT2N0 | 3 cm × 1.8 cm | Positive | Negative |
| F | 67 | IB | pT2N0 | 1.5 cm × 1.0 cm | Negative | Negative |
| M | 71 | IB | pT1bN1MX | 2 cm × 1.8 cm | Positive | Negative |
| M | 69 | IB | pT2N0 | 2.2 cm × 2.1 cm | Positive | Negative |
| F | 86 | IIA | pT3N0Mx | 5 cm × 4.5 cm | Positive | Positive |
| F | 87 | IIA | pT3N0Mx | 7.5 cm × 7.0 cm | Negative | Positive |
| M | 68 | IIA | pT3N0 | 4 cm × 3.5 cm | Positive | Positive |
| M | 58 | IIA | pT2N1 | 7 cm × 6 cm | Positive | Negative |
| M | 50 | IIA | pT2N1MX | 3.5 cm × 3.5 cm | Negative | Negative |
| M | 71 | IIA | pT3N0 | 1 cm × 1 cm | Positive | Negative |
| F | 69 | IIB | pT3N1 | 1.2 cm × 1.8 cm | Positive | Positive |
| M | 61 | IIB | pT4aN0 | 2 cm × 1.8 cm | Positive | Positive |
| M | 74 | IIB | pT2N2 | 4.5 cm × 2.0 cm | Positive | Negative |
| M | 64 | IIB | pT4aN0 | 2.6 cm × 2.0 cm | Positive | Negative |
| M | 75 | IIB | pT2N2 | 4.2 cm × 4.0 cm | Negative | Negative |
| M | 76 | IIIA | pT3N2MX | 5 cm × 4.5 cm | Positive | Positive |
| M | 79 | IIIA | pT3N2Mx | 3.5 cm × 3.5 cm | Negative | Negative |
| M | 51 | IIIA | pT4N1 | 8 cm × 6 cm | Positive | Negative |
| M | 63 | IIIA | pT3N2 | 3.2 cm × 2.2 cm | Negative | Negative |
| M | 87 | IIIB | pT3N3b | 6.5 cm × 5 cm | Positive | Positive |
| F | 64 | IIIB | pT3N3 | 7 cm × 5 cm | Positive | Negative |
| F | 65 | IIIB | pT4aN2 | 12 cm × 10 cm | Negative | Negative |
| F | 78 | IIIB | pT3N3b | 3 cm × 2 cm | Positive | Negative |
| M | 59 | IIIB | pT4bN1Mx | 8.5 cm × 7.5 cm | Positive | Negative |
| M | 58 | IIIB | pT3N3aMX | 3 cm × 3 cm | Negative | Negative |
| M | 87 | IIIB | pT4aN2 | 3 cm × 2 cm | Positive | Negative |
| M | 83 | IIIB | pT4aN2 | 6 cm × 5 cm | Negative | Negative |
| M | 69 | IIIB | pT4aN2 | 6 cm × 5 cm | Positive | Negative |
| M | 65 | IIIB | pT3N3b | 5.5 cm × 4 cm | Positive | Negative |
| F | 73 | IIIC | pT4N3a | 4.5 cm × 4 cm | Positive | Positive |
| F | 74 | IIIC | pT4a N3b | 2.0 cm × 2.0 cm | Positive | Positive |
| M | 57 | IIIC | pT4bN2Mx | 4 cm × 4 cm | Positive | Positive |
| M | 67 | IIIC | pT4aN3b | 8 cm × 7.5 cm | Positive | Positive |
| F | 53 | IIIC | pT4bN3a | 6 cm × 5 cm | Negative | Negative |
| F | 72 | IIIC | pT4aN3a | 7 cm × 2 cm | Negative | Negative |
| F | 79 | IIIC | pT4aN3a | 4.5 cm × 2.5 cm | Negative | Negative |
| M | 52 | IIIC | pT4bN3aMX | 8 cm × 6 cm | Positive | Negative |
| M | 76 | IIIC | pT4aN3a | 2.0 cm × 1.8 cm | Positive | Negative |
| M | 71 | IIIC | pT4aN3a | 4.8 cm × 4.5 cm | Positive | Negative |
| F | 47 | IV | pT4aN3aM1 | 4.5 cm × 4.5 cm | Positive | Positive |
| M | 47 | IV | pT4bN3aM1 | 6 cm × 3 cm | Negative | Positive |
| M | 69 | IV | pT4N3M1 | 5 cm × 4.5 cm | Positive | Positive |
| M | 73 | IV | pT3N3bM1 | 12 cm × 10.5 cm | Positive | Positive |
| M | 75 | IV | pT1bN1M1 | 2.2 cm × 2.0 cm | Positive | Negative |
| M | 66 | IV | pT4aN3bM1b | 7 cm × 4 cm | Positive | Negative |
| M | 75 | IV | pT4bN3bM1 | 3 cm × 3 cm | Positive | Negative |
| M | 70 | IV | pT3N3aM1 | 9 cm × 8 cm × 2.5 cm | Negative | Negative |
To determine the presence of F. nucleatum in the specimens, we employed a nested PCR method with increased detection sensitivity and specificity. In a previous study utilizing conventional PCR, F. nucleatum was found at relatively low frequencies in gastric cancer specimens[28]. Our results obtained by metagenomic profiling indicated that the F. nucleatum frequency is much higher. The discrepancy could account for the higher frequency of F. nucleatum in the tested cohort, suggesting that the observed phenomenon is specific for this region of Taiwan. Nonetheless, the previously observed low frequency might be associated with methodological limitations resulting in the detection of lower amounts of F. nucleatum. This is a likely scenario, considering that resected cancer tissues may contain less surface mucosa. Nested PCR allowed for higher F. nucleatum detection sensitivity.
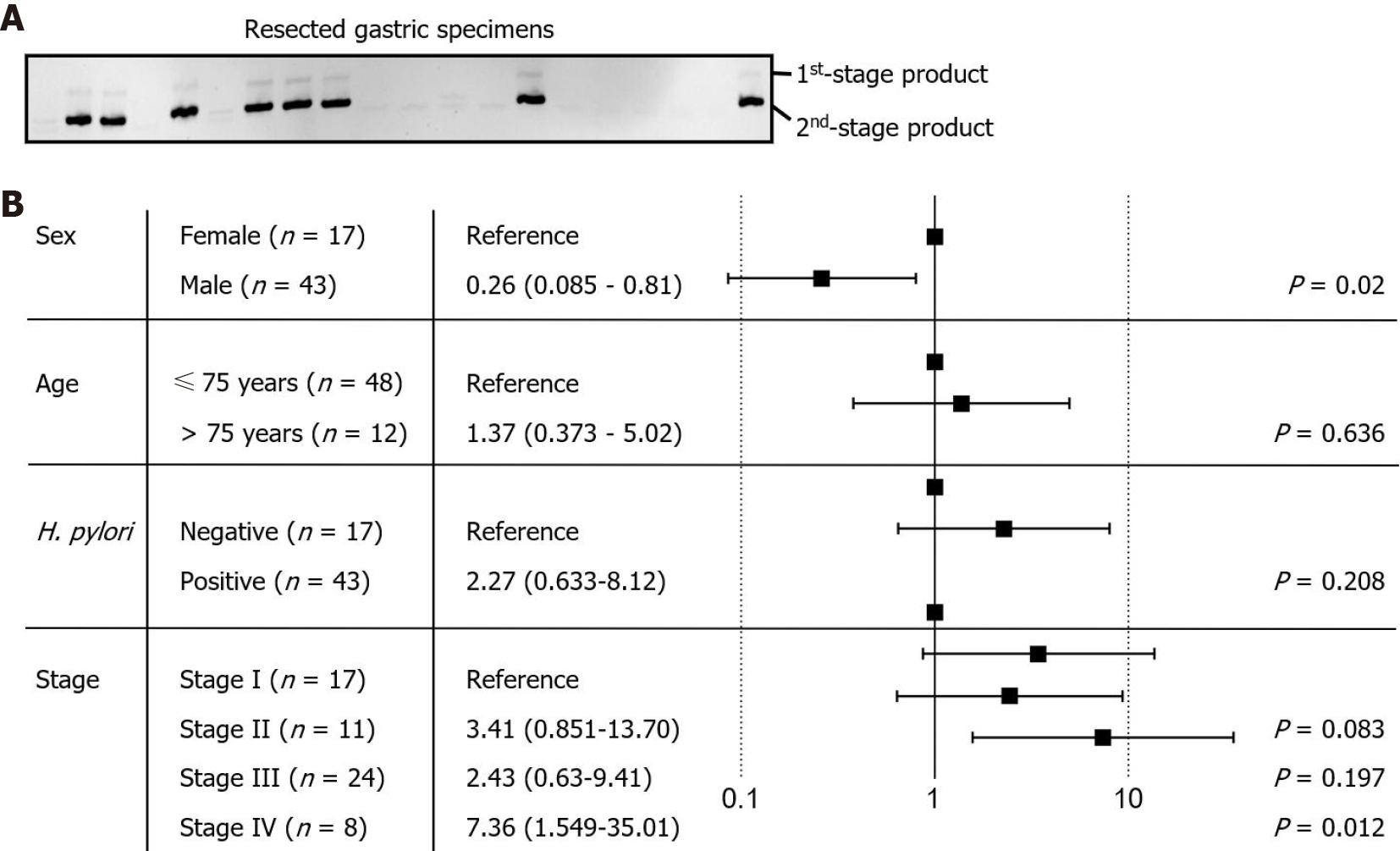
The detection target in this study was the highly conserved NusG gene of F. nucleatum[28,29]. The sizes of the first- and second-stage PCR products were expected to be 175 bp and 124 bp, respectively. The PCR product was resolved by acrylamide electrophoresis, and identity was confirmed by dideoxynucleotide sequencing. To validate the nested PCR protocol, we tested DNA specimens from our previous study. The method allowed us to identify the majority of F. nucleatum-positive specimens, except for those with an exceptionally low bacterial load. The positive identification rate was 90%, with no false-positives. Thus, we used nested PCR to assess the presence of F. nucleatum in the resected gastric cancer tissues used in this study (Figure 3A). The results showed 19 of the 60 examined specimens to be positive for F. nucleatum. The other 41 specimens were negative or showed only marginal PCR product levels, all of which were defined as F. nucleatum negative. Accordingly, the proportion of F. nucleatum-positive patients was approximately one-third of the cohort. The results are consistent with our previous study examining upper endoscopic-collected gastric biopsies.
The risk of F. nucleatum colonization was analyzed against clinical characteristics and cancer stages (Figure 3B), indicating that older age (≥ 75) is not associated with the risk of F. nucleatum colonization (P value = 0.636). Although H. pylori colonization appears to be associated with a higher risk of F. nucleatum colonization, no statistical significance was obtained (P value = 0.208). The risk of F. nucleatum colonization was increased in late-stage gastric cancer patients, suggesting that the microenvironment during cancer progression is more suitable for colonization and growth of F. nucleatum (P value = 0.012). Interestingly, we found that the risk of F. nucleatum colonization was significantly decreased in male patients (P value = 0.02), which may be associated with the lifestyle or hormonal status of the patients and requires confirmation in larger and/or independent gastric cancer patient cohorts.
We then analyzed the impact of F. nucleatum on the survival of gastric cancer patients. Although results showed that F. nucleatum-positive patients had a lower survival rate, this was not significant (Figure 4A). Stage I cancer patients with a nearly 100% 5-year survival rate were then excluded from the survival analysis, but significance was still not reached while poorer survival outcomes were maintained (Figure 4A). As noted above, patients who undergo gastrectomy have good treatment outcomes, and it is likely that oncogenic factors, including pathogenic gastric microbiota, are reduced after surgery. Despite the lack of statistical significance, our analysis shows that F. nucleatum colonization may adversely impact treatment outcome.
Approximately one-third of the patients in our cohort tested positive for H. pylori colonization, and our previous study demonstrated that F. nucleatum is likely a secondary settler of the gastric microbiota after H. pylori colonizes the gastric epithelium. Consecutive or simultaneous colonization of H. pylori and F. nucleatum might have synergistic effects and promote cancer progression. To test this hypothesis, we analyzed the impact of F. nucleatum on the survival of H. pylori-positive patients. Our data demonstrate that patients positive for both H. pylori and F. nucleatum had a poorer survival outcome than those who were positive for H. pylori alone (Figure 4B). Similar results were observed when stage I cancer patients were excluded from the analysis (Figure 4B). Therefore, the presence of F. nucleatum colonization negatively impacts gastric cancer prognosis. Our analysis confirms that F. nucleatum colonization is frequent in patients with advanced-stage gastric cancer, with an unfavorable effect on patient survival.
F. nucleatum is an opportunistic pathogen mainly residing in the oral cavity[30]. Although it is likely that oral-resident microbes are regularly passed to the stomach through dietary intake, they are unable to colonize the rest of the gastric microenvironment under normal physiological conditions. However, H. pylori colonization and the growth of carcinoma cells create a suitable microenvironment[31,32] and likely allow F. nucleatum invasion. Our previous study, which utilized 16S metagenomic analysis, showed that F. nucleatum can be present in the gastric cancer-associated microbiota[17]. In the present study, we employed nested PCR to determine the existence of F. nucleatum in resected gastric cancer tissues. The results confirmed our previous finding, showing F. nucleatum colonization in approximately one-third of gastric cancer patients in southwestern Taiwan. Whether F. nucleatum colonization is common remains to be investigated.
Analysis of our in vitro coculture experiment indicated that F. nucleatum evokes two distinct cellular responses. Based on dosage dependence and expression pattern, these two responses are likely activated through independent signaling pathways. One is an immediate response that peaked at 24 h to 48 h of incubation and declined to a near-unstimulated level after 72 h. This immediate response was induced only by a high amount of F. nucleatum and marked by activation of interferon response genes, antiviral genes, cytokines, and chemokines. Other inflammation-inducing pathogens, such as H. pylori, may collaborate with F. nucleatum to sustain expression of these genes. The second response involved activation of actin and genes that regulate cell mobility. High expression of these gene products has multiple effects in promoting cancer progression, especially in increasing cell mobility and promoting distant metastasis[33-34]. Overall, our experiment demonstrates that colonization by F. nucleatum activates specific signaling pathways and promotes aggressiveness in gastric cancer cells.
After determining the prevalence of F. nucleatum colonization in gastric cancer patients, we evaluated its clinical impact. All enrolled patients underwent gastrectomy, in which the main cancerous lesions were completely removed. Gastrectomy also results in extraction of cancer-associated microbiota, minimizing the negative impact of pathogenic microbes. The average 5-year survival in our study cohort was approximately 60%, indicating the effectiveness of treatment. Notably, survival analysis showed that F. nucleatum colonization negatively impacts the survival outcome of H. pylori-positive patients. It is possible that F. nucleatum sequentially and/or synergistically cooperates with H. pylori to promote gastric cancer progression. Our analysis indicates a complex interaction between multiple pathogens and gastric cancer cells. Moreover, our data suggest that F. nucleatum colonization may serve as a prognostic biomarker for H. pylori-positive patients.
Our experimental findings show that F. nucleatum colonization in the gastric microbiota is a common event in gastric cancer patients. The risk of colonization appears to increase toward the later stages of gastric cancer progression. Cocolonization of F. nucleatum with H. pylori results in poorer survival than that observed for patients with H. pylori colonization alone. As the majority of gastric microbiota components are presumably removed during gastrectomy, preliminary exposure to F. nucleatum may exert a long-term impact on the aggressiveness of cancer cells. Our findings indicate that F. nucleatum may precondition and promote gastric cancer progression.
In this study, we found that F. nucleatum is able to alter actin filament dynamics to promote cell mobility. Additionally, the prevalence of F. nucleatum colonization in gastric cancer tissues is much higher than previously thought. Importantly, cocolonization of F. nucleatum with H. pylori was found to lead to reduced survival in gastric cancer patients. Our study suggests that F. nucleatum increases the aggressiveness of gastric cancer and negatively impacts the prognosis of gastric cancer patients.
Our previous research identified Fusobacterium nucleatum (F. nucleatum) as an opportunistic pathogen frequently found in the gastric cancer-associated microbiota. F. nucleatum has been demonstrated to promote carcinogenesis and metastasis of colorectal cancer. However, the role of F. nucleatum in gastric cancer remains unclear.
It is our goal to determine the impact of F. nucleatum colonization to progression of gastric cancer.
The objective of current study is to identify the impact of F. nucleatum to the cellular function of gastric cancers and to the prognosis of gastric cancer patients.
F. nucleatum-induced expression change of a patient-derived gastric cancer cell line was profiled by RNA sequencing and ontological analysis. The presence of F. nucleatum in patients' tumor tissue was determined by nested polymerase chain reaction. Statistical analysis of F. nucleatum colonization status was performed to determine the correlation with clinical characterization and patients' survival.
F. nucleatum induces a drastic but temporary interferon response and prolonged deregulation of actin and its regulators from gastric cancer cells. A survey of clinical specimens showed that approximately one-third of gastric cancer patients are positive for F. nucleatum. Survival analysis showed that the combined colonization of Helicobacter pylori (H. pylori) and F. nucleatum leads to poorer survival of late-stage patients.
The actin filament dynamic change caused by F. nucleatum colonization likely promotes cell mobility and cancer metastasis. This observation is correlated with the finding that F. nucleatum colonization leads to poor survival of H. pylori-positive late-stage patients.
F. nucleatum colonization leads to poorer survival of gastrectomy-received patients. Our findings indicate the importance of tumor-associated microbiota to the progression of gastric cancer.
Manuscript source: Unsolicited manuscript
Specialty type: Gastroenterology and hepatology
Country/Territory of origin: Taiwan
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): B
Grade C (Good): C
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Krzyżek P S-Editor: Wu YXJ L-Editor: Filipodia P-Editor: Liu JH
| 1. | Shah MA. Gastric cancer: The gastric microbiota - bacterial diversity and implications. Nat Rev Gastroenterol Hepatol. 2017;14:692-693. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 19] [Cited by in F6Publishing: 18] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 2. | Holleczek B, Schöttker B, Brenner H. Helicobacter pylori infection, chronic atrophic gastritis and risk of stomach and esophagus cancer: Results from the prospective population-based ESTHER cohort study. Int J Cancer. 2020;146:2773-2783. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 17] [Cited by in F6Publishing: 19] [Article Influence: 4.8] [Reference Citation Analysis (0)] |
| 3. | Yong X, Tang B, Li BS, Xie R, Hu CJ, Luo G, Qin Y, Dong H, Yang SM. Helicobacter pylori virulence factor CagA promotes tumorigenesis of gastric cancer via multiple signaling pathways. Cell Commun Signal. 2015;13:30. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 105] [Cited by in F6Publishing: 132] [Article Influence: 14.7] [Reference Citation Analysis (0)] |
| 4. | Uemura N, Okamoto S, Yamamoto S, Matsumura N, Yamaguchi S, Yamakido M, Taniyama K, Sasaki N, Schlemper RJ. Helicobacter pylori infection and the development of gastric cancer. N Engl J Med. 2001;345:784-789. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 3126] [Cited by in F6Publishing: 3021] [Article Influence: 131.3] [Reference Citation Analysis (0)] |
| 5. | Ferreira RM, Pereira-Marques J, Pinto-Ribeiro I, Costa JL, Carneiro F, Machado JC, Figueiredo C. Gastric microbial community profiling reveals a dysbiotic cancer-associated microbiota. Gut. 2018;67:226-236. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 341] [Cited by in F6Publishing: 388] [Article Influence: 64.7] [Reference Citation Analysis (1)] |
| 6. | Liu X, Shao L, Liu X, Ji F, Mei Y, Cheng Y, Liu F, Yan C, Li L, Ling Z. Alterations of gastric mucosal microbiota across different stomach microhabitats in a cohort of 276 patients with gastric cancer. EBioMedicine. 2019;40:336-348. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 92] [Cited by in F6Publishing: 160] [Article Influence: 26.7] [Reference Citation Analysis (0)] |
| 7. | Espinoza JL, Matsumoto A, Tanaka H, Matsumura I. Gastric microbiota: An emerging player in Helicobacter pylori-induced gastric malignancies. Cancer Lett. 2018;414:147-152. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 55] [Cited by in F6Publishing: 58] [Article Influence: 8.3] [Reference Citation Analysis (0)] |
| 8. | Mima K, Nishihara R, Qian ZR, Cao Y, Sukawa Y, Nowak JA, Yang J, Dou R, Masugi Y, Song M, Kostic AD, Giannakis M, Bullman S, Milner DA, Baba H, Giovannucci EL, Garraway LA, Freeman GJ, Dranoff G, Garrett WS, Huttenhower C, Meyerson M, Meyerhardt JA, Chan AT, Fuchs CS, Ogino S. Fusobacterium nucleatum in colorectal carcinoma tissue and patient prognosis. Gut. 2016;65:1973-1980. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 530] [Cited by in F6Publishing: 615] [Article Influence: 76.9] [Reference Citation Analysis (0)] |
| 9. | Castellarin M, Warren RL, Freeman JD, Dreolini L, Krzywinski M, Strauss J, Barnes R, Watson P, Allen-Vercoe E, Moore RA, Holt RA. Fusobacterium nucleatum infection is prevalent in human colorectal carcinoma. Genome Res. 2012;22:299-306. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1164] [Cited by in F6Publishing: 1328] [Article Influence: 102.2] [Reference Citation Analysis (0)] |
| 10. | Oh HJ, Kim JH, Bae JM, Kim HJ, Cho NY, Kang GH. Prognostic Impact of Fusobacterium nucleatum Depends on Combined Tumor Location and Microsatellite Instability Status in Stage II/III Colorectal Cancers Treated with Adjuvant Chemotherapy. J Pathol Transl Med. 2019;53:40-49. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 23] [Cited by in F6Publishing: 36] [Article Influence: 6.0] [Reference Citation Analysis (0)] |
| 11. | Yoshida Y, Suwabe K, Nagano K, Kezuka Y, Kato H, Yoshimura F. Identification and enzymic analysis of a novel protein associated with production of hydrogen sulfide and L-serine from L-cysteine in Fusobacterium nucleatum subsp. nucleatum ATCC 25586. Microbiology (Reading). 2011;157:2164-2171. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 14] [Cited by in F6Publishing: 14] [Article Influence: 1.1] [Reference Citation Analysis (0)] |
| 12. | Chen Y, Peng Y, Yu J, Chen T, Wu Y, Shi L, Li Q, Wu J, Fu X. Invasive Fusobacterium nucleatum activates beta-catenin signaling in colorectal cancer via a TLR4/P-PAK1 cascade. Oncotarget. 2017;8:31802-31814. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 89] [Cited by in F6Publishing: 110] [Article Influence: 18.3] [Reference Citation Analysis (0)] |
| 13. | Rubinstein MR, Baik JE, Lagana SM, Han RP, Raab WJ, Sahoo D, Dalerba P, Wang TC, Han YW. Fusobacterium nucleatum promotes colorectal cancer by inducing Wnt/β-catenin modulator Annexin A1. EMBO Rep. 2019;20. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 160] [Cited by in F6Publishing: 254] [Article Influence: 50.8] [Reference Citation Analysis (0)] |
| 14. | Chen Y, Lu Y, Ke Y, Li Y. Prognostic impact of the Fusobacterium nucleatum status in colorectal cancers. Medicine (Baltimore). 2019;98:e17221. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 20] [Cited by in F6Publishing: 24] [Article Influence: 4.8] [Reference Citation Analysis (0)] |
| 15. | Wong SH, Kwong TNY, Chow TC, Luk AKC, Dai RZW, Nakatsu G, Lam TYT, Zhang L, Wu JCY, Chan FKL, Ng SSM, Wong MCS, Ng SC, Wu WKK, Yu J, Sung JJY. Quantitation of faecal Fusobacterium improves faecal immunochemical test in detecting advanced colorectal neoplasia. Gut. 2017;66:1441-1448. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 157] [Cited by in F6Publishing: 174] [Article Influence: 24.9] [Reference Citation Analysis (0)] |
| 16. | Kapatral V, Anderson I, Ivanova N, Reznik G, Los T, Lykidis A, Bhattacharyya A, Bartman A, Gardner W, Grechkin G, Zhu L, Vasieva O, Chu L, Kogan Y, Chaga O, Goltsman E, Bernal A, Larsen N, D'Souza M, Walunas T, Pusch G, Haselkorn R, Fonstein M, Kyrpides N, Overbeek R. Genome sequence and analysis of the oral bacterium Fusobacterium nucleatum strain ATCC 25586. J Bacteriol. 2002;184:2005-2018. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 261] [Cited by in F6Publishing: 272] [Article Influence: 12.4] [Reference Citation Analysis (0)] |
| 17. | Hsieh YY, Tung SY, Pan HY, Yen CW, Xu HW, Lin YJ, Deng YF, Hsu WT, Wu CS, Li C. Increased Abundance of Clostridium and Fusobacterium in Gastric Microbiota of Patients with Gastric Cancer in Taiwan. Sci Rep. 2018;8:158. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 125] [Cited by in F6Publishing: 160] [Article Influence: 26.7] [Reference Citation Analysis (0)] |
| 18. | Schoggins JW. Interferon-Stimulated Genes: What Do They All Do? Annu Rev Virol. 2019;6:567-584. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 310] [Cited by in F6Publishing: 493] [Article Influence: 98.6] [Reference Citation Analysis (0)] |
| 19. | Chen M, Meng Q, Qin Y, Liang P, Tan P, He L, Zhou Y, Chen Y, Huang J, Wang RF, Cui J. TRIM14 Inhibits cGAS Degradation Mediated by Selective Autophagy Receptor p62 to Promote Innate Immune Responses. Mol Cell. 2016;64:105-119. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 198] [Cited by in F6Publishing: 249] [Article Influence: 31.1] [Reference Citation Analysis (0)] |
| 20. | Tan P, He L, Cui J, Qian C, Cao X, Lin M, Zhu Q, Li Y, Xing C, Yu X, Wang HY, Wang RF. Assembly of the WHIP-TRIM14-PPP6C Mitochondrial Complex Promotes RIG-I-Mediated Antiviral Signaling. Mol Cell. 2017;68:293-307.e5. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 63] [Cited by in F6Publishing: 52] [Article Influence: 7.4] [Reference Citation Analysis (0)] |
| 21. | Zhao C, Collins MN, Hsiang TY, Krug RM. Interferon-induced ISG15 pathway: an ongoing virus-host battle. Trends Microbiol. 2013;21:181-186. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 93] [Cited by in F6Publishing: 86] [Article Influence: 7.8] [Reference Citation Analysis (0)] |
| 22. | Basters A, Knobeloch KP, Fritz G. USP18 - a multifunctional component in the interferon response. Biosci Rep. 2018;38. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 41] [Cited by in F6Publishing: 50] [Article Influence: 8.3] [Reference Citation Analysis (0)] |
| 23. | Hornung V, Hartmann R, Ablasser A, Hopfner KP. OAS proteins and cGAS: unifying concepts in sensing and responding to cytosolic nucleic acids. Nat Rev Immunol. 2014;14:521-528. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 179] [Cited by in F6Publishing: 211] [Article Influence: 21.1] [Reference Citation Analysis (0)] |
| 24. | Wang Y, Holleufer A, Gad HH, Hartmann R. Length dependent activation of OAS proteins by dsRNA. Cytokine. 2020;126:154867. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 9] [Cited by in F6Publishing: 9] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 25. | de Neergaard M, Kim J, Villadsen R, Fridriksdottir AJ, Rank F, Timmermans-Wielenga V, Langerød A, Børresen-Dale AL, Petersen OW, Rønnov-Jessen L. Epithelial-stromal interaction 1 (EPSTI1) substitutes for peritumoral fibroblasts in the tumor microenvironment. Am J Pathol. 2010;176:1229-1240. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 25] [Cited by in F6Publishing: 31] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 26. | Nielsen HL, Rønnov-Jessen L, Villadsen R, Petersen OW. Identification of EPSTI1, a novel gene induced by epithelial-stromal interaction in human breast cancer. Genomics. 2002;79:703-710. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 44] [Cited by in F6Publishing: 46] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 27. | Chen S, Pan S, Wu H, Zhou J, Huang Y, Wang S, Liu A. ICAM1 Regulates the Development of Gastric Cancer and May Be a Potential Biomarker for the Early Diagnosis and Prognosis of Gastric Cancer. Cancer Manag Res. 2020;12:1523-1534. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 7] [Cited by in F6Publishing: 5] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 28. | Huang S, Yang Z, Zou D, Dong D, Liu A, Liu W, Huang L. Rapid detection of nusG and fadA in Fusobacterium nucleatum by loop-mediated isothermal amplification. J Med Microbiol. 2016;65:760-769. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 10] [Cited by in F6Publishing: 11] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 29. | Yamamura K, Baba Y, Miyake K, Nakamura K, Shigaki H, Mima K, Kurashige J, Ishimoto T, Iwatsuki M, Sakamoto Y, Yamashita Y, Yoshida N, Watanabe M, Baba H. Fusobacterium nucleatum in gastroenterological cancer: Evaluation of measurement methods using quantitative polymerase chain reaction and a literature review. Oncol Lett. 2017;14:6373-6378. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 12] [Cited by in F6Publishing: 30] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 30. | Brennan CA, Garrett WS. Fusobacterium nucleatum - symbiont, opportunist and oncobacterium. Nat Rev Microbiol. 2019;17:156-166. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 359] [Cited by in F6Publishing: 528] [Article Influence: 132.0] [Reference Citation Analysis (0)] |
| 31. | Bravo D, Hoare A, Soto C, Valenzuela MA, Quest AF. Helicobacter pylori in human health and disease: Mechanisms for local gastric and systemic effects. World J Gastroenterol. 2018;24:3071-3089. [PubMed] [DOI] [Cited in This Article: ] [Cited by in CrossRef: 109] [Cited by in F6Publishing: 103] [Article Influence: 17.2] [Reference Citation Analysis (4)] |
| 32. | Coker OO, Dai Z, Nie Y, Zhao G, Cao L, Nakatsu G, Wu WK, Wong SH, Chen Z, Sung JJY, Yu J. Mucosal microbiome dysbiosis in gastric carcinogenesis. Gut. 2018;67:1024-1032. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 304] [Cited by in F6Publishing: 358] [Article Influence: 59.7] [Reference Citation Analysis (0)] |
| 33. | Izdebska M, Zielińska W, Grzanka D, Gagat M. The Role of Actin Dynamics and Actin-Binding Proteins Expression in Epithelial-to-Mesenchymal Transition and Its Association with Cancer Progression and Evaluation of Possible Therapeutic Targets. Biomed Res Int. 2018;2018:4578373. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 64] [Cited by in F6Publishing: 83] [Article Influence: 13.8] [Reference Citation Analysis (0)] |
| 34. | Ohtaki S, Wanibuchi M, Kataoka-Sasaki Y, Sasaki M, Oka S, Noshiro S, Akiyama Y, Mikami T, Mikuni N, Kocsis JD, Honmou O. ACTC1 as an invasion and prognosis marker in glioma. J Neurosurg. 2017;126:467-475. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 20] [Cited by in F6Publishing: 24] [Article Influence: 3.0] [Reference Citation Analysis (0)] |