Copyright
©The Author(s) 2020.
World J Gastroenterol. Sep 21, 2020; 26(35): 5223-5247
Published online Sep 21, 2020. doi: 10.3748/wjg.v26.i35.5223
Published online Sep 21, 2020. doi: 10.3748/wjg.v26.i35.5223
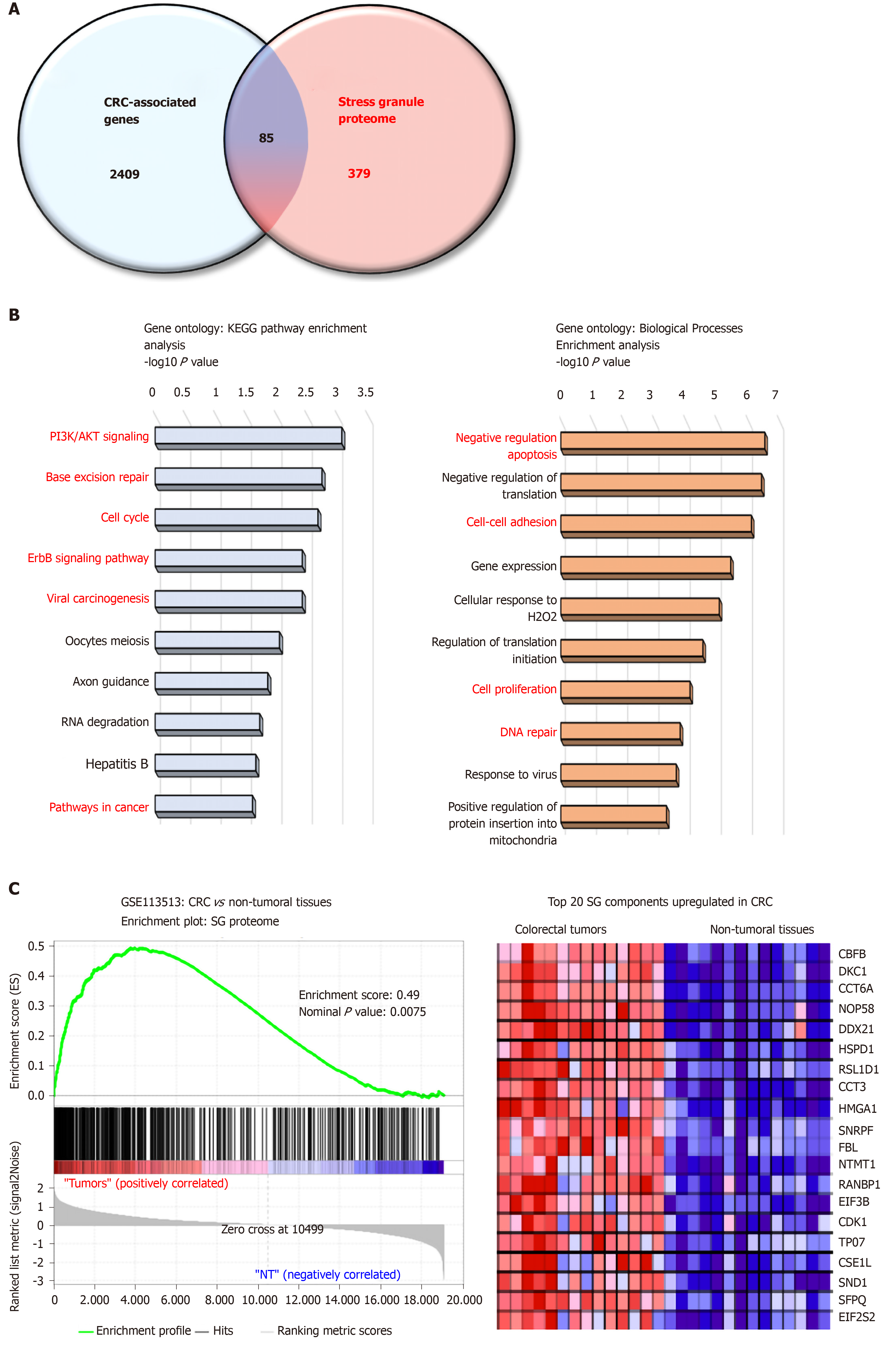
Figure 1 The stress granule proteome contains several colorectal cancer-associated proteins.
A: Venn diagram merging a list of colorectal cancer (CRC) -associated genes (retrieved from Metacore software) and the mammalian stress granule (SG) proteome from https://msgp.pt; B: Gene ontology analysis of SG proteome using KEGG pathway and biological processes analysis. Enrichment is represented with a-log10 P value. Processes and pathways in red are those involved in cancer development; C: Gene-Set Enrichment Analysis (version 3.0, Broad Institute, Cambridge, MA, United States) of the SG proteome on CRC patients (GSE113513). The top 20 genes upregulated in CRC patients as compared to non-tumoral tissues are represented in a heatmap. The enrichment score was calculated using the number of genes ranking at the top or the bottom of the gene list (permutation type: Phenotype; with 1000 permutations). The Signal2Noise was used for ranking genes. A nominal P value < 0.05 and an FDR < 0.2 were considered significant.
- Citation: Legrand N, Dixon DA, Sobolewski C. Stress granules in colorectal cancer: Current knowledge and potential therapeutic applications. World J Gastroenterol 2020; 26(35): 5223-5247
- URL: https://www.wjgnet.com/1007-9327/full/v26/i35/5223.htm
- DOI: https://dx.doi.org/10.3748/wjg.v26.i35.5223