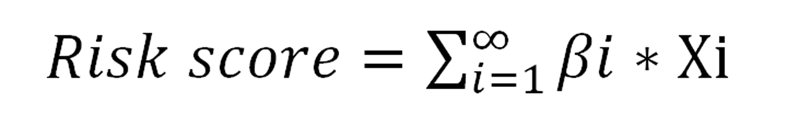Published online Mar 28, 2020. doi: 10.3748/wjg.v26.i12.1298
Peer-review started: December 1, 2019
First decision: December 23, 2019
Revised: January 8, 2020
Accepted: March 9, 2020
Article in press: March 9, 2020
Published online: March 28, 2020
Colorectal cancer (CRC) is one of the most prevalent tumors worldwide. Recently, long noncoding RNAs (lncRNAs) have been shown to influence tumorigenesis and tumor progression by acting as competing endogenous RNAs (ceRNAs). It is difficult to extract prognostic lncRNAs and useful bioinformation from most ceRNA networks constructed previously.
To construct a prognostic related ceRNA regulatory network and lncRNA related signature based on risk score in CRC.
RNA transcriptome profile and clinical information of 506 CRC patients were downloaded from the Cancer Genome Atlas database. R packages and Perl program were used for data processing. Cox regression analysis was used for prognostic model construction. Quantitative real-time polymerase chain reaction was used to detect the expression of lncRNAs.
A prognostic-related ceRNA network was constructed, including 9 lncRNAs, 44 mRNAs, and 30 miRNAs. In addition, a four-lncRNA model was constructed using multivariate Cox regression analysis, which could be an independent prognostic model in CRC. The risk score for each patient was calculated, and the 506 patients were divided into high and low-risk groups (253 for each group) based on the median risk score. The results of the survival analysis showed that patients with a high-risk score had a poor survival rate. Furthermore, the predictive value of the four-lncRNA model was evaluated in GSE38832. Patient survival probabilities could be better predicted when combing the risk score and clinical features. Gene Set Enrichment Analysis results verified that a number of cancer-related signaling pathways were enriched with a high-risk score in CRC. Finally, we validated a novel lncRNA (LINC00488) using quantitative real-time polymerase chain reaction in 22 paired CRC patient tumor tissues compared to adjacent non-tumor tissues.
The four-lncRNA model could give better predictive value for CRC patients. Our understanding of the lncRNA-related ceRNA regulatory mechanism could provide a potential diagnostic indicator for CRC patients.
Core tip: Although there have been improvements in the treatment of colorectal cancer, a number of patients still lost the chance for survival because of a shortage of useful biomarkers. In this study, we systematically analyzed RNA transcriptome profile and constructed a prognostic-related competing endogenous RNA network by a comprehensive analysis, constructed a four-lncRNA model, and evaluated the predictive value of this model in the Gene Expression Omnibus and the Cancer Genome Atlas databases. Our understanding of the long noncoding RNA-related competing endogenous RNA regulatory mechanism could provide a potential diagnostic indicator for colorectal cancer patients.
- Citation: Yang ZD, Kang H. Exploring prognostic potential of long noncoding RNAs in colorectal cancer based on a competing endogenous RNA network. World J Gastroenterol 2020; 26(12): 1298-1316
- URL: https://www.wjgnet.com/1007-9327/full/v26/i12/1298.htm
- DOI: https://dx.doi.org/10.3748/wjg.v26.i12.1298
Colorectal cancer (CRC) is one of the three most common cancers worldwide[1]. Although there have been improvements in the treatment of this disease, numerous patients are diagnosed at an advanced stage of the disease because of a shortage of useful biomarkers[2]. Advanced stage disease with distant metastasis is the main reason for the poor therapeutic efficacy of surgery in CRC. Therefore, discovering highly effective diagnostic biomarkers and understanding the molecular mechanisms underlying CRC tumorigenesis are needed to significantly increase the survival rate of CRC patients.
Long noncoding RNAs (lncRNAs) are non-coding RNAs of over 200 nucleotides that are located in the nucleus or cytoplasm. Several studies have demonstrated that lncRNAs play a significant role in the development of malignant tumors[3,4]. Salmena et al[5] hypothesized that lncRNAs and mRNAs could interact with each other via microRNAs (miRNAs). This hypothesis was built on the fact that lncRNAs contain miRNA response elements (MREs) and, thus, could competitively bind to the core seed regions of miRNAs to further regulate the expression of target genes. Increasing evidence in recent years has supported this theory by demonstrating that lncRNAs can act as competing endogenous RNAs (ceRNAs), causing decreased miRNA binding to mRNA targets[6]. Indeed, an increasing number of papers have implicated ceRNAs in cancer development[7].
Recently, studies have confirmed that the regulatory networks of lncRNA, miRNA, mRNA, and ceRNA play significant roles in the development of gastric cancer[8], cholangiocarcinoma[9], and CRC[10-12]. However, most of these generated ceRNA networks contain a large number of lncRNAs, and it is difficult to extract the lncRNAs closely related to survival prognosis. In this paper, univariate Cox regression analysis was performed on all the differentially expressed lncRNAs before a ceRNA network was constructed. The RNA expression profiles were combined with patient clinical features to generate the ceRNA network. Through the multivariate Cox regression model, a four-lncRNA model based on the risk score of the ceRNA network was developed. Furthermore, the association between this model, clinical information, and survival prognosis was analyzed. Key lncRNAs in the ceRNA network were validated in clinical CRC tissues by quantitative real-time polymerase chain reaction (qRT-PCR). Our study provided a deep understanding of ceRNAs, and the four-lncRNA model could identify novel candidate biomarkers for CRC.
The raw RNA sequences of 506 colorectal cancer patient samples (534 tumor and 41 non-tumor tissues) were retrieved from the Cancer Genome Atlas (TCGA) database (https://portal.gdc.cancer.gov/). The criteria of inclusion were: (1) Clinical information (age, gender, pathological stage, survival time, and status) should be complete; (2) Sequencing data should be complete, including RNA-Seq data for both mRNAs and lncRNAs; and (3) The follow-up survival time should be ≥ 30 d. Our study followed the TCGA publication guidelines.
We used the Ensemble annotation file (http://www.ensembl.org/index.html) to define and annotate lncRNAs and mRNAs. We identified differentially expressed mRNAs (DEmRNAs), lncRNAs (DElncRNAs), and miRNAs (DEmiRNAs) using program codes written with R and Perl software. A log2 absolute of the fold change ≥ 2 and P < 0.01 were considered significant. The “heatmap” and “gplots” functions of the R software package were used to generate a volcano map of the differentially expressed RNAs. Functional enrichment analysis was carried out for the DEmRNAs using the “clusterProfiler” package in the R software. Transcription factors (TFs) were identified amongst the clustered gene sets according to the ENCODE CHIP-seq data at the University of California Santa Cruz Genome Browser.
Univariate Cox regression was performed for all the aberrantly expressed lncRNAs to evaluate the correlation between expression and overall survival (OS). Only DElncRNAs correlated with prognosis were used for the analysis.
The ceRNA network of CRC was constructed following the steps listed below: (1) The miRcode database (http://www.mircode.org) was used to scientifically retrieve DElncRNA-miRNA interactions and matched DEmiRNA interactions were performed; (2) DEmiRNA-targeted mRNAs were predicted using miRTarBase (http://www.targetscan.org/), miRDB (http://www.mirdb.org/), and TargetScan (http://www.microrna.org/microrna/home.do) databases. Only the mRNAs presented in all three databases and overlapping with the DEmRNAs were used to construct the ceRNA network; and (3) The resulting map was generated using Cytoscape 3.5.1.
A prognostic predictive model based on the risk score was applied using multivariate Cox regression analysis for all the DElncRNAs involved in the ceRNA network. As shown the formula in the picture, where βi stands for the coefficient for each gene and Xi represents the z-score. Kaplan-Meier (K-M) survival analysis and receiver operating characteristic (ROC) curve analysis were used to assess the predictive value of this four-lncRNA model. Univariate and multivariable Cox regression analyses were used to evaluate whether the risk score was an independent predictor when compared with the clinical information.
The raw data was downloaded from the Gene Expression Omnibus (GEO) database. Expression profiles were processed with R software. The risk scores of the patients were calculated using the Four-lncRNA model. K-M and ROC curve analyses were carried out to assess the predictive value of the four-lncRNA model.
Total RNA was isolated from 22 paired CRC patient tissues using TRIzol reagent (Invitrogen, CA), cDNA was synthesized using the Transcriptor First Strand cDNA Synthesis Kit (Roche) following the manufacturer’s instructions. qRT-PCR was performed using FastStart Universal SYBR Green Master (ROX) (Roche) on the LightCycler 480. The primers used for target amplification are presented in Table 1. Relative expression was determined using the 2-ΔΔCt method.
| Gene | Sequence |
| LINC00488 | Forward: 5’ – GTTCACGGTGCTTCCTCAGA - 3’ |
| Reverse: 5’ – GGGCGACAGAACAAGACTCA - 3’ | |
| GAPDH | Forward: 5’ – GCACCGTCAAGGCTGAGAAC - 3’ |
| Reverse: 5’ – TGGTGAAGACGCCAGTGGA - 3’ |
The prognostic value of American Joint Committee on Cancer (AJCC) tumor invasion depth (T), lymph node metastasis (N), distant metastasis (M), and TNM stage was assessed through ROC curve and survival analyses. In addition, risk score and clinical features were combined, and the prediction value was evaluated using a K-M estimator.
Gene Set Enrichment Analysis (GSEA) v4.0.1 was carried out using the JAVA program with enrichment analysis performed on normalized mRNA expression profiles. Random sample permutations (n = 1000) were performed to calculate the P value. A false discovery rate < 0.01 and a nominal P < 0.05 were considered significant.
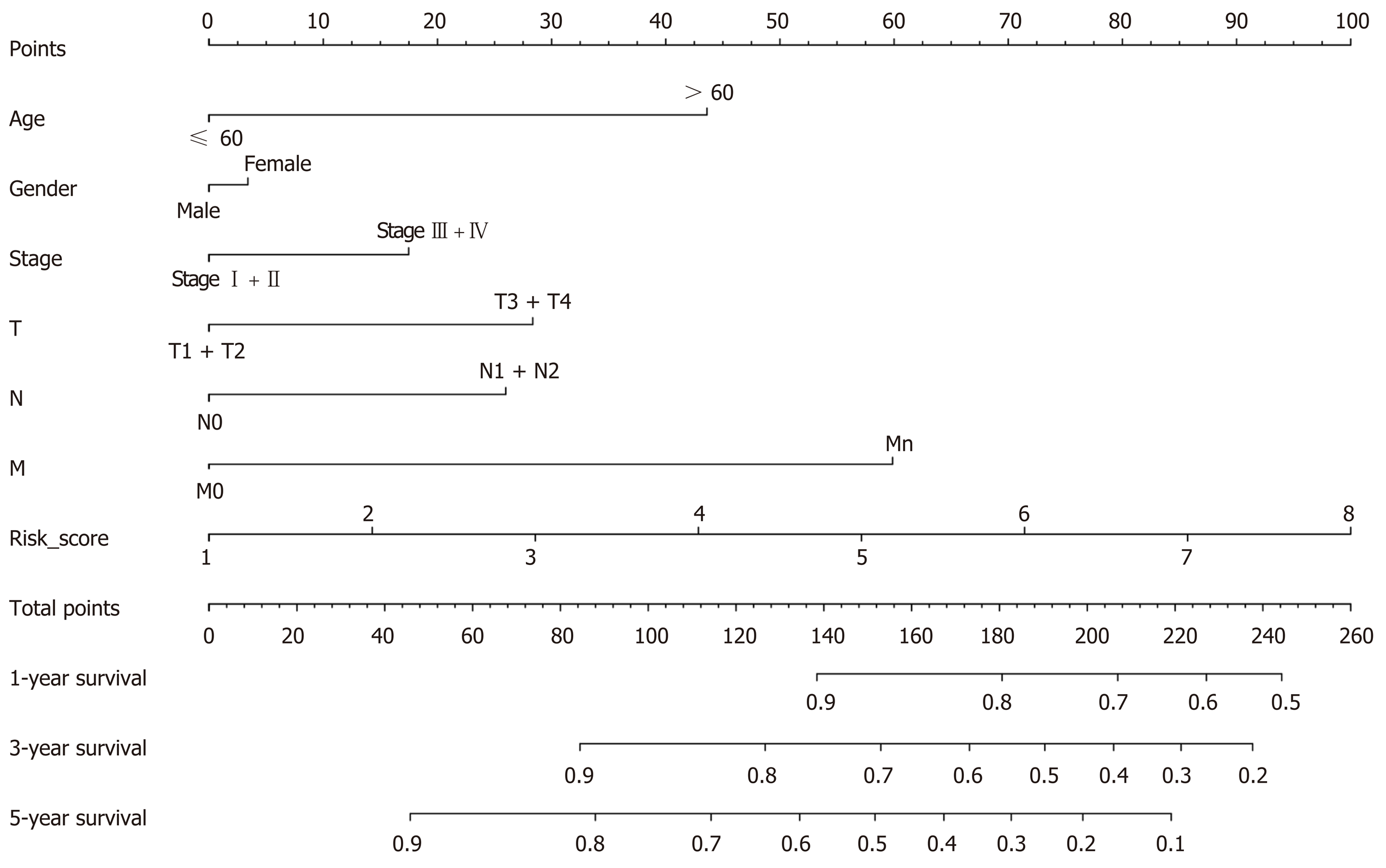
The risk score and clinical features were combined, and a nomogram was generated using the “rms” package of the R software.
R software and relevant R packages were used for all statistical analyses. Survival analysis was performed using the “survival” package. ROC curves were plotted using the “pROC” package. Pearson correlation analysis was carried out to determine correlations among DERNAs.
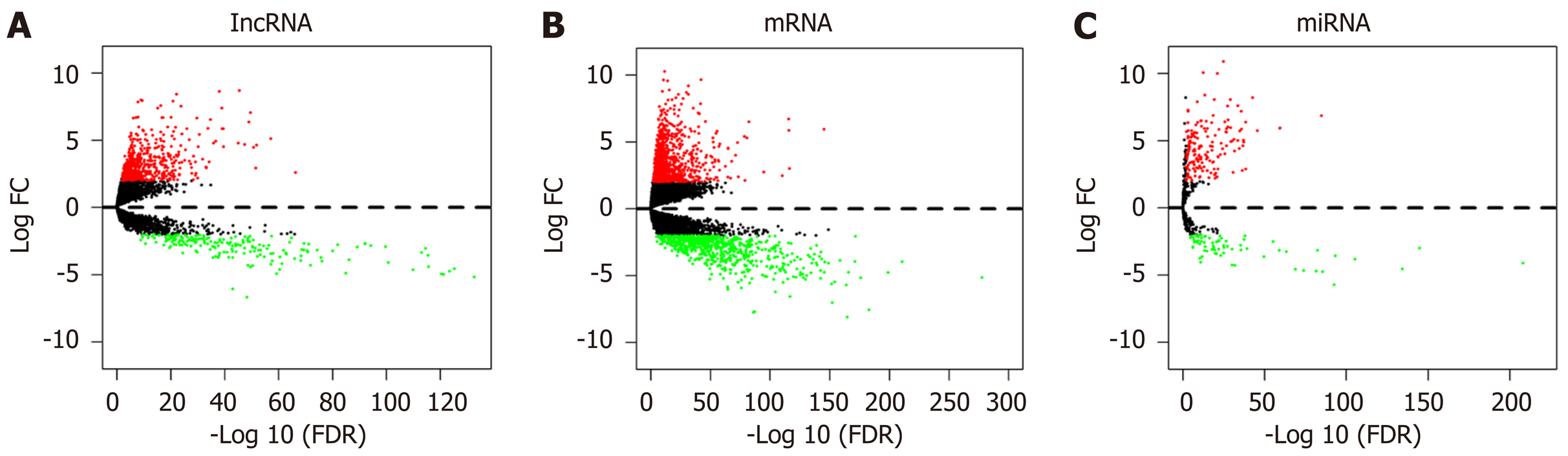
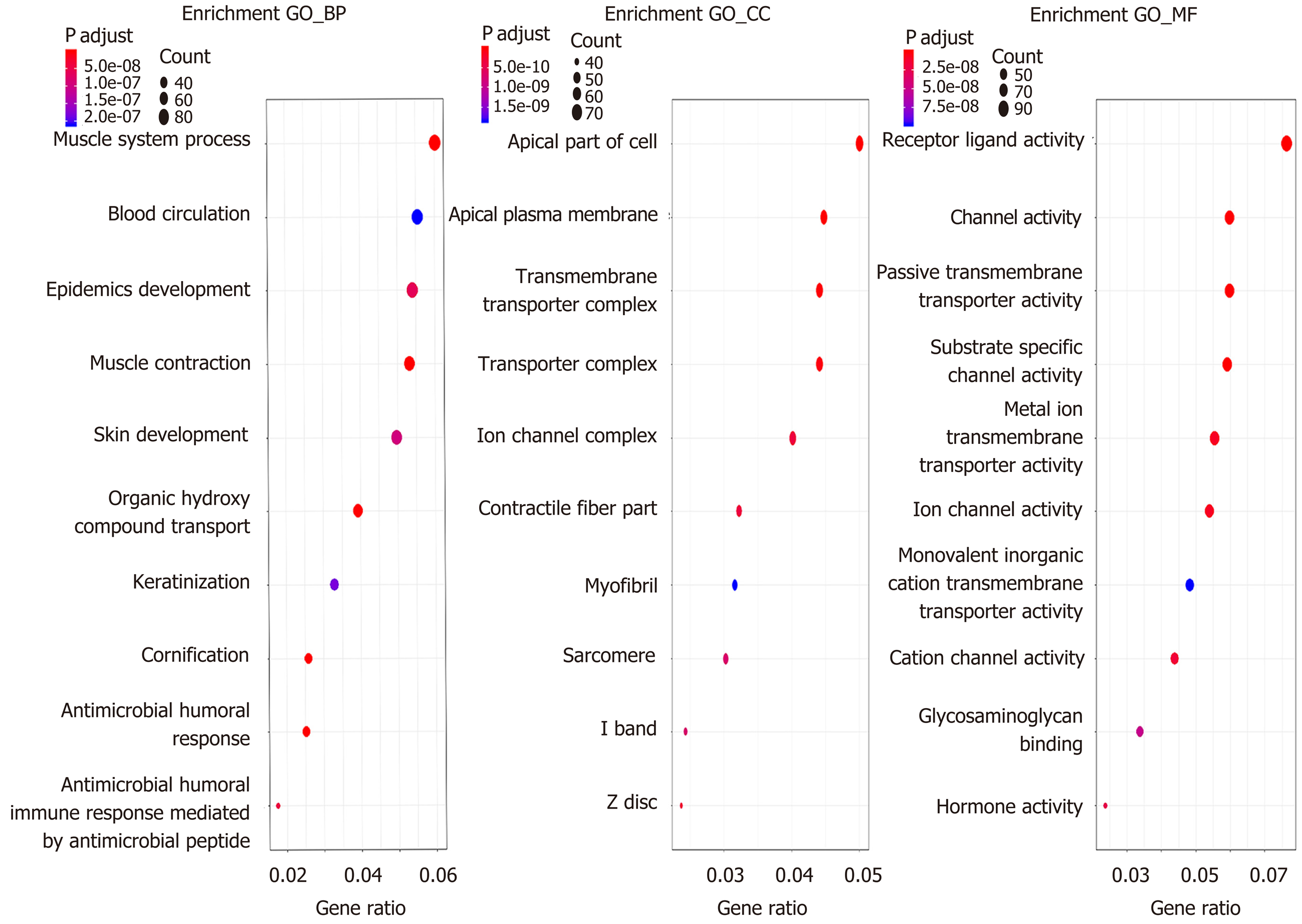
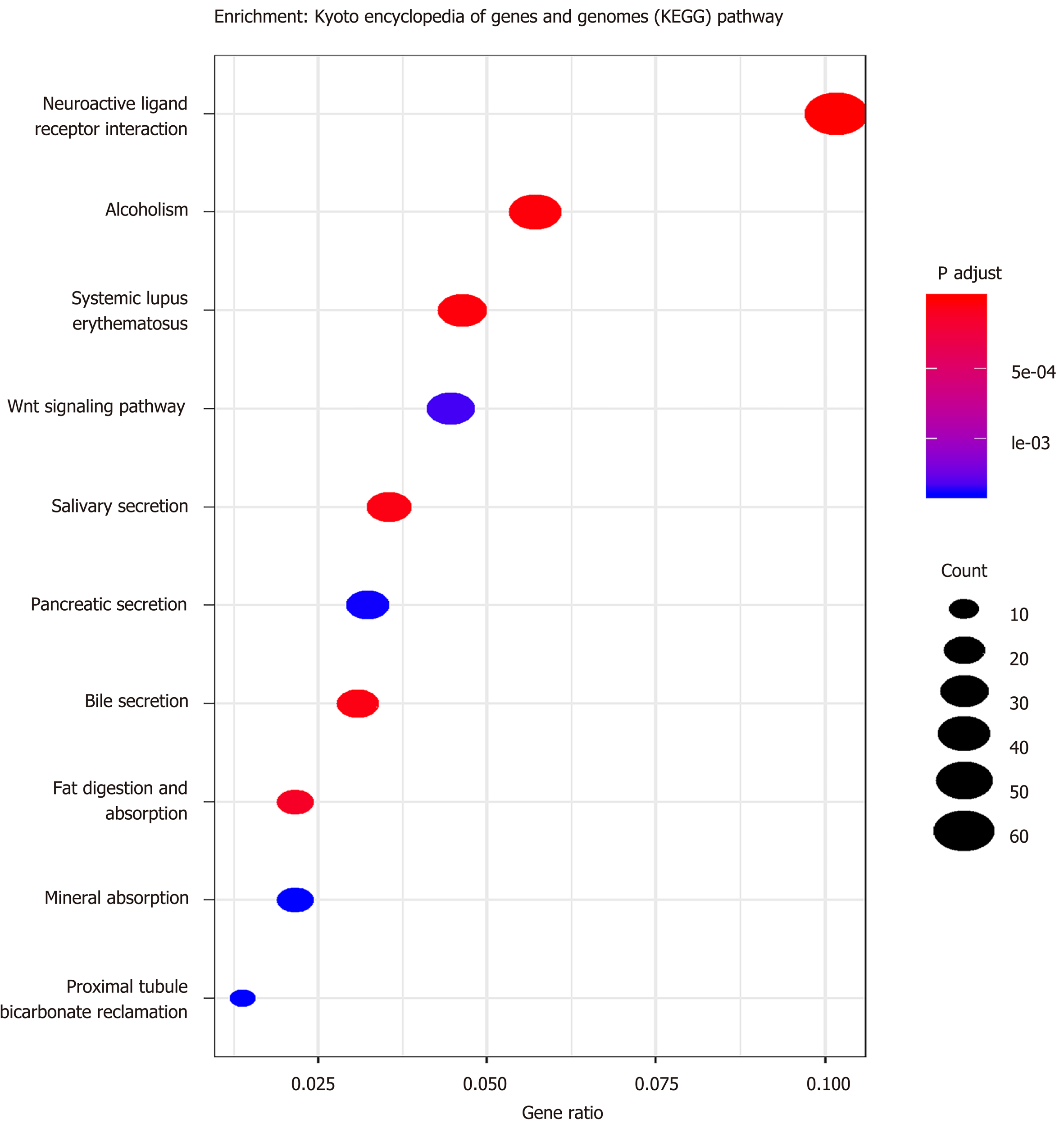
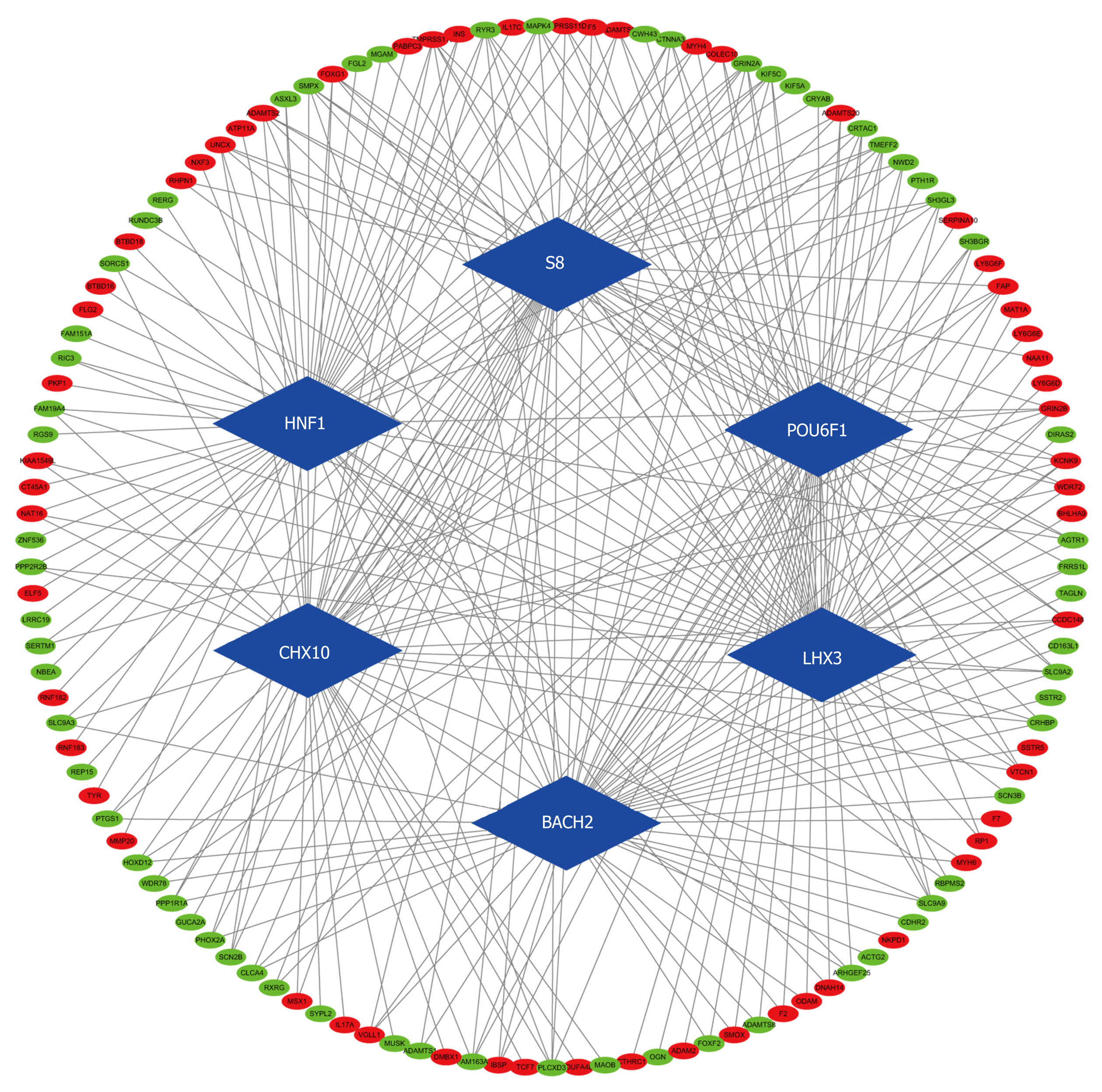
According to the cutoff threshold, 823 lncRNAs were differentially expressed (622 upregulated and 201 downregulated) (Figure 1A). We identified 95 lncRNAs that correlated with OS using univariate Cox regression analysis (Supplementary Table S1). In addition, we identified 869 upregulated and 756 downregulated mRNAs. Furthermore, 258 miRNAs (186 upregulated and 72 downregulated) were differentially expressed in the CRC tumors compared to the adjacent non-tumor tissues. The volcano maps of the RNAs are presented in Figure 1B and C. To further understand the molecular mechanisms involved in CRC development, functional enrichment analyses of the Kyoto Encyclopedia of Genes and Genomes pathway and Gene Ontology were performed for the DEmRNAs. The results are presented in Figures 2 and 3. TF-mRNA analysis was performed using a false discovery rate < 0.05. The most significant TFs and their links are shown in Figure 4.
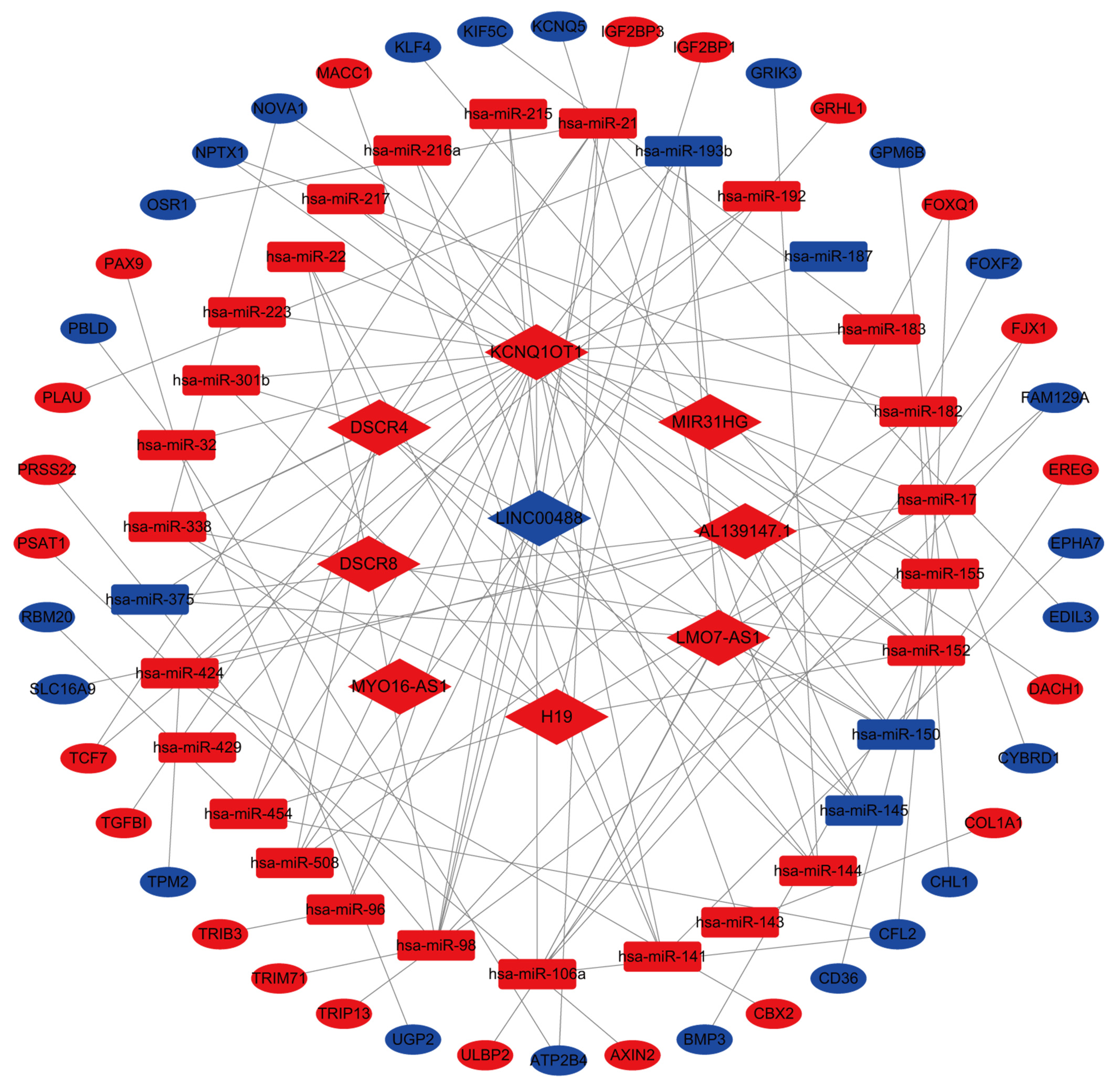
To understand the interactions between the DElncRNAs, DEmRNAs, and DEmiRNAs, 72 paired lncRNA-miRNA interactions (9 lncRNAs and 30 miRNAs) were constructed between the aberrantly expressed lncRNAs and miRNAs through the miRcode database. We found that 1419 mRNAs were targeted by these 30 miRNAs in the miRTarBase, miRDB, and TargetScan databases. The possible interactions between these mRNAs and 1625 DEmRNAs were cross-checked. As a result, 44 mRNAs were involved in the network. The final CRC ceRNA network was constructed by the incorporation of nine DElncRNAs, 44 DEmRNAs, and 30 DEmiRNAs (Figure 5).
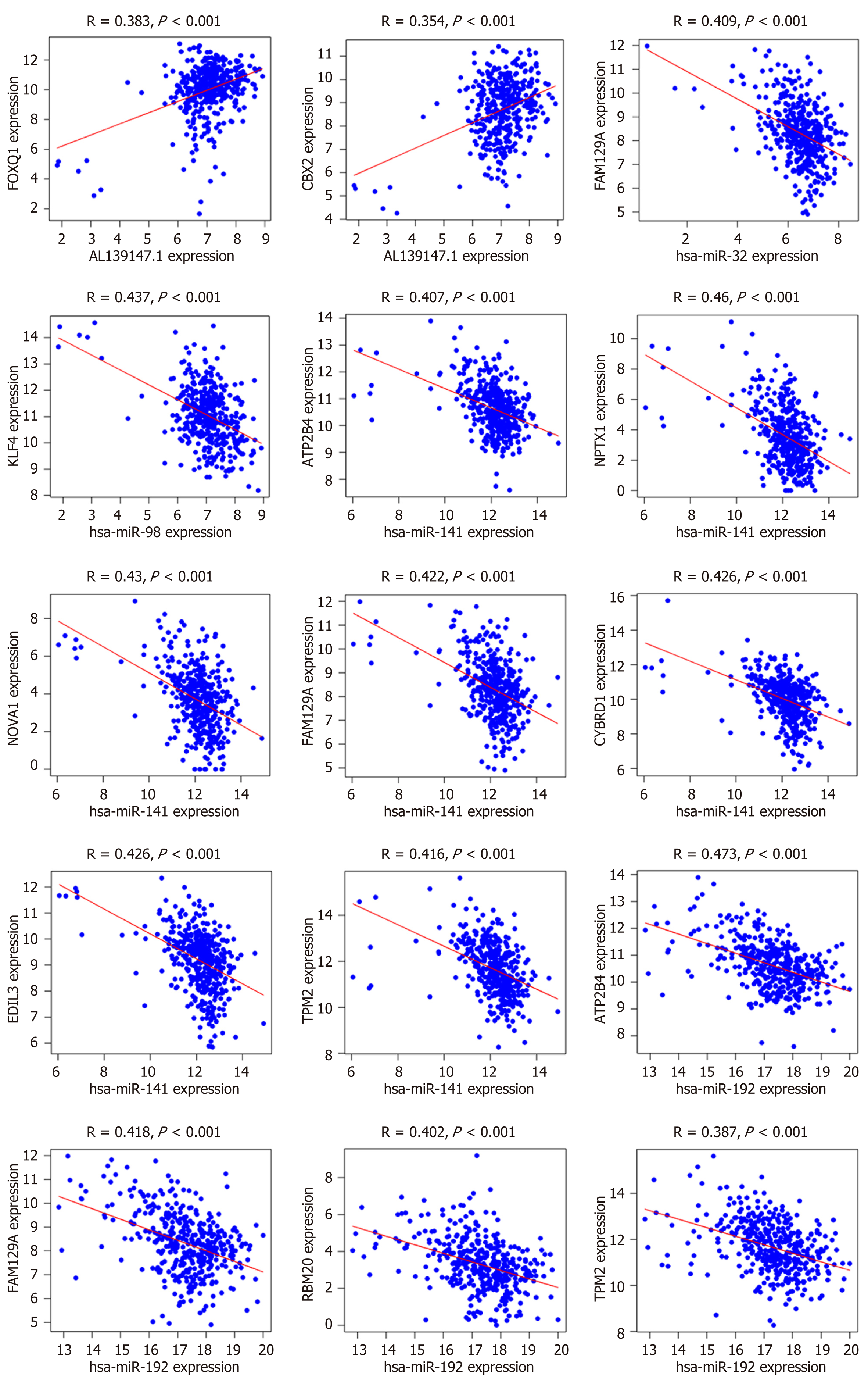
The ceRNA network could indicate possible interactions between the differentially expressed RNAs in CRC. To validate this hypothesis, Pearson correlation analysis was carried out on the mRNA expression levels of the RNAs included in the network. The results showed that only lncRNA AL139147.1 had a direct positive linear correlation with FOXQ1 and CBX2, whereas four miRNAs (has-miR-32, has-miR-98, has-miR-141, has-miR-192) had negative linear correlations with the DEmRNAs (R > 0.35, P < 0.01, Figure 6).
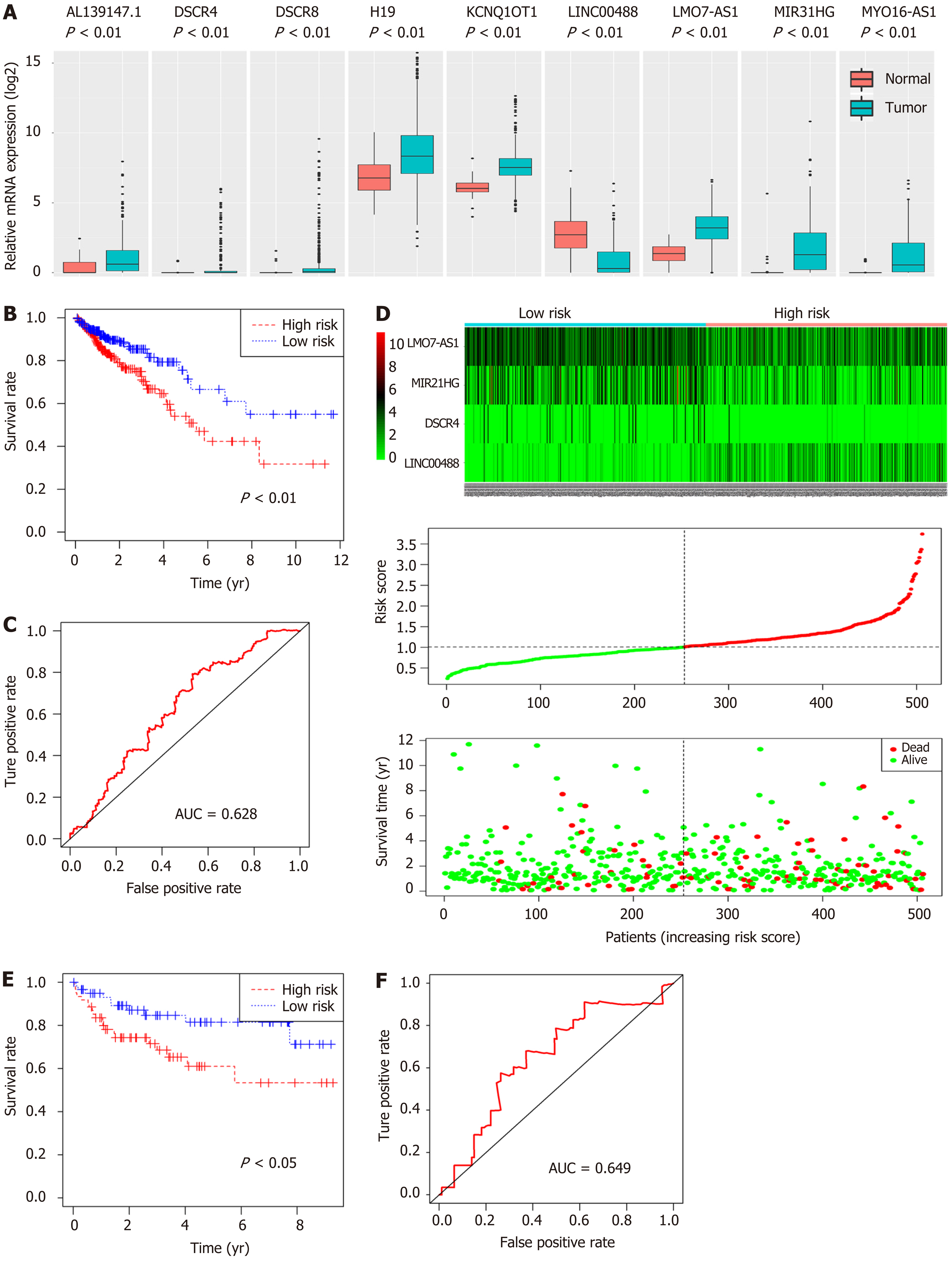
In order to identify potential prognostic markers, the expression levels of nine lncRNAs involved in the constructed ceRNA network were measured. The results showed that LINC00488 was downregulated whereas AL139147.1, DSCR4, DSCR8, H19, KCNQ1OT1, LMO7-AS1, MIR31HG, and MYO16-AS1 were upregulated in the CRC tumors compared to the non-tumor tissues (P < 0.05, Figure 7A).
A four-lncRNA multivariate Cox regression model was made for these nine lncRNAs of the ceRNA network. The risk scores were identified for each patient using the following formula: Risk score = (0.20671 * DSCR4) + (-0.27339 * LINC00488) + (0.09205 * MIR31HG) + (0.17811 * LMO7-AS1). The 506 patients were categorized into a high- or low-risk group (253 patients in each group) using the median risk score as the cutoff. Furthermore, the K-M method with the log-rank statistical test was used to evaluate survival between the two groups. We found that the high-risk group had a worse OS (P = 0.003, Figure 7B). Moreover, the area under the ROC curve (AUC) for the model was 0.628 (P < 0.05, Figure 7C), indicating the prognostic value of the four-lncRNA model based on the risk score. The heatmap, risk score distribution, and survival status for the expression profiles are presented in Figure 7D.
To demonstrate the predictive value of the four-lncRNA model for CRC patients, we downloaded GSE38832 from the GEO database, including the gene profiles and clinical features. The risk scores were calculated for 122 patients in this cohort. Interestingly, the survival analysis demonstrated that the patients with a high-risk score had a shorter survival time than those with a low-risk score (P = 0.024; Figure 7E), and the area under the ROC curve was 0.649 (Figure 7F).
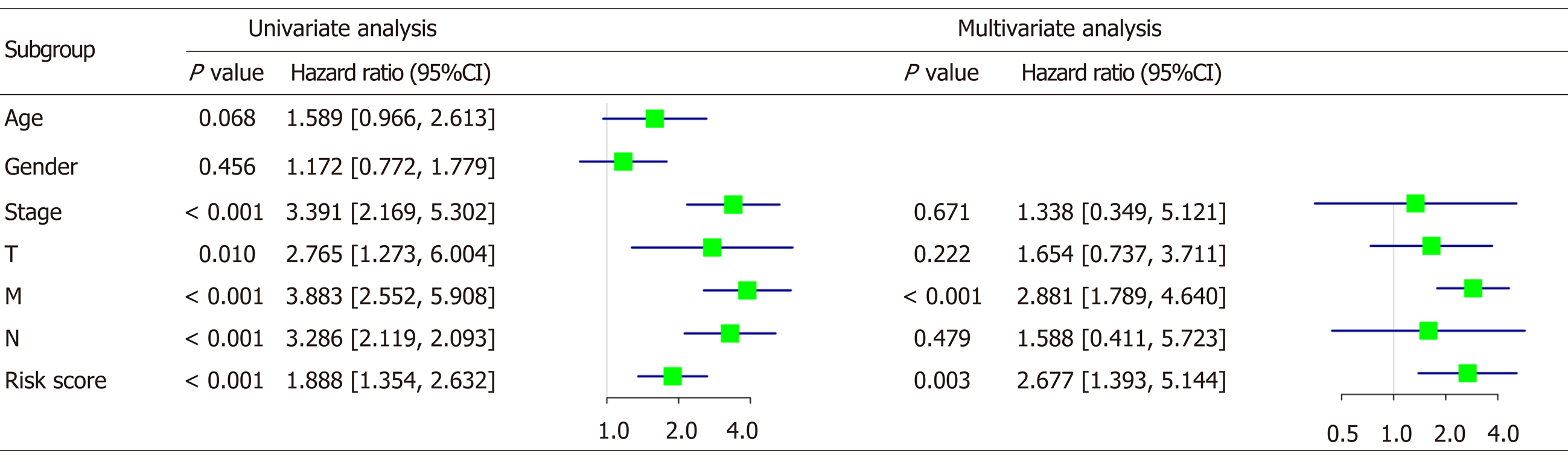
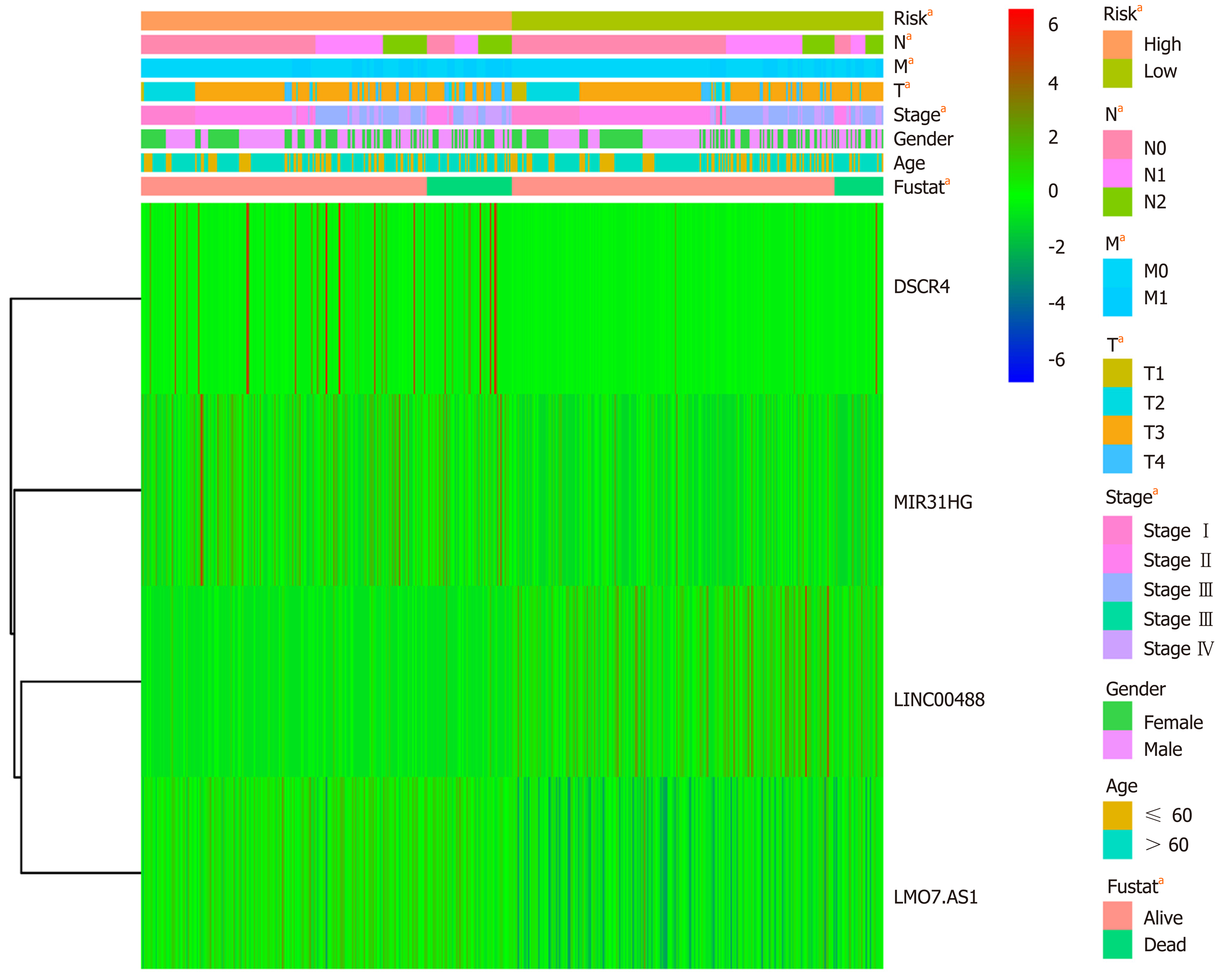
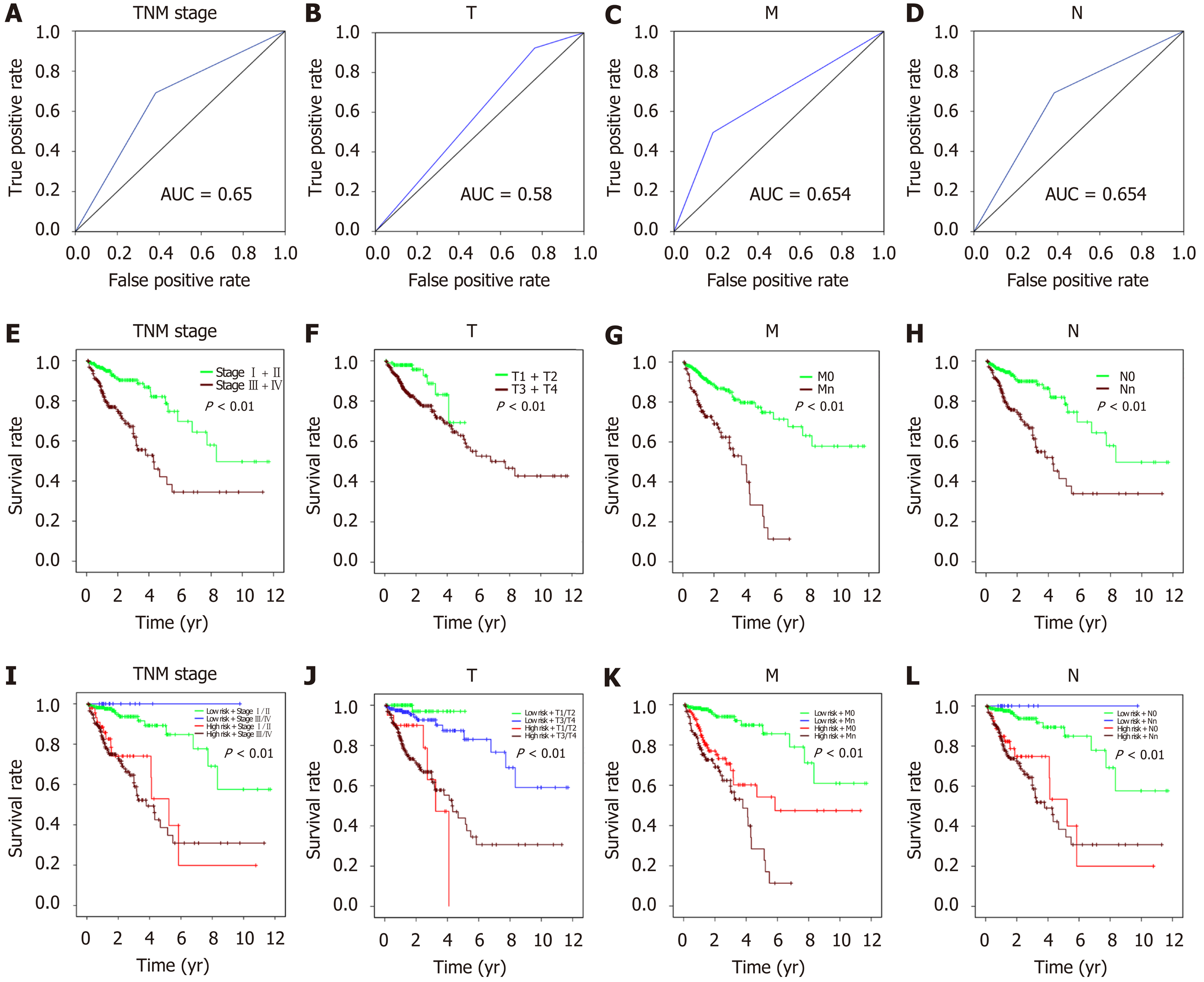
Correlation analysis between the risk score and patient clinical features revealed that the risk score was associated with tumor invasion depth (T), lymph node metastasis (N), and distant metastasis (M). However, no association was observed with other parameters (e.g., gender and age) (Table 2). Furthermore, univariate and multivariate Cox regression analyses were carried out to assess whether the four-lncRNA model and clinical features correlated with the OS (P < 0.05, Figure 8). Univariate analysis showed that TNM stage (P < 0.001), T (P = 0.01), N (P < 0.001), M (P < 0.001), and risk score of the four-lncRNA model (P < 0.001) were significantly associated with the OS. In addition, we found that the four-lncRNA model could be an independent prognostic factor for CRC patients using multivariate analysis (P = 0.003). The four-lncRNA model risk score with clinical features combined with mRNA expression levels is comprehensively presented in the heatmap (P < 0.05, Figure 9).
| Characteristic | Number of cases | Risk | P value | |
| Low (n = 253) | High (n = 253) | |||
| Age (yr) | 0.057 | |||
| ≤ 60 | 163 | 71 | 92 | |
| > 60 | 343 | 182 | 161 | |
| Gender | ||||
| Female | 230 | 112 | 118 | 0.655 |
| Male | 276 | 141 | 135 | |
| Stage | ||||
| Stage I + II | 285 | 238 | 47 | < 0.011 |
| Stage III + IV | 221 | 15 | 206 | |
| Tumor invasion depth | ||||
| T1 + T2 | 105 | 85 | 20 | < 0.011 |
| T3 + T4 | 401 | 168 | 233 | |
| Lymph node metastasis | ||||
| N0 | 295 | 238 | 57 | < 0.011 |
| N1 + N2 | 211 | 15 | 196 | |
| Distant metastasis | ||||
| M0 | 384 | 253 | 131 | < 0.011 |
| M1 + M2 | 122 | 0 | 122 | |
Interestingly, the ROC and survival analyses for the AJCC stage could effectively predict the prognosis of CRC patients. For the integrated analysis of risk score and AJCC stage, the results showed that patients with a high-risk score and tumor grade had a significantly worse prognosis (P < 0.001, Figure 10).
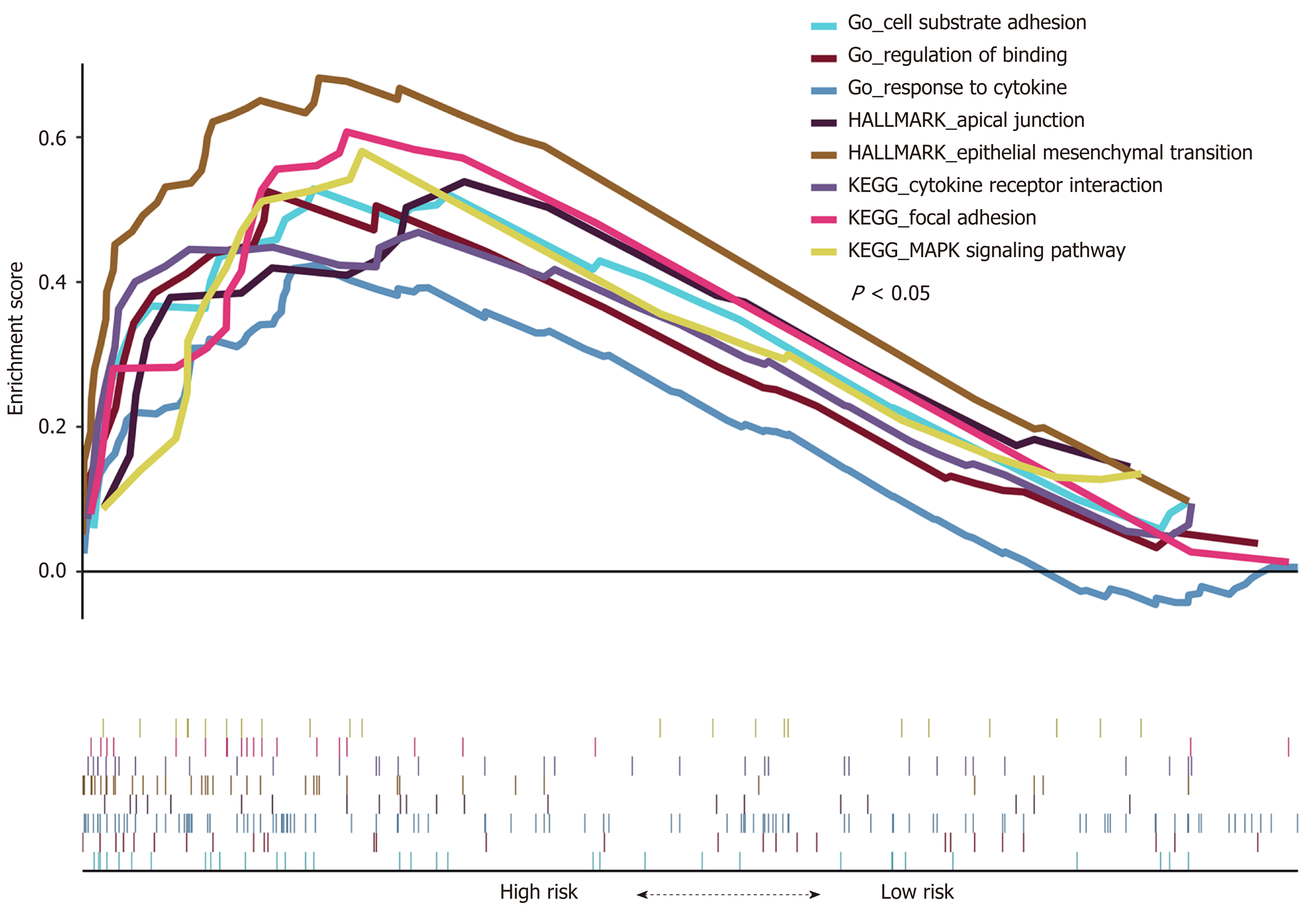
GSEA was carried out between the high- and low-risk groups (253 for each group) in the TCGA cohort to identify the high-risk-associated pathways. We found that “epithelial-mesenchymal transition (EMT),” “MAPK signaling pathway,” and some other cancer-related biological pathways were significantly enriched (P < 0.05, Figure 11). The results suggest that the four-lncRNA model may represent the status of these biological processes to further predict the OS of CRC patients.
Based on all the analyses, a comprehensive nomogram that integrated the risk scores and clinical information was constructed to predict the OS of CRC patients. Using the nomogram results, we could better predict the 1-, 3-, and 5-year survival probabilities of CRC patients (P < 0.05, Figure 12).
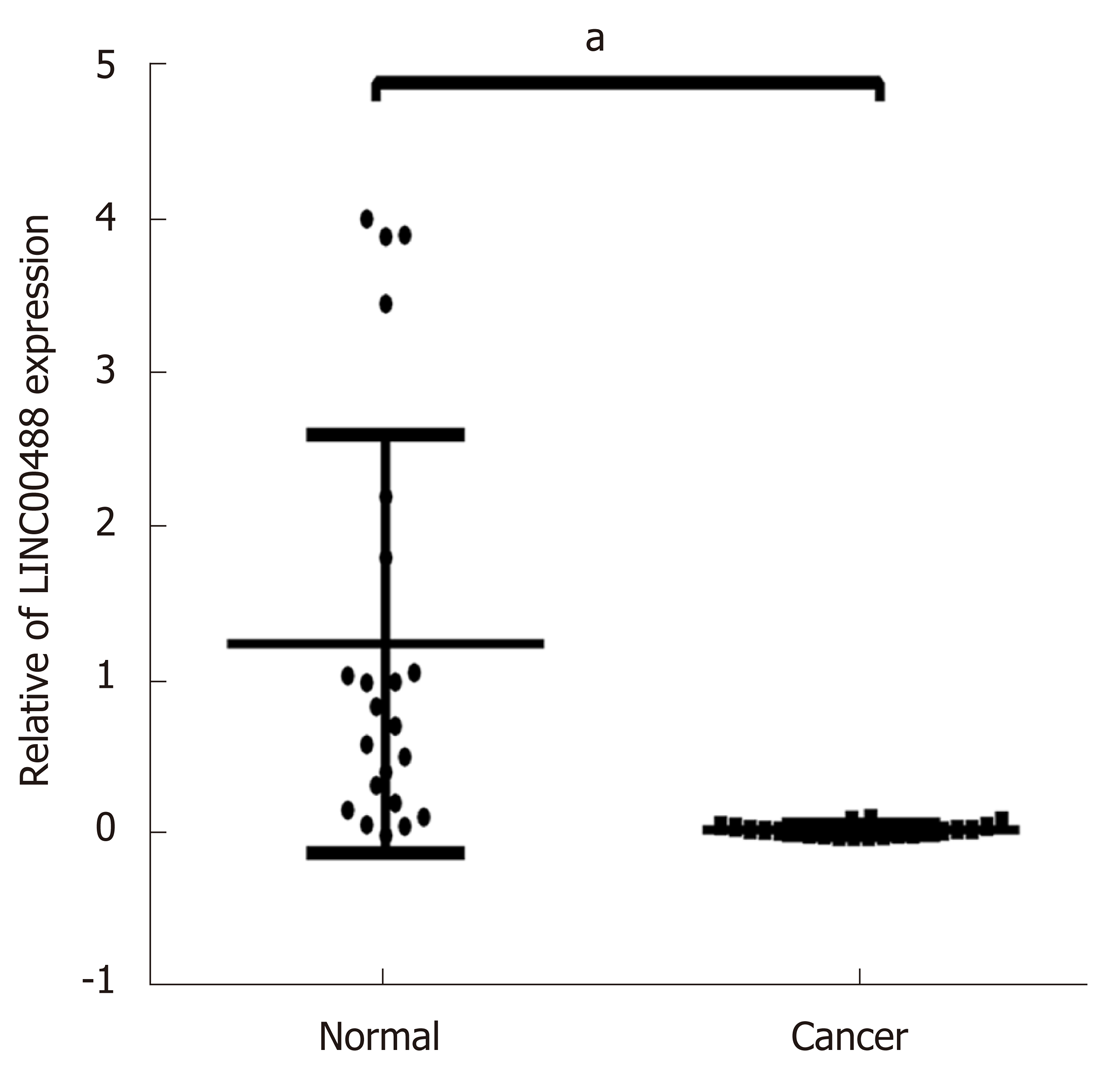
Based on the results of the bioinformatics analysis, we found that lncRNA LINC00488 involved in the ceRNA network was a novel lncRNA. Therefore, we evaluated the mRNA expression levels of LINC00488 by qRT-PCR in 22 paired patient samples (CRC and normal tissues). The results showed that LINC00488 was downregulated in the CRC tumors compared to the corresponding non-tumor tissues (P < 0.05, Figure 13). These results demonstrated that our bioinformatics analysis was reliable.
CRC is the third leading cause of cancer mortality[1,13]. Despite significant advances in the diagnosis and treatment of cancer, progress in improving the OS of CRC patients has been slow. LncRNAs have been the focus of the search for new biomarkers to improve the prognosis of these patients due to the accumulating evidence that lncRNAs have an essential regulatory function in tumorigenesis[14]. LncRNAs can act as ceRNAs in both tumorigenesis and biological processes. Unlike other methods for ceRNA network construction, we performed univariate regression analysis on all the aberrantly expressed lncRNAs and screened those closely related to survival prognosis to subsequently construct a ceRNA network and prognostic model. This method effectively filtered out a large number of lncRNAs that were unrelated to survival, which provides an effective basis for survival prognosis evaluation for CRC patients.
In recent years, several lncRNAs have been identified as diagnostic and prognostic biomarkers[15,16]. Our ceRNA network was constructed from 9 lncRNAs, 30 miRNAs, and 44 mRNAs. We found that LINC00488 was downregulated while the other eight lncRNAs (AL139147.1, DSCR4, DSCR8, H19, KCNQ1OT1, LMO7-AS1, MIR31HG, and MYO16-AS1) were upregulated in CRC tumors compared to non-tumor tissues. Ding et al[17] found that H19 was upregulated in tumors from 185 CRC patients compared to non-tumor tissues and promoted EMT through the H19/miR-29b/PGRN axis in CRC. Wang et al[18] demonstrated that high H19 expression was associated with 5-fluorouracil resistance. Eide et al[19] investigated the relationship between miR-31 and its host transcript lncRNA MIR31HG and found a strong correlation. In particular, their results showed that high lncRNA MIR31HG expression correlated with a shorter relapse-free survival in CRC patients. Numerous studies have demonstrated that lncRNAs DSCR8 and DSCR4 are involved in the progression of multiple cancers[20]. Moreover, Li et al[21] demonstrated that lncRNA KCNQ1OT1 was upregulated in colon cancer and regulates oxaliplatin chemoresistance through the miR-34a/ATG4B pathway. Enrico et al[22] also demonstrated that lncRNA KCNQ1OT1 could be a predictive clinical biomarker for CRC patients. All these results are consistent with our data presented in this study.
We found a novel lncRNA (LINC00488) in our ceRNA network. Expression analysis of LINC00488 in 22 paired CRC tumor tissues and adjacent non-tumor tissues confirmed the bioinformatics results. Correlation regression analysis revealed that AL139147.1 had direct linear correlations with forkhead box protein Q1 (FOXQ1) (R = 0.383, P < 0.001) and protein chromobox 2 (CBX2) (R = 0.354, P < 0.001). Several studies have demonstrated that many lncRNAs can interact with these two proteins to regulate cancer prognosis[23,24].
Gene expression profile-based prognostic lncRNAs, miRNAs, and mRNA signatures for prognosis prediction in patients with cancer have been investigated[10,25]. Seeking more reliable prognostic biomarkers for patient survival seems necessary to provide more personalized cancer management strategies for CRC patients. The contribution of mRNAs to CRC clinical prognosis has been demonstrated in the past decade. Several studies have also suggested the potential utilization of lncRNAs as prognostic markers and therapeutic targets[26-28]. However, the lncRNAs involved in the ceRNA network have not yet been reported for prognosis. Through multivariate Cox regression analysis, we identified four putatively informative genes (LINC00488, LMO7-AS1, MIR31HG, and DSCR4), which were used for the establishment of the four-lncRNA model. Based on this model, patients in the high-risk group survived significantly shorter than those in the low-risk group.
The AJCC stage provides important clinical information and is preferred for the management of CRC[29]. Interestingly, the prognostic prediction was more effective by combining the risk score and AJCC stage, which indicates that the four-lncRNA model based on risk score could improve the accuracy of prognostic prediction in CRC. Furthermore, nomogram, a statistical model based on the combination of risk score and clinical features, provides a more extensive prediction for each patient of CRC. Based on the nomogram, we could predict the 1-, 3-, and 5-year survival probabilities.
EMT is a major player in modulating tumor metastasis[30]. Many signaling pathways contribute to the regulation of EMT, such as Wnt/β-catenin and Notch[31,32]. The GSEA results showed that the “EMT” was significantly enriched. In this study, we found that COL1A1 and TPM2, which are key genes in EMT, were significantly expressed in the ceRNA network. COL1A1 encodes alpha 1 type I collagen, which is a target protein for TGFβ1. Pudova et al[33] found that COL1A1 is positively correlated with hexokinase 3 (HK3), and HK3 upregulation is associated with EMT. The study showed that TPM2 K152Ac was the most significantly downregulated protein in liver metastases in CRC[34]. We also found other signaling pathways (e.g., MAPK signaling pathway[35], regulation of binding[36], and focal adhesion[37]) that correlated with cancer. These results revealed a more extensive perspective in understanding the signaling pathways involved in CRC.
The major findings reported in this study could be considered a preliminary attempt to explore the prognostic lncRNAs, miRNAs, and mRNAs in CRC. However, this study has some limitations: All CRC patient data used to construct the prognostic signature were retrieved from the TCGA database. We only used one dataset in GEO to validate the predictive value of our model. Further validation based on other independent datasets is required to evaluate the general applicability of the signature in CRC. In addition, only LINC00488 related to OS was investigated for its expression pattern using qRT-PCR in 22 paired CRC patient samples. Additional studies are needed to confirm the biological functions of the lncRNAs, miRNAs, and mRNAs of the ceRNA network in CRC.
Colorectal cancer (CRC) is one of the most prevalent tumors worldwide. Recently, long noncoding RNAs (lncRNAs) have been identified to influence tumorigenesis and tumor progression through acting as competing endogenous RNAs (ceRNAs). It is difficult to extract prognostic lncRNAs and useful bioinformation in most of ceRNA networks constructed before.
To construct a prognostic related ceRNA regulatory network and lncRNA related signature based on risk score in CRC.
We systematically analyzed RNA transcriptome profile and clinical information of 506 CRC patients in the Cancer Genome Atlas database and constructed a prognostic related ceRNA network including 9 lncRNAs, 44 mRNAs, and 30 miRNAs. In addition, a four-lncRNA model was constructed by multivariate Cox regression analysis, which could be an independent prognostic model in CRC. The risk score for each patient was calculated and 506 patients were divided into high/low-risk groups (253 for each group) based on the median risk score.
The results of survival analysis showed that patients with a high risk score had a poor survival rate. In addition, the predictive value of the four-lncRNA model was evaluated in GSE38832. Furthermore, the patients’ survival probabilities could be better predicted when combing the risk score and clinical features. GSEA results verified that a number of cancer-related signaling pathways were definitely enriched with high risk score in CRC. Finally, we validated a novel lncRNA (LINC00488) that has not been reported before by using qRT-PCR in 22 paired CRC patient’s tumor tissues compared with adjacent non-tumor tissues.
The major findings reported in this study could be considered a preliminary attempt to explore the prognostic lncRNAs, miRNAs, and mRNAs in CRC. Therefore, our understanding of lncRNA related ceRNA regulatory mechanism could provide a potential diagnostic target for CRC patient.
Further validation in other independent datasets is required to evaluate the general applicability of the signature in CRC. Additional studies are needed to confirm the biological functions of the lncRNAs, miRNAs, and mRNAs of the ceRNA network in CRC.
Manuscript source: Unsolicited manuscript
Specialty type: Gastroenterology and hepatology
Country of origin: China
Peer-review report classification
Grade A (Excellent): A
Grade B (Very good): B
Grade C (Good): 0
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Chandrakesan P, Yoo CG S-Editor: Wang JL L-Editor: Wang TQ E-Editor: Zhang YL
| 1. | Siegel RL, Miller KD, Fedewa SA, Ahnen DJ, Meester RGS, Barzi A, Jemal A. Colorectal cancer statistics, 2017. CA Cancer J Clin. 2017;67:177-193. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 2526] [Cited by in F6Publishing: 2757] [Article Influence: 393.9] [Reference Citation Analysis (2)] |
| 2. | Huber MA, Kraut N, Beug H. Molecular requirements for epithelial-mesenchymal transition during tumor progression. Curr Opin Cell Biol. 2005;17:548-558. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1362] [Cited by in F6Publishing: 1408] [Article Influence: 74.1] [Reference Citation Analysis (0)] |
| 3. | Wang W, Xie Y, Chen F, Liu X, Zhong LL, Wang HQ, Li QC. LncRNA MEG3 acts a biomarker and regulates cell functions by targeting ADAR1 in colorectal cancer. World J Gastroenterol. 2019;25:3972-3984. [PubMed] [DOI] [Cited in This Article: ] [Cited by in CrossRef: 32] [Cited by in F6Publishing: 33] [Article Influence: 6.6] [Reference Citation Analysis (0)] |
| 4. | Wang CZ, Yan GX, Dong DS, Xin H, Liu ZY. LncRNA-ATB promotes autophagy by activating Yes-associated protein and inducing autophagy-related protein 5 expression in hepatocellular carcinoma. World J Gastroenterol. 2019;25:5310-5322. [PubMed] [DOI] [Cited in This Article: ] [Cited by in CrossRef: 47] [Cited by in F6Publishing: 50] [Article Influence: 10.0] [Reference Citation Analysis (0)] |
| 5. | Salmena L, Poliseno L, Tay Y, Kats L, Pandolfi PP. A ceRNA hypothesis: the Rosetta Stone of a hidden RNA language? Cell. 2011;146:353-358. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 4127] [Cited by in F6Publishing: 5106] [Article Influence: 392.8] [Reference Citation Analysis (0)] |
| 6. | Fang L, Du WW, Yang X, Chen K, Ghanekar A, Levy G, Yang W, Yee AJ, Lu WY, Xuan JW, Gao Z, Xie F, He C, Deng Z, Yang BB. Versican 3'-untranslated region (3'-UTR) functions as a ceRNA in inducing the development of hepatocellular carcinoma by regulating miRNA activity. FASEB J. 2013;27:907-919. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 94] [Cited by in F6Publishing: 106] [Article Influence: 8.8] [Reference Citation Analysis (0)] |
| 7. | Feng C, Song C, Ning Z, Ai B, Wang Q, Xu Y, Li M, Bai X, Zhao J, Liu Y, Li X, Zhang J, Li C. ce-Subpathway: Identification of ceRNA-mediated subpathways via joint power of ceRNAs and pathway topologies. J Cell Mol Med. 2019;23:967-984. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 9] [Cited by in F6Publishing: 10] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 8. | Arun K, Arunkumar G, Bennet D, Chandramohan SM, Murugan AK, Munirajan AK. Comprehensive analysis of aberrantly expressed lncRNAs and construction of ceRNA network in gastric cancer. Oncotarget. 2018;9:18386-18399. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 37] [Cited by in F6Publishing: 38] [Article Influence: 6.3] [Reference Citation Analysis (0)] |
| 9. | Li G, Liu T, Zhang B, Chen W, Ding Z. Genome-wide identification of a competing endogenous RNA network in cholangiocarcinoma. J Cell Biochem. 2019;120:18995-19003. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 11] [Cited by in F6Publishing: 14] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 10. | Xing Y, Zhao Z, Zhu Y, Zhao L, Zhu A, Piao D. Comprehensive analysis of differential expression profiles of mRNAs and lncRNAs and identification of a 14-lncRNA prognostic signature for patients with colon adenocarcinoma. Oncol Rep. 2018;39:2365-2375. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 15] [Cited by in F6Publishing: 34] [Article Influence: 5.7] [Reference Citation Analysis (0)] |
| 11. | Qiu G, Zhang XB, Zhang SQ, Liu PL, Wu W, Zhang JY, Dai SR. Dysregulation of MALAT1 and miR-619-5p as a prognostic indicator in advanced colorectal carcinoma. Oncol Lett. 2016;12:5036-5042. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 19] [Cited by in F6Publishing: 22] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 12. | He F, Song Z, Chen H, Chen Z, Yang P, Li W, Yang Z, Zhang T, Wang F, Wei J, Wei F, Wang Q, Cao J. Long noncoding RNA PVT1-214 promotes proliferation and invasion of colorectal cancer by stabilizing Lin28 and interacting with miR-128. Oncogene. 2019;38:164-179. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 68] [Cited by in F6Publishing: 79] [Article Influence: 13.2] [Reference Citation Analysis (0)] |
| 13. | Siegel RL, Miller KD, Jemal A. Cancer Statistics, 2017. CA Cancer J Clin. 2017;67:7-30. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 11065] [Cited by in F6Publishing: 11895] [Article Influence: 1699.3] [Reference Citation Analysis (3)] |
| 14. | Mao Y, Fu Z, Zhang Y, Dong L, Zhang Y, Zhang Q, Li X, Liu J. A seven-lncRNA signature predicts overall survival in esophageal squamous cell carcinoma. Sci Rep. 2018;8:8823. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 49] [Cited by in F6Publishing: 47] [Article Influence: 7.8] [Reference Citation Analysis (0)] |
| 15. | Du Z, Sun T, Hacisuleyman E, Fei T, Wang X, Brown M, Rinn JL, Lee MG, Chen Y, Kantoff PW, Liu XS. Integrative analyses reveal a long noncoding RNA-mediated sponge regulatory network in prostate cancer. Nat Commun. 2016;7:10982. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 216] [Cited by in F6Publishing: 240] [Article Influence: 30.0] [Reference Citation Analysis (0)] |
| 16. | Ye J, Zhang J, Lv Y, Wei J, Shen X, Huang J, Wu S, Luo X. Integrated analysis of a competing endogenous RNA network reveals key long noncoding RNAs as potential prognostic biomarkers for hepatocellular carcinoma. J Cell Biochem. 2019;120:13810-13825. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 27] [Cited by in F6Publishing: 32] [Article Influence: 6.4] [Reference Citation Analysis (0)] |
| 17. | Ding D, Li C, Zhao T, Li D, Yang L, Zhang B. LncRNA H19/miR-29b-3p/PGRN Axis Promoted Epithelial-Mesenchymal Transition of Colorectal Cancer Cells by Acting on Wnt Signaling. Mol Cells. 2018;41:423-435. [PubMed] [DOI] [Cited in This Article: ] [Cited by in F6Publishing: 90] [Reference Citation Analysis (0)] |
| 18. | Wang M, Han D, Yuan Z, Hu H, Zhao Z, Yang R, Jin Y, Zou C, Chen Y, Wang G, Gao X, Wang X. Long non-coding RNA H19 confers 5-Fu resistance in colorectal cancer by promoting SIRT1-mediated autophagy. Cell Death Dis. 2018;9:1149. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 103] [Cited by in F6Publishing: 129] [Article Influence: 21.5] [Reference Citation Analysis (0)] |
| 19. | Eide PW, Eilertsen IA, Sveen A, Lothe RA. Long noncoding RNA MIR31HG is a bona fide prognostic marker with colorectal cancer cell-intrinsic properties. Int J Cancer. 2019;144:2843-2853. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 25] [Cited by in F6Publishing: 23] [Article Influence: 4.6] [Reference Citation Analysis (0)] |
| 20. | Wang Y, Sun L, Wang L, Liu Z, Li Q, Yao B, Wang C, Chen T, Tu K, Liu Q. Long non-coding RNA DSCR8 acts as a molecular sponge for miR-485-5p to activate Wnt/β-catenin signal pathway in hepatocellular carcinoma. Cell Death Dis. 2018;9:851. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 81] [Cited by in F6Publishing: 96] [Article Influence: 16.0] [Reference Citation Analysis (0)] |
| 21. | Li Y, Li C, Li D, Yang L, Jin J, Zhang B. lncRNA KCNQ1OT1 enhances the chemoresistance of oxaliplatin in colon cancer by targeting the miR-34a/ATG4B pathway. Onco Targets Ther. 2019;12:2649-2660. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 61] [Cited by in F6Publishing: 85] [Article Influence: 17.0] [Reference Citation Analysis (0)] |
| 22. | Mini E, Lapucci A, Perrone G, D'Aurizio R, Napoli C, Brugia M, Landini I, Tassi R, Picariello L, Simi L, Mancini I, Messerini L, Magi A, Pinzani P, Mazzei T, Tonelli F, Nobili S. RNA sequencing reveals PNN and KCNQ1OT1 as predictive biomarkers of clinical outcome in stage III colorectal cancer patients treated with adjuvant chemotherapy. Int J Cancer. 2019;145:2580-2593. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 17] [Cited by in F6Publishing: 22] [Article Influence: 4.4] [Reference Citation Analysis (0)] |
| 23. | Ma P, Li Y, Zhang W, Fang F, Sun J, Liu M, Li K, Dong L. Long Non-coding RNA MALAT1 Inhibits Neuron Apoptosis and Neuroinflammation While Stimulates Neurite Outgrowth and Its Correlation With MiR-125b Mediates PTGS2, CDK5 and FOXQ1 in Alzheimer's Disease. Curr Alzheimer Res. 2019;16:596-612. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 54] [Cited by in F6Publishing: 74] [Article Influence: 18.5] [Reference Citation Analysis (0)] |
| 24. | Jiao D, Li Z, Zhu M, Wang Y, Wu G, Han X. LncRNA MALAT1 promotes tumor growth and metastasis by targeting miR-124/foxq1 in bladder transitional cell carcinoma (BTCC). Am J Cancer Res. 2018;8:748-760. [PubMed] [Cited in This Article: ] |
| 25. | Zhang Y, Xu Y, Feng L, Li F, Sun Z, Wu T, Shi X, Li J, Li X. Comprehensive characterization of lncRNA-mRNA related ceRNA network across 12 major cancers. Oncotarget. 2016;7:64148-64167. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 128] [Cited by in F6Publishing: 152] [Article Influence: 25.3] [Reference Citation Analysis (1)] |
| 26. | Tian X, Xu G. Clinical value of lncRNA MALAT1 as a prognostic marker in human cancer: systematic review and meta-analysis. BMJ Open. 2015;5:e008653. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 83] [Cited by in F6Publishing: 99] [Article Influence: 11.0] [Reference Citation Analysis (0)] |
| 27. | Mehrad-Majd H, Ravanshad S, Moradi A, Khansalar N, Sheikhi M, Akhtari J. Decreased expression of lncRNA loc285194 as an independent prognostic marker in cancer: A systematic review and meta-analysis. Pathol Res Pract. 2019;215:152426. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 8] [Cited by in F6Publishing: 6] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 28. | Liu Z, Mi M, Li X, Zheng X, Wu G, Zhang L. lncRNA OSTN-AS1 May Represent a Novel Immune-Related Prognostic Marker for Triple-Negative Breast Cancer Based on Integrated Analysis of a ceRNA Network. Front Genet. 2019;10:850. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 30] [Cited by in F6Publishing: 33] [Article Influence: 6.6] [Reference Citation Analysis (0)] |
| 29. | Weiser MR. AJCC 8th Edition: Colorectal Cancer. Ann Surg Oncol. 2018;25:1454-1455. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 294] [Cited by in F6Publishing: 479] [Article Influence: 79.8] [Reference Citation Analysis (0)] |
| 30. | Larue L, Bellacosa A. Epithelial-mesenchymal transition in development and cancer: role of phosphatidylinositol 3' kinase/AKT pathways. Oncogene. 2005;24:7443-7454. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 884] [Cited by in F6Publishing: 923] [Article Influence: 48.6] [Reference Citation Analysis (0)] |
| 31. | Jin M, Wang J, Ji X, Cao H, Zhu J, Chen Y, Yang J, Zhao Z, Ren T, Xing J. MCUR1 facilitates epithelial-mesenchymal transition and metastasis via the mitochondrial calcium dependent ROS/Nrf2/Notch pathway in hepatocellular carcinoma. J Exp Clin Cancer Res. 2019;38:136. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 69] [Cited by in F6Publishing: 98] [Article Influence: 19.6] [Reference Citation Analysis (0)] |
| 32. | Zhang J, Cai H, Sun L, Zhan P, Chen M, Zhang F, Ran Y, Wan J. LGR5, a novel functional glioma stem cell marker, promotes EMT by activating the Wnt/β-catenin pathway and predicts poor survival of glioma patients. J Exp Clin Cancer Res. 2018;37:225. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 69] [Cited by in F6Publishing: 106] [Article Influence: 17.7] [Reference Citation Analysis (0)] |
| 33. | Pudova EA, Kudryavtseva AV, Fedorova MS, Zaretsky AR, Shcherbo DS, Lukyanova EN, Popov AY, Sadritdinova AF, Abramov IS, Kharitonov SL, Krasnov GS, Klimina KM, Koroban NV, Volchenko NN, Nyushko KM, Melnikova NV, Chernichenko MA, Sidorov DV, Alekseev BY, Kiseleva MV, Kaprin AD, Dmitriev AA, Snezhkina AV. HK3 overexpression associated with epithelial-mesenchymal transition in colorectal cancer. BMC Genomics. 2018;19:113. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 41] [Cited by in F6Publishing: 39] [Article Influence: 6.5] [Reference Citation Analysis (0)] |
| 34. | Shen Z, Wang B, Luo J, Jiang K, Zhang H, Mustonen H, Puolakkainen P, Zhu J, Ye Y, Wang S. Global-scale profiling of differential expressed lysine acetylated proteins in colorectal cancer tumors and paired liver metastases. J Proteomics. 2016;142:24-32. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 24] [Cited by in F6Publishing: 26] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 35. | Fang JY, Richardson BC. The MAPK signalling pathways and colorectal cancer. Lancet Oncol. 2005;6:322-327. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 577] [Cited by in F6Publishing: 729] [Article Influence: 38.4] [Reference Citation Analysis (0)] |
| 36. | García-Cárdenas JM, Guerrero S, López-Cortés A, Armendáriz-Castillo I, Guevara-Ramírez P, Pérez-Villa A, Yumiceba V, Zambrano AK, Leone PE, Paz-Y-Miño C. Post-transcriptional Regulation of Colorectal Cancer: A Focus on RNA-Binding Proteins. Front Mol Biosci. 2019;6:65. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 29] [Cited by in F6Publishing: 26] [Article Influence: 5.2] [Reference Citation Analysis (0)] |
| 37. | Jeong KY. Inhibiting focal adhesion kinase: A potential target for enhancing therapeutic efficacy in colorectal cancer therapy. World J Gastrointest Oncol. 2018;10:290-292. [PubMed] [DOI] [Cited in This Article: ] [Cited by in CrossRef: 15] [Cited by in F6Publishing: 20] [Article Influence: 3.3] [Reference Citation Analysis (0)] |