Published online Dec 7, 2019. doi: 10.3748/wjg.v25.i45.6619
Peer-review started: September 5, 2019
First decision: October 14, 2019
Revised: October 29, 2019
Accepted: November 13, 2019
Article in press: November 13, 2019
Published online: December 7, 2019
Gastric cancer (GC) has become a serious threat to people's health. Accumulative evidence reveals that dysregulation of numerous microRNAs (miRNAs) has been found during malignant formation. So far, the role of microRNA-760 (miR-760) in the development of GC is largely unknown.
To measure the expression level of miR-760 in GC and investigate its role in gastric tumorigenesis.
Real-time quantitative polymerase chain reaction and Western blot analysis were used to measure the expression of miR-760 and G-protein-coupled receptor kinase interacting protein-1 (GIT1). Cell growth was detected by 3-(4,5-dimethyl-2-thiazolyl)-2,5-diphenyl-2-H-tetrazolium bromide (MTT) and cell colony formation assays. Apoptosis was assessed by flow cytometric analysis. The relationship between miR-760 and GIT1 was verified by luciferase reporter assay.
The results showed that the expression of miR-760 was decreased in GC and associated with poor clinical outcomes in GC patients. Furthermore, miR-760 restrained cell proliferation and cell colony formation and induced apoptosis in GC cells. In addition, miR-760 directly targeted GIT1 and negatively regulated its expression in GC. GIT1 was upregulated in GC and predicted a worse prognosis in GC patients. We also found that upregulation of GIT1 weakened the inhibitory effect of miR-760 in GC.
In conclusion, miR-760 targets GIT1 to inhibit cell growth and promote apoptosis in GC cells. Our data demonstrate that miR-760 may be a potential target for the treatment of GC.
Core tip: The expression of microRNA-760 (miR-760) was decreased in gastric cancer (GC), which was related to poor clinical outcomes in GC patients. MiR-760 restrained cell proliferation and cell colony formation and induced cell apoptosis in GC cells. MiR-760 directly targets G-protein-coupled receptor kinase interacting protein-1 that is involved in tumorigenesis of GC.
- Citation: Ge L, Wang Y, Duan QH, Liu SS, Liu GJ. MicroRNA-760 acts as a tumor suppressor in gastric cancer development via inhibiting G-protein-coupled receptor kinase interacting protein-1 transcription. World J Gastroenterol 2019; 25(45): 6619-6633
- URL: https://www.wjgnet.com/1007-9327/full/v25/i45/6619.htm
- DOI: https://dx.doi.org/10.3748/wjg.v25.i45.6619
The stomach is an essential organ of the digestive system and is vital to our body. Gastric cancer (GC) is one of the most common malignant tumors in China[1]. In recent years, the incidence of GC in China has been rising, indicating that GC is indeed a serious threat to people's health[2]. About 80% of patients with early GC are asymptomatic. In addition, a small number of GC patients are very susceptible to gastritis, stomach ulcers, and other stomach diseases. Therefore, most patients with GC have reached the advanced stage when they are diagnosed[3]. Despite advances in current medical technology, the prognosis of GC patients remains poor. The pathological stage is very important for the prognosis of GC patients[4]. Therefore, it is necessary to explore new methods for early diagnosis of GC.
Currently, microRNAs (miRNAs) are known to be modulators of human diseases and cancers. In GC, the molecular mechanisms of many miRNAs have been investigated. For example, overexpression of miR-375 inhibited cell proliferation and migration in GC[5]. Li et al[6] demonstrated that miR-217 was involved in the carcinogenesis of GC by downregulating CDH1 expression. Recently, the dysregulation of microRNA-760 (miR-760) has caught our attention. Upregulation of miR-760 was observed in ovarian cancer and promoted ovarian cancer cell proliferation by downregulation of PHLPP2[7]. However, downregulation of miR-760 was found in colorectal cancer. Overexpression of miR-760 suppressed the growth of human colorectal cancer cells by targeting BATF3/AP-1/cyclinD1 signaling[8]. Previous studies have shown that the expression of miR-760 is tissue specific. Especially, the expression of miR-760 has been found to be reduced in primary tumor in patients with advanced GC, which was associated with tumor stage[9]. However, the function of miR-760 has not been investigated in previous studies and needs to be illuminated.
TargetScan (http://www.targetscan.org/) predicts that miR-760 has a binding site with the 3’ untranslated region (3’UTR) of G-protein-coupled receptor kinase interacting protein-1 (GIT1). Furthermore, GIT1 has been found to be stably expressed in many diseases and is involved in tumorigenesis. For example, chondrocyte proliferation has been reported to be mediated by GIT1[10]. In addition, upregulation of GIT1 can also regulate chondrocyte apoptosis[11]. GIT1, a motile, multi-molecular signaling complex, can regulate protrusive activity and cell migration[12]. The role of GIT1 has also been reported in human cancers. For example, GIT1 was found to promote lung cancer cell metastasis and be associated with a poor prognosis[13]. In oral squamous cell carcinoma, GIT1 served as a modulator and biomarker for invasion and metastasis[14]. Moreover, the interaction between GIT1 and miRNAs has been found in cancers. Dong et al[15] proposed that miR-149 suppressed cell proliferation, metastasis, and tumor growth in breast cancer by targeting GIT1. However, the role of GIT1 and the regulatory mechanism of the miR-760/GIT1 axis have not been investigated in GC.
In our study, the dysregulation of miR-760 was found in GC. In the meanwhile, how miR-760 regulates cell proliferation, colony formation, and apoptosis was assessed in GC. Additionally, the target gene of miR-760 and their relevant molecular mechanism were also explored. This study may help us better understand the pathogenesis of GC.
Eighty-two GC patients from the Affiliated Hospital of Weifang Medical University participated in this study between July 2014 and July 2015 (median follow-up time, 28 mo). Table 1 summarizes the clinicopathological characteristics of these patients. Prior to the start of the experiment, we obtained informed consent from all GC patients. Experimental GC and normal tissues were obtained from these patients. These patients with GC did not receive any treatment except surgery. This study was approved by the Institutional Ethics Committee of the Affiliated Hospital of Weifang Medical University.
The human GC cell lines MKN-45 (gastric adenocarcinoma cells, BNCC337682) and AGS (gastric adenocarcinoma epithelial cells, ATCC® CRL-1739™) and the normal human gastric epithelial cell line GES-1 (pSC1021) were purchased from the American Type Culture Collection (Manassas, VA, United States). These cells were cultured in a medium consisting of 90% RPMI-1640 and 10% fetal bovine serum (FBS) in an incubator with 5% CO2 at 37 °C. Next, MKN-45 and AGS cells were transfected with miR-149 mimic and inhibitor or GIT1 vector (RiboBio, Guangzhou, China), respectively.
Total RNA extraction was performed with TRIzol reagent (Sigma, United States). cDNA was synthesized using the First-Strand cDNA Synthesis kit (Promega, United States). Real-time quantitative polymerase chain reaction (RT-qPCR) assay was performed using the miScript SYBR®Green PCR kit (Qiagen, Germany) according to the manufacturer’s instructions. MiR-760 or GIT1 was normalized to U6 or glyceraldehyde-3-phosphate dehydrogenase (GAPDH) using the 2−△△ct method.
RIPA lysis buffer was used to obtain protein samples. Next, 25 μg of protein was separated by 10% sodium dodecyl sulfate-polyacrylamide gel electrophoresis. After the protein was transferred to a polyvinylidene fluoride membrane, the membrane was blocked with 5% non-fat milk and incubated with GIT1 and GAPDH primary antibodies (Abcam, Cambridge, MA, United States) overnight at 4°C, followed by incubation with horseradish peroxidase-conjugated secondary antibodies (Abcam, United States) for 1 hour. Finally, chemiluminescence (ECL, Pierce) was used to detect protein expression levels.
The pmirGLO luciferase vector (RiboBio, Guangzhou, China) with wild-type-GIT-3’UTR (Wt-GIT-3’UTR) or mutant (Mut)-GIT-3’UTR and miR-760 mimic was transfected into MKN-45 and AGS cells. Next, the transfected cells were incubated for 20 min at room temperature. After 48 h, we discarded the medium and washed it with phosphate buffer saline (PBS). Finally, luciferase activity was assessed using a dual luciferase assay system (Promega, United States).
Transfected MKN-45 and AGS cells (4 × 103 cells/well) were seeded in RPMI-1640 medium (10% FBS) for 24, 48, 72, or 96 h. Next, MKN-45 and AGS cell suspensions were incubated with 20 μL of MTT for 4 h. After removing the MTT solution, 150 μL of dimethyl sulfoxide was added. Finally, cell viability was assessed using a microplate reader to determine the optical density at 490 nm.
The transfected MKN-45 and AGS cells (50 cells/well) were seeded into plates containing 10 mL of RPMI-1640, and gently rotated to uniformly disperse the cells. The cells were then cultured at 37 °C in a cell incubator with 5% CO2 and saturated humidity for 2 wk. When macroscopic clones appeared in plates, the culture was terminated. The supernatant was discarded, and the cells were carefully immersed twice with PBS. Next, we added 4% paraformaldehyde to fix the cells for 15 min. Then, the fixing solution was removed, and Giemsa stain was added to dye cells for 15 min. The staining solution was slowly washed with running water and air-dried. The plate was inverted and the number of cell colonies was counted under a microscope (Olympus, Tokyo, Japan).
Flow cytometric analysis was used to detect apoptosis in GC cells. First, MKN-45 and AGS cells (2 × 103 cell/well) transfected with miR-760 mimic, miR-760 inhibitor, or miR-760 mimic + GIT1 vector were seed in 6-well plates. After 48 h, MKN-45 and AGS cells were collected by digestion with trypsin (without EDTA). We then suspended the collected MKN-45 and AGS cells in PBS at 4 °C. After PBS was discarded, Annexin V-FITC from the Annexin V-FITC Apoptosis Detection Kit (Biovision, K101) was used to stain MKN-45 and AGS cells for 15 mins. MKN-45 and AGS cells were then stained with PI (Sigma-Aldrich, Shanghai, China). Finally, we observed apoptotic MKN-45 cells using a flow cytometer (BD Biosciences).
Kaplan-Meier Plotter (http://kmplot.com/analysis/), an online database including gene expression data and clinical data, was used to analyze the prognostic significance of miR-760 or GIT1 in GC. Log rank P-value and hazard ratio (HR) with 95% confidence interval (CI) were calculated. The expression level of GIT1 in GC was assessed using the Ge-mini database (gemini.cancer-pku.cn/). TCGA database and GO database (http://www.geneontology.org/) were used to explore genes associated with GIT1.
Data are shown as the mean ± standard deviation and analyzed with SPSS 18.0 or Graphpad Prism 6. Statistical methods included Chi-squared test, one-way ANOVA with Bonferroni post hoc test, and univariate Kaplan-Meier method with log-rank test. Overall survival and disease-free survival were analyzed in GC patients (high miR-760 expression group, n = 23; low miR-760 expression group, n = 58). Statistical differences were defined at P < 0.05.
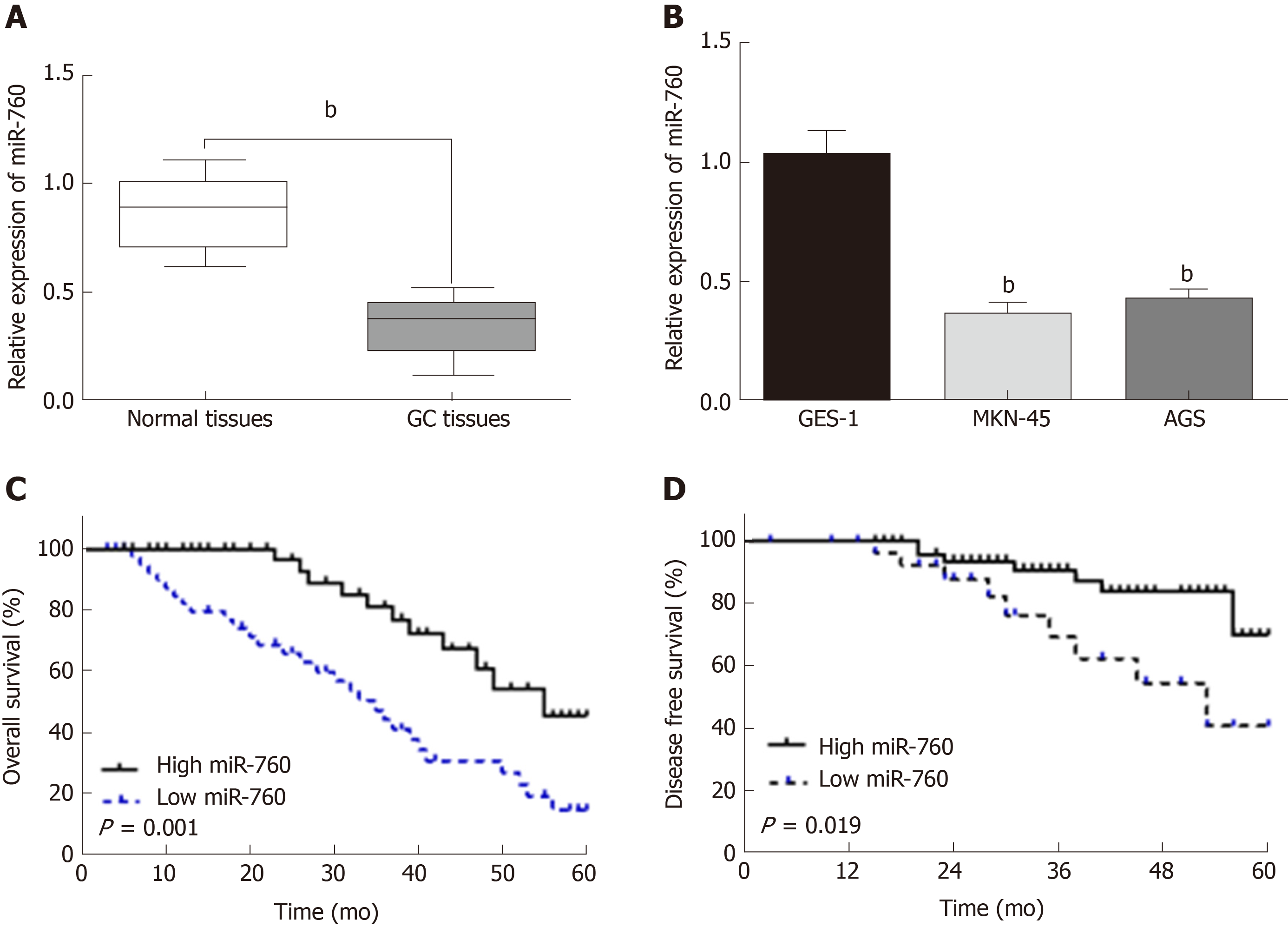
First, miR-760 expression was observed in GC tissues. RT-qPCR showed that the expression of miR-760 in GC tissues was lower than that in normal tissues (P < 0.01, Figure 1A). The median relative expression level of miR-760 in these tissues was taken as a cut-off point to separate low and high expression of miR‑760 (low, 71.6%; high, 28.4%). Similarly, miR-760 was downregulated in MKN-45 and AGS cells compared to GES-1 cells (P = 0.004, Figure 1B). Downregulation of miR-760 was associated with a shorter overall survival (P = 0.001, Figure 1C) and disease-free survival (P = 0.019, Figure 1D) in GC patients. Additionally, the dysregulation of miR-760 was also associated with clinical features in GC patients, including TNM stage (P = 0.01, Table 1) and lymph nodes metastasis (P = 0.02, Table 1). Combining these results, we conclude that miR-760 may regulate the biological processes of GC.
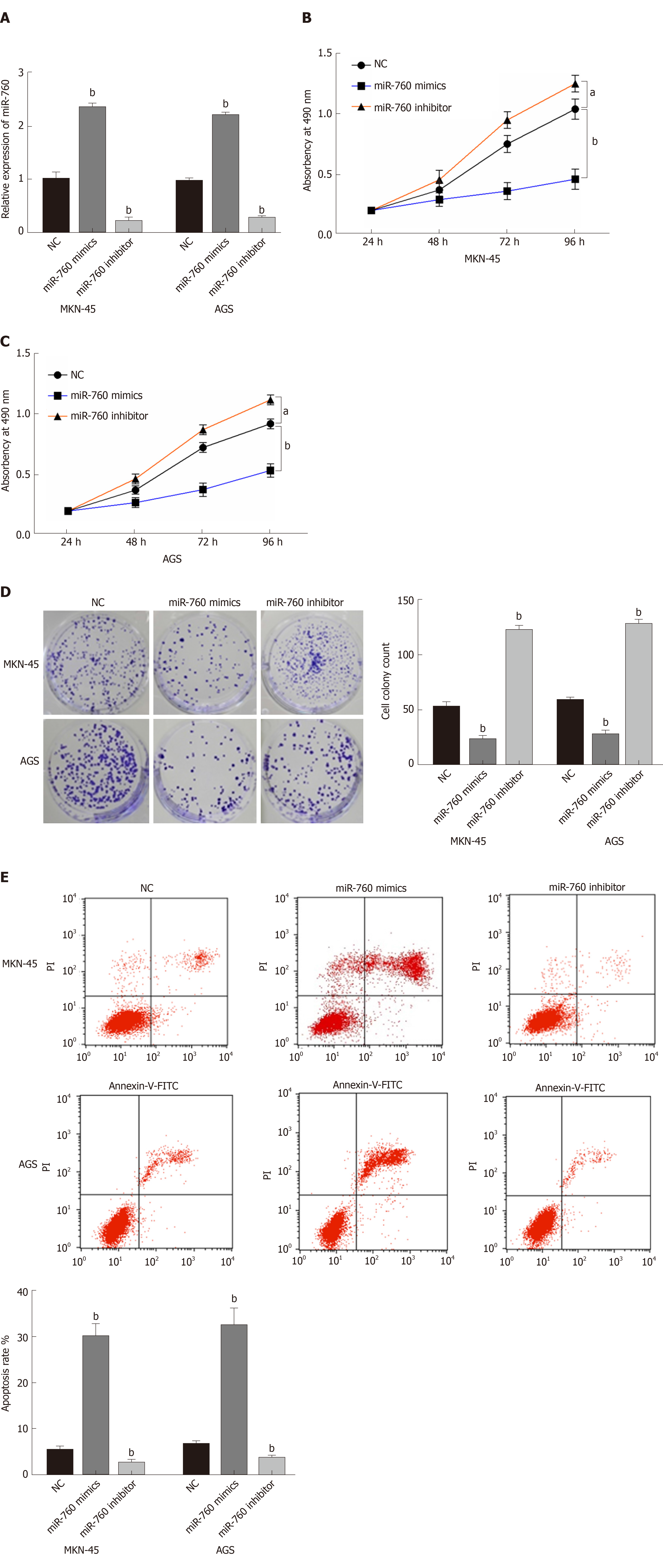
To explore the role of miR-760 in GC, miR-760 mimic or inhibitor was transfected to MKN-45 and AGS cells. First, RT-qPCR assay was performed to assess transfection efficiency. In MKN-45 and AGS cells, miR-760 expression was found to be upregulated by its mimic and downregulated by its inhibitor (P = 0.0056, Figure 2A). Next, MTT assay indicated that overexpression of miR-760 restrained the proliferation of MKN-45 cells (P = 0.02, Figure 2B). But downregulation of miR-760 promoted GC cell proliferation (P = 0.009, Figure 2B). The same results were also identified in AGS cells (P = 0.04 or 0.008, Figure 2C). In addition, miR-760 mimic was found to hinder the formation of cell colony, whereas miR-760 inhibitor promoted cell colony formation of MKN-45 (P = 0.007 and 0.005, respectively, Figure 2D) and AGS cells (P = 0.003 and 0.004, respectively, Figure 2D). Flow cytometric analysis suggested that miR-760 overexpression promoted apoptosis, while downregulation of miR-760 blocked apoptosis of MKN-45 (P = 0.001 or 0.004, Figure 2E) and AGS (P = 0.002 or 0.004, Figure 2E) cells. Taken together, miR-760 is involved in GC development by suppressing cell proliferation and promoting apoptosis.
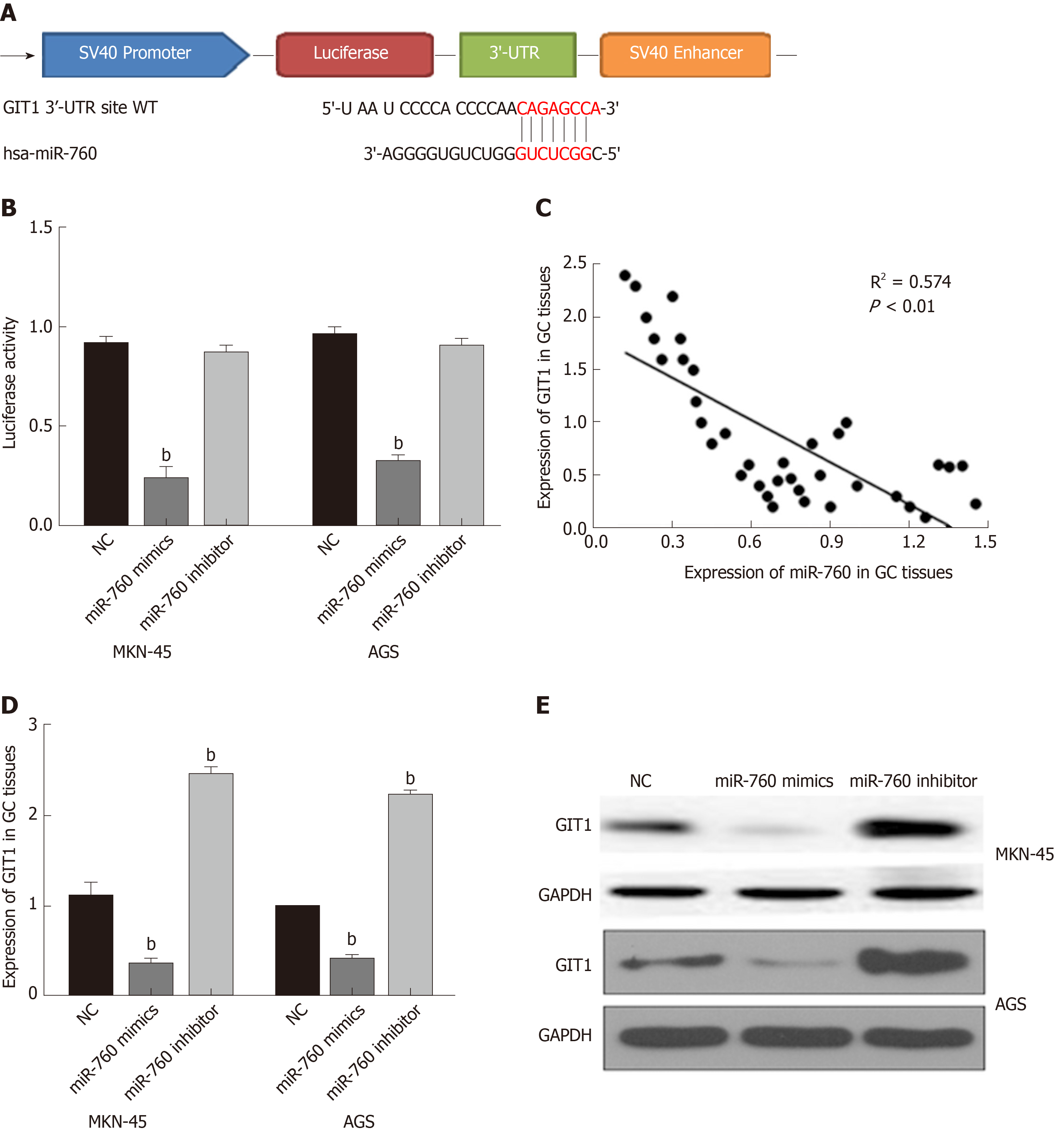
TargetScan (http://www.targetscan.org/) shows that miR-760 has a binding site to GIT1 (Figure 3A). Next, luciferase reporter assay was designed to verify the relationship between miR-760 and GIT1. MiR-760 mimic was found to reduce Wt-GIT luciferase activity, but had no effect on Mut-GIT1 luciferase activity (P = 0.003, Figure 3B). Next, a negative correlation between miR-760 and GIT1 expression was identified in GC tissues (P = 0.0001, R2 = 0.574; Figure 3C). To further verify the above results, we detected how miR-760 regulates GIT1 expression in MKN-45 and AGS cells. The mRNA and protein expression of GIT1 was restrained by miR-760 mimic and enhanced by miR-760 inhibitor (P = 0.004, Figure 3D and E). These results imply that miR-760 directly targets GIT1 and negatively regulates its expression in GC.
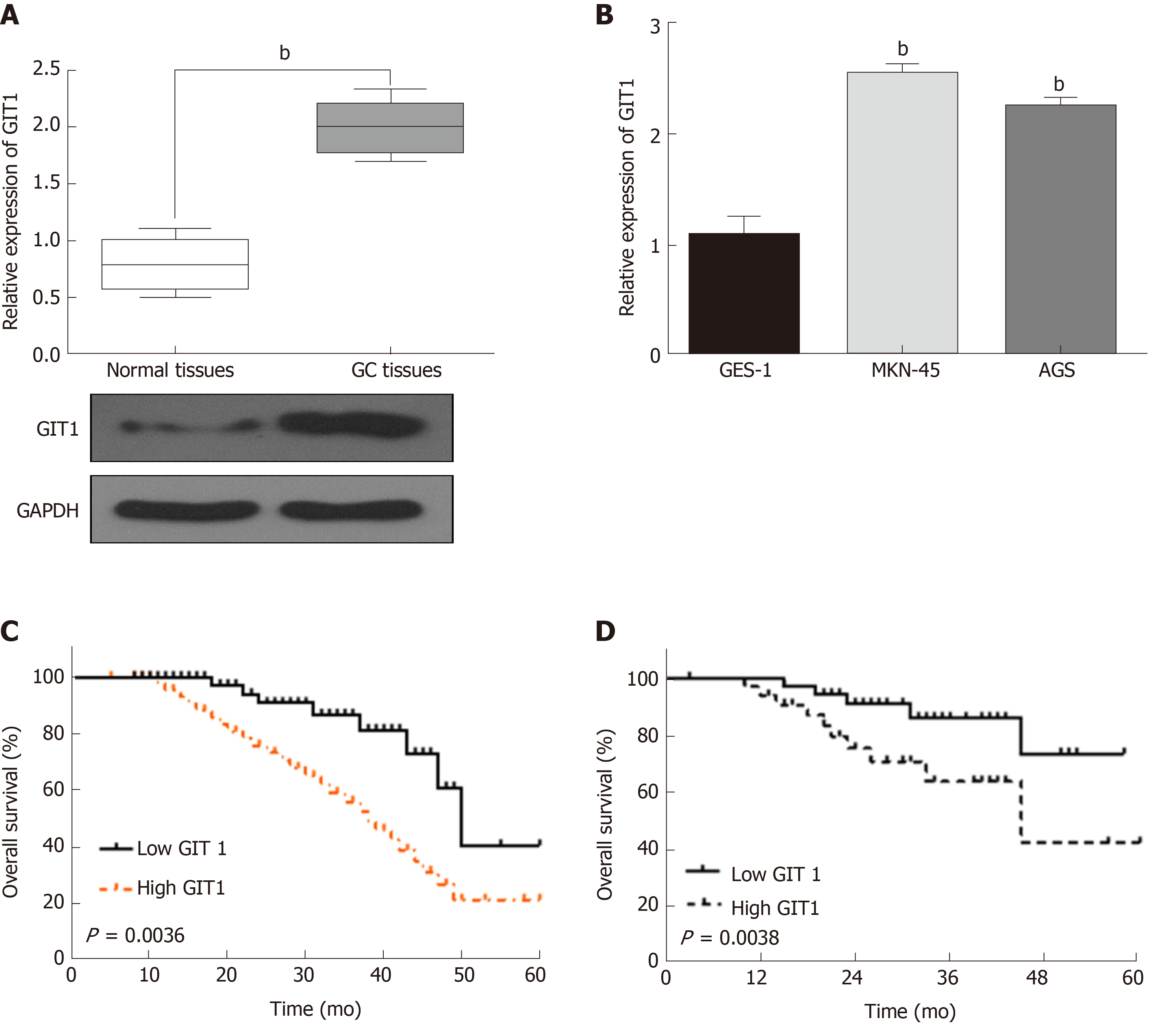
In order to confirm whether GIT1 is involved in tumorigenesis of GC, the expression of GIT1 was detected in GC. We found that GIT1 was upregulated in GC tissues compared to normal tissues (P = 0.002, Figure 4A). GIT1 expression was also higher in MKN-45 and AGS cells than in GES-1 cells (P = 0.003, Figure 4B). Next, the association between GIT1 expression and prognosis in GC patients was analyzed. Upregulation of GIT1 was found to be associated with a shorter overall survival (P = 0.0036, Figure 4C) and disease-free survival in GC patients (P = 0.038, Figure 4D). Based on these results, we consider that GIT1 is involved in the tumorigenesis of GC.
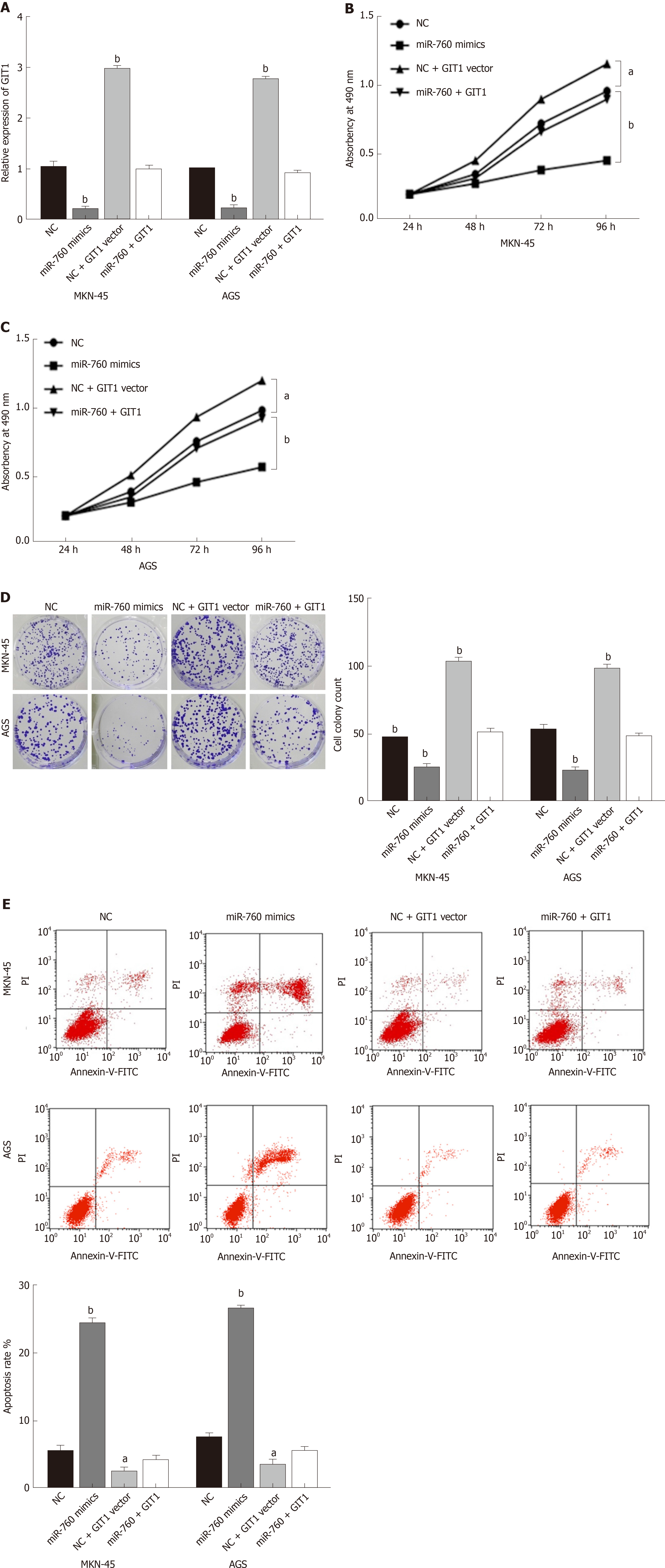
Finally, the GIT1 vector was transfected into MKN-45 and AGS cells with miR-760 mimic to investigate their interaction. First, the decreased expression of GIT1 mediated by miR-760 mimic was restored by GIT1 vector (Figure 5A). Next, we found that upregulation of GIT1 attenuated miR-760 mediated inhibition of cell proliferation in MKN-45 and AGS cells (Figure 5B and C). Similarly, upregulation of GIT1 also abolished the inhibitory effect of miR-760 on cell colony formation (Figure 5D). Furthermore, the promotion of apoptosis induced by miR-760 mimic was also weakened by upregulation of GIT1 (Figure 5E). Briefly, upregulation of GIT1 reverses the effect of miR-760 in GC cells.
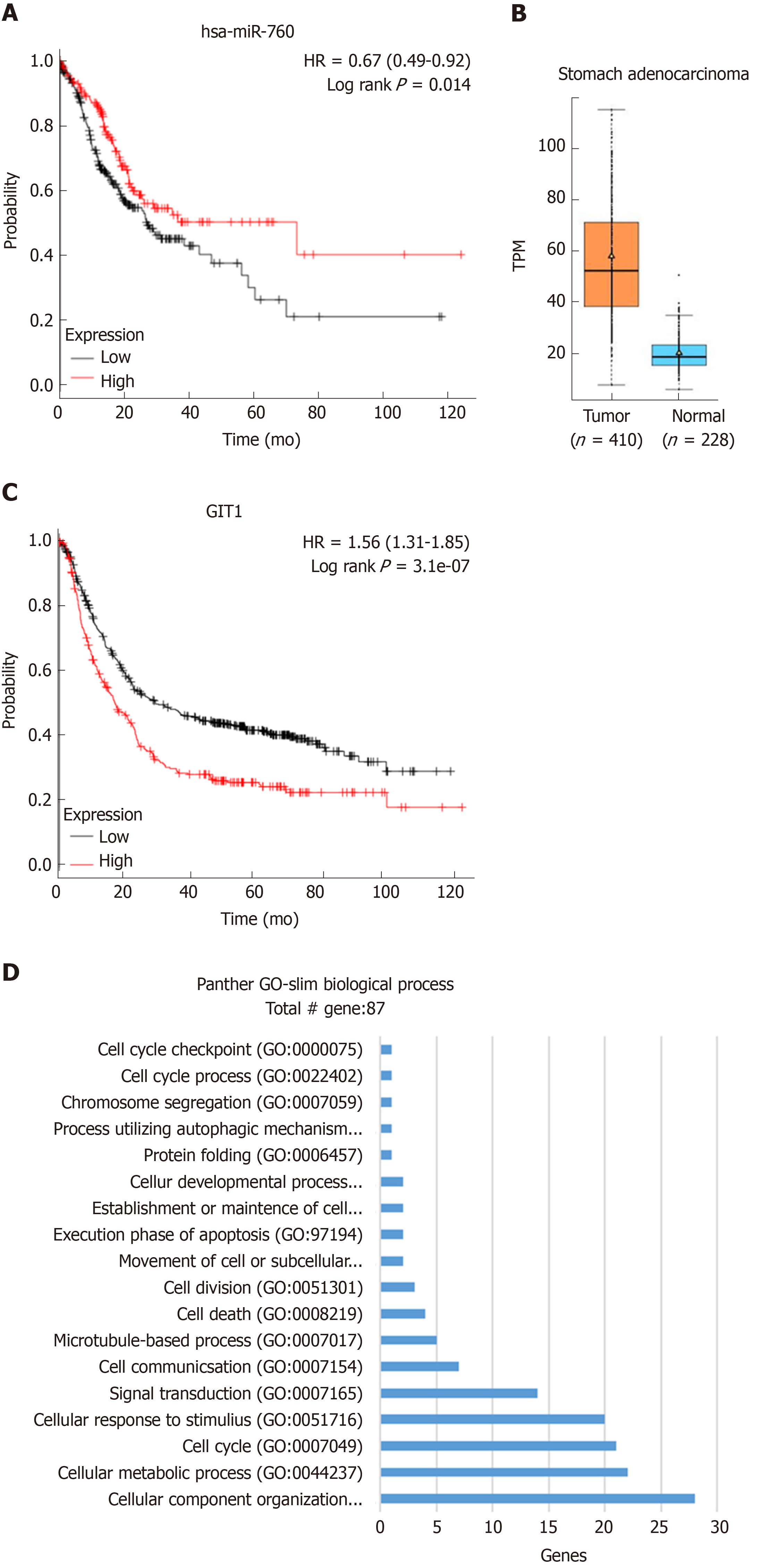
In addition to the use of clinical samples for experimental validation, we also investigated the relationship between miR-760/GIT1 expression and survival rate of GC patients using Kaplan-Meier plotter. It was found that high expression of miR-760 was closely associated with a good prognosis in GC patients (Figure 6A). In addition, the expression level of GIT1 in GC tissues was found to be higher than that in normal tissues (Figure 6B). And GC patients with high expression of GIT1 had a poor prognosis (Figure 6C). To further examine the function of GIT1 during GC development, we analyzed the genes highly associated with GIT1 in GC patients (Spearman's correlation coefficient > 0.4) using the TCGA database. Through analysis of the GO database (http://www.geneontology.org/), GIT1 was found to be closely associated with genes regulating cell cycle (GO: 0007049, Figure 6D), suggesting that upregulation of GIT1 is closely related to cell proliferation and apoptosis in GC cells.
As important regulators, the role of miRNAs has been extensively investigated in GC. For example, miR-5590-3p was downregulated in GC and inhibited tumor growth by regulating the DDX5/AKT/m-TOR pathway[16]. Here, downregulation of miR-760 was found in GC and was associated with poor clinical features and prognosis in GC patients. Functionally, miR-760 restrained cell proliferation, cell colony formation, and promoted apoptosis in GC cells. Furthermore, miR-760 acted as an inhibitor in GC cells by suppressing GIT1 expression.
Consistent with our results, downregulation of miR-760 was also detected in colorectal cancer and non-small cell lung cancer[17,18]. In addition, the prognostic role of miR-760 was found in hepatocellular carcinoma. Contrary to our results, the low expression of miR-760 predicted a good prognosis in patients with hepatocellular carcinoma[19]. These results suggest that the dysregulation and effects of miR-760 vary with the type of cancer. In most cancers, miR-760 functions as a tumor suppressor. For example, miR-760 suppressed the proliferation and metastasis of non-small cell lung cancer cells by targeting ROS1[20]. Han et al[21] demonstrated that miR-760 suppressed breast cancer cell proliferation and metastasis by downregulating NANOG. Here, overexpression of miR-760 was also found to restrain cell proliferation and colony formation in GC cells. In addition, overexpression of miR-760 promoted apoptosis in GC cells, which has not been reported in previous studies. In most previous studies, miR-760 was found to exert an effect in cancers by regulating target genes, such as FOXC1 and PTEN[22,23]. In this study, GIT1 was confirmed to be a direct target of miR-760. Upregulation of GIT1 weakened the inhibitory effect of miR-760 on the development of GC.
As a multifunctional protein, GIT1 has been shown to regulate the formation of lamellipodia, focal adhesion, and cell migration[24]. It has been reported that GIT1 is involved in the development of human cancers by interacting with miRNAs, such as miR-195 and miR-149[25,26]. In our study, miR-760 inhibited the development of GC by suppressing GIT1 expression. We also found that GIT1 was upregulated and predicted a worse prognosis in GC patients. Upregulation of GIT1 was also observed in medullary thyroid carcinoma, which was correlated with a shorter overall survival[27]. Zhao et al[28] reported that miR-638 suppressed GC cell proliferation by upregulating GIT1. Similarly, we found that miR-760 inhibited cell growth by downregulating GIT1 in GC. Additionally, miR-760 promoted apoptosis in GC cells by targeting GIT1, which has not been investigated in previous studies.
Briefly, downregulation of miR-760 was measured in GC, which was associated with poor clinical outcomes in GC patients. Functionally, miR-760 restrained cell growth and promoted apoptosis in GC cells by suppressing GIT1 expression. Based on these findings, miR-760 may be a promising therapeutic target for GC treatment. However, this study has some drawbacks, such as the small number of samples and the absence of in vivo experiments. In the future, we will increase the number of samples and perform in vivo experiments.
An increasing number of studies have revealed that microRNAs are the main drivers of carcinogenesis including progression to later stages of gastric cancer (GC). Recently, microRNA-760 (miR-760) was identified as a cancer-related miRNA in ovarian cancer and colorectal cancer. Nonetheless, the expression pattern and specific roles of miR-760 in GC have not yet been clarified.
More biomarkers are required for the diagnosis and treatment of GC.
To investigate the expression and molecular mechanism of miR-760 in GC.
MiR-760 expression was analyzed by real-time quantitative polymerase chain reaction in GC tissue and cell lines, and the clinicopathological significance of the miR-760 expression level in GC patients was investigated. Cellular experiments were performed to explore the functions of miR-760 in GC cells. Moreover, the regulatory effects of miR-760/G-protein-coupled receptor kinase interacting protein-1 (GIT1) were investigated using luciferase reporter assay, MTT assay, flow cytometric analysis, and transwell assay. At last, the relationship between the miR-760/GIT1 axis and GC was further confirmed through database analysis.
We found that miR-760 was upregulated in GC tissues and cell lines, and had a significant positive relationship with lymph nodes metastasis and TNM stage. Cellular experiments showed that miR-760 increased the proliferation and invasion capacity of GC cells but promoted apoptosis. Furthermore, miR-760 directly targeted GIT1 and downregulated its expression in GC cells. Moreover, miR-760 increased GIT1 expression to promote GC progression.
Our study demonstrated that the miR-760/GIT1 axis can significantly increase the growth, migration, and invasion of GC cells. This study provides the functional mechanism of new diagnostic biomarkers for GC.
In the future, more in-depth research will be carried out to reveal the important role of miR-760 to enhance the sensitivity of GC detection and to develop novel anti-cancer treatments targeting miR-760 and GIT1. The identification of the miR-760/GIT1 molecular axis may further provide new strategies for GC prevention and treatment.
Manuscript source: Unsolicited manuscript
Specialty type: Gastroenterology and hepatology
Country of origin: China
Peer-review report classification
Grade A (Excellent): 0
Grade B (Very good): B
Grade C (Good): C
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Lazăr DC, Nakajima N S-Editor: Tang JZ L-Editor: Wang TQ E-Editor: Ma YJ
| 1. | Van Cutsem E, Sagaert X, Topal B, Haustermans K, Prenen H. Gastric cancer. Lancet. 2016;388:2654-2664. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1282] [Cited by in F6Publishing: 1367] [Article Influence: 170.9] [Reference Citation Analysis (0)] |
| 2. | Smith MG, Hold GL, Tahara E, El-Omar EM. Cellular and molecular aspects of gastric cancer. World J Gastroenterol. 2006;12:2979-2990. [PubMed] [DOI] [Cited in This Article: ] [Cited by in CrossRef: 152] [Cited by in F6Publishing: 173] [Article Influence: 9.6] [Reference Citation Analysis (0)] |
| 3. | Ishihara R. Infrared endoscopy in the diagnosis and treatment of early gastric cancer. Endoscopy. 2010;42:672-676. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 14] [Cited by in F6Publishing: 19] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 4. | Turaga KK, Gamblin TC, Pappas S. Surgical treatment of peritoneal carcinomatosis from gastric cancer. Int J Surg Oncol. 2012;2012:405652. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 2] [Cited by in F6Publishing: 5] [Article Influence: 0.4] [Reference Citation Analysis (0)] |
| 5. | Yuan KT, Li BX, Yuan YJ, Tan M, Tan JF, Dai WG, Feng WD, Zuo JD. Deregulation of MicroRNA-375 Inhibits Proliferation and Migration in Gastric Cancer in Association With Autophagy-Mediated AKT/mTOR Signaling Pathways. Technol Cancer Res Treat. 2018;17:1533033818806499. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 13] [Cited by in F6Publishing: 21] [Article Influence: 4.2] [Reference Citation Analysis (0)] |
| 6. | Li W, Gao YQ. MiR-217 is involved in the carcinogenesis of gastric cancer by down-regulating CDH1 expression. Kaohsiung J Med Sci. 2018;34:377-384. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 25] [Cited by in F6Publishing: 30] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 7. | Liao Y, Deng Y, Liu J, Ye Z, You Z, Yao S, He S. MiR-760 overexpression promotes proliferation in ovarian cancer by downregulation of PHLPP2 expression. Gynecol Oncol. 2016;143:655-663. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 41] [Cited by in F6Publishing: 47] [Article Influence: 5.9] [Reference Citation Analysis (0)] |
| 8. | Cao L, Liu Y, Wang D, Huang L, Li F, Liu J, Zhang C, Shen Z, Gao Q, Yuan W, Zhang Y. MiR-760 suppresses human colorectal cancer growth by targeting BATF3/AP-1/cyclinD1 signaling. J Exp Clin Cancer Res. 2018;37:83. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 51] [Cited by in F6Publishing: 61] [Article Influence: 10.2] [Reference Citation Analysis (0)] |
| 9. | Iwaya T, Fukagawa T, Suzuki Y, Takahashi Y, Sawada G, Ishibashi M, Kurashige J, Sudo T, Tanaka F, Shibata K, Endo F, Katagiri H, Ishida K, Kume K, Nishizuka S, Iinuma H, Wakabayashi G, Mori M, Sasako M, Mimori K. Contrasting expression patterns of histone mRNA and microRNA 760 in patients with gastric cancer. Clin Cancer Res. 2013;19:6438-6449. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 32] [Cited by in F6Publishing: 32] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 10. | Xiao J, Chen X, Xu L, Zhang Y, Yin Q, Wang F. Regulation of chondrocyte proliferation through GIT1-Rac1-mediated ERK1/2 pathway by PDGF. Cell Biol Int. 2014;38:695-701. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 8] [Cited by in F6Publishing: 8] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 11. | Zhang LQ, Zhao GZ, Xu XY, Fang J, Chen JM, Li JW, Gao XJ, Hao LJ, Chen YZ. Integrin-β1 regulates chondrocyte proliferation and apoptosis through the upregulation of GIT1 expression. Int J Mol Med. 2015;35:1074-1080. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 31] [Cited by in F6Publishing: 33] [Article Influence: 3.7] [Reference Citation Analysis (0)] |
| 12. | Manabe R, Kovalenko M, Webb DJ, Horwitz AR. GIT1 functions in a motile, multi-molecular signaling complex that regulates protrusive activity and cell migration. J Cell Sci. 2002;115:1497-1510. [PubMed] [Cited in This Article: ] |
| 13. | Chang JS, Su CY, Yu WH, Lee WJ, Liu YP, Lai TC, Jan YH, Yang YF, Shen CN, Shew JY, Lu J, Yang CJ, Huang MS, Lu PJ, Lin YF, Kuo ML, Hua KT, Hsiao M. GIT1 promotes lung cancer cell metastasis through modulating Rac1/Cdc42 activity and is associated with poor prognosis. Oncotarget. 2015;6:36278-36291. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 32] [Cited by in F6Publishing: 36] [Article Influence: 4.5] [Reference Citation Analysis (0)] |
| 14. | Huang WC, Chan SH, Jang TH, Chang JW, Ko YC, Yen TC, Chiang SL, Chiang WF, Shieh TY, Liao CT, Juang JL, Wang HC, Cheng AJ, Lu YC, Wang LH. miRNA-491-5p and GIT1 serve as modulators and biomarkers for oral squamous cell carcinoma invasion and metastasis. Cancer Res. 2014;74:751-764. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 119] [Cited by in F6Publishing: 129] [Article Influence: 11.7] [Reference Citation Analysis (0)] |
| 15. | Dong Y, Chang C, Liu J, Qiang J. Targeting of GIT1 by miR-149* in breast cancer suppresses cell proliferation and metastasis in vitro and tumor growth in vivo. Onco Targets Ther. 2017;10:5873-5882. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 17] [Cited by in F6Publishing: 18] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 16. | Wu N, Han Y, Liu H, Jiang M, Chu Y, Cao J, Lin J, Liu Y, Xu B, Xie X. miR-5590-3p inhibited tumor growth in gastric cancer by targeting DDX5/AKT/m-TOR pathway. Biochem Biophys Res Commun. 2018;503:1491-1497. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 24] [Cited by in F6Publishing: 28] [Article Influence: 4.7] [Reference Citation Analysis (0)] |
| 17. | Zhang X, Wang L, Liu Y, Huang W, Cheng D. MiR-760 enhances TRAIL sensitivity in non-small cell lung cancer via targeting the protein FOXA1. Biomed Pharmacother. 2018;99:523-529. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 20] [Cited by in F6Publishing: 20] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 18. | Elshafei A, Shaker O, Abd El-Motaal O, Salman T. The expression profiling of serum miR-92a, miR-375, and miR-760 in colorectal cancer: An Egyptian study. Tumour Biol. 2017;39:1010428317705765. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 28] [Cited by in F6Publishing: 33] [Article Influence: 4.7] [Reference Citation Analysis (0)] |
| 19. | Sun D, Lu J, Hu C, Zhang Q, Wang X, Zhang Z, Hu S. Prognostic role of miR-760 in hepatocellular carcinoma. Oncol Lett. 2018;16:7239-7244. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 5] [Cited by in F6Publishing: 9] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 20. | Yan C, Zhang W, Shi X, Zheng J, Jin X, Huo J. MiR-760 suppresses non-small cell lung cancer proliferation and metastasis by targeting ROS1. Environ Sci Pollut Res Int. 2018;25:18385-18391. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 32] [Cited by in F6Publishing: 31] [Article Influence: 5.2] [Reference Citation Analysis (0)] |
| 21. | Han ML, Wang F, Gu YT, Pei XH, Ge X, Guo GC, Li L, Duan X, Zhu MZ, Wang YM. MicroR-760 suppresses cancer stem cell subpopulation and breast cancer cell proliferation and metastasis: By down-regulating NANOG. Biomed Pharmacother. 2016;80:304-310. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 30] [Cited by in F6Publishing: 29] [Article Influence: 3.6] [Reference Citation Analysis (0)] |
| 22. | Zhu Y, Xing Y, Chi F, Sun W, Zhang Z, Piao D. Long noncoding RNA SNHG6 promotes the progression of colorectal cancer through sponging miR-760 and activation of FOXC1. Onco Targets Ther. 2018;11:5743-5752. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 26] [Cited by in F6Publishing: 34] [Article Influence: 5.7] [Reference Citation Analysis (0)] |
| 23. | Tian T, Fu X, Lu J, Ruan Z, Nan K, Yao Y, Yang Y. MicroRNA-760 Inhibits Doxorubicin Resistance in Hepatocellular Carcinoma through Regulating Notch1/Hes1-PTEN/Akt Signaling Pathway. J Biochem Mol Toxicol. 2018;32:e22167. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 28] [Cited by in F6Publishing: 30] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 24. | Hsu RM, Tsai MH, Hsieh YJ, Lyu PC, Yu JS. Identification of MYO18A as a novel interacting partner of the PAK2/betaPIX/GIT1 complex and its potential function in modulating epithelial cell migration. Mol Biol Cell. 2010;21:287-301. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 58] [Cited by in F6Publishing: 58] [Article Influence: 3.9] [Reference Citation Analysis (0)] |
| 25. | Chan SH, Huang WC, Chang JW, Chang KJ, Kuo WH, Wang MY, Lin KY, Uen YH, Hou MF, Lin CM, Jang TH, Tu CW, Lee YR, Lee YH, Tien MT, Wang LH. MicroRNA-149 targets GIT1 to suppress integrin signaling and breast cancer metastasis. Oncogene. 2014;33:4496-4507. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 92] [Cited by in F6Publishing: 100] [Article Influence: 10.0] [Reference Citation Analysis (0)] |
| 26. | Gu YL, Rong XX, Wen LT, Zhu GX, Qian MQ. miR-195 inhibits the proliferation and migration of chondrocytes by targeting GIT1. Mol Med Rep. 2017;15:194-200. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 15] [Cited by in F6Publishing: 16] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 27. | Ye X, Chen X. miR-149-5p inhibits cell proliferation and invasion through targeting GIT1 in medullary thyroid carcinoma. Oncol Lett. 2019;17:372-378. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 13] [Cited by in F6Publishing: 20] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 28. | Zhao LY, Tong DD, Xue M, Ma HL, Liu SY, Yang J, Liu YX, Guo B, Ni L, Liu LY, Qin YN, Wang LM, Zhao XG, Huang C. MeCP2, a target of miR-638, facilitates gastric cancer cell proliferation through activation of the MEK1/2-ERK1/2 signaling pathway by upregulating GIT1. Oncogenesis. 2017;6:e368. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 40] [Cited by in F6Publishing: 43] [Article Influence: 6.1] [Reference Citation Analysis (0)] |