Copyright
©The Author(s) 2019.
World J Gastroenterol. Aug 28, 2019; 25(32): 4661-4672
Published online Aug 28, 2019. doi: 10.3748/wjg.v25.i32.4661
Published online Aug 28, 2019. doi: 10.3748/wjg.v25.i32.4661
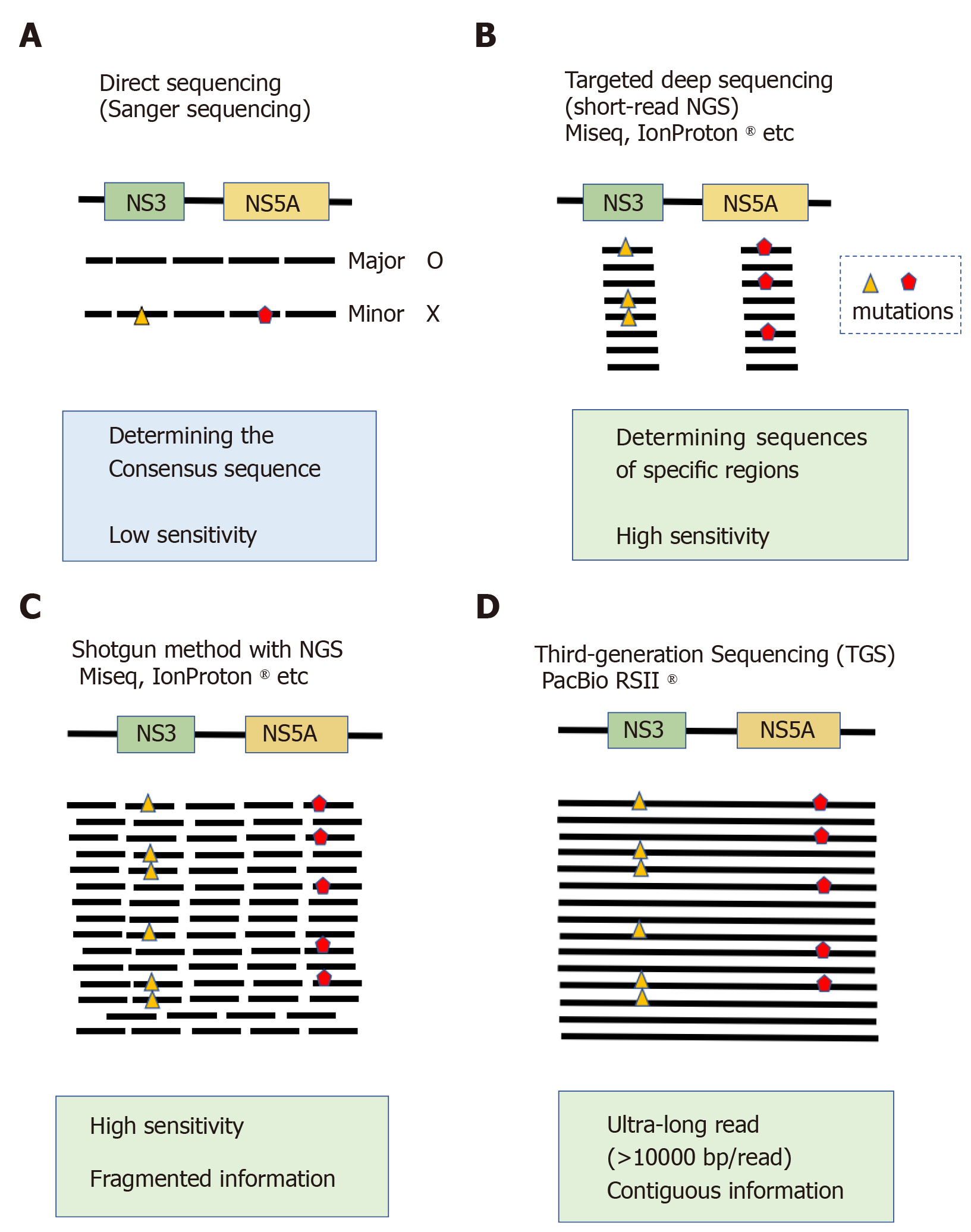
Figure 1 Comparison of sequencing platforms.
A: Direct sequencing (Sanger sequencing). This conventional sequencing method determines the consensus sequence of target regions. Nucleotide variants with allele frequencies of approximately 15% can be detected; B: Targeted deep sequencing using conventional short-read next-generation sequencing (NGS) can detect low abundance variants making up approximately 1% of total mapped reads; C: When long PCR products are used as templates for conventional short-read NGS, they are first fragmented into 100-200 bp segments, ligated to sequence adapters, amplified and then sequenced. The sequenced reads are mapped to a reference sequence using the shotgun method. One of the limitations of this technique is a lack of information regarding whether two distant mutations co-exist on a single template molecule; D: Third-generation sequencing methods represented by single-molecular real-time sequencing can generate ultra-long reads of more than 10000 bp, and contiguous sequence information can be obtained. NGS: Next-generation sequencing.
- Citation: Takeda H, Yamashita T, Ueda Y, Sekine A. Exploring the hepatitis C virus genome using single molecule real-time sequencing. World J Gastroenterol 2019; 25(32): 4661-4672
- URL: https://www.wjgnet.com/1007-9327/full/v25/i32/4661.htm
- DOI: https://dx.doi.org/10.3748/wjg.v25.i32.4661