Published online Aug 14, 2018. doi: 10.3748/wjg.v24.i30.3313
Peer-review started: April 4, 2018
First decision: May 30, 2018
Revised: June 10, 2018
Accepted: June 28, 2018
Article in press: June 28, 2018
Published online: August 14, 2018
Processing time: 131 Days and 2.7 Hours
Non-invasive diagnostic biomarkers may contribute to an early identification of gastric cancer (GC) and improve the clinical management. Unfortunately, no sensitive and specific screening biomarkers are available yet and the currently available approaches are limited by the nature of the disease. GC is a heterogenic disease with various distinct genetic and epigenetic events that occur during the multifactorial cascade of carcinogenesis. MicroRNAs (miRNAs) are commonly deregulated in gastric mucosa during the Helicobacter pylori infection and in stepwise manner from chronic gastritis, through preneoplastic conditions such as atrophic gastritis and intestinal metaplasia, to early dysplasia and invasive cancer. Identification of miRNAs in blood in 2008 led to a great interest on miRNA-based diagnostic, prognostic biomarkers in GC. In this review, we provide the most recent systematic review on the existing studies related to miRNAs as diagnostic biomarkers for GC. Here, we systematically evaluate 75 studies related to differential expression of circulating miRNAs in GC patients and provide novel view on various heterogenic aspects of the existing data and summarize the methodological differences. Finally, we highlight several important aspects crucial to improve the future translational and clinical research in the field.
Core tip: Over the past years, large amount of data to microRNAs (miRNAs) in gastric cancer (GC) has been published. We aimed to provide the critical, first of its kind in depth overview of existing studies to the miRNAs diagnostic biomarkers in GC. For this, we systematically reviewed published literature and identified 75 studies related specifically to microRNAs as blood-related non-invasive diagnostic biomarker in GC. This work provides a critical compendium to 106 studied microRNAs and summarizes the technical and methodological differences in reported studies. Furthermore, we highlight several aspects that need careful attention in future studies.
- Citation: Link A, Kupcinskas J. MicroRNAs as non-invasive diagnostic biomarkers for gastric cancer: Current insights and future perspectives. World J Gastroenterol 2018; 24(30): 3313-3329
- URL: https://www.wjgnet.com/1007-9327/full/v24/i30/3313.htm
- DOI: https://dx.doi.org/10.3748/wjg.v24.i30.3313
Gastric cancer (GC) is a deadly disease with a great challenge in clinical management despite of a steady decline of the cancer incidence[1,2]. Despite of increasing understanding of genetic and epigenetic cancer events, the absence of non-invasive methods or biomarkers for early identification of GC is one of the biggest difficulties in GC. With an ongoing technical revolution, there is a great hope to find an appropriate way to solve this limitation. Non-coding RNAs (ncRNAs) and specifically microRNAs (miRNAs) have entered the “cancer-arena” for now more than 10 years ago[3] and much research has been done over the past decade. For several years we performed systematic analysis and reviewed the role of miRNAs in GC and potential of miRNAs as a biomarker in gastrointestinal cancers[4,5]. Since then, huge amount of data has been gained, making impossible to keep an overview of existing research. In this work, we performed a systematic search and reviewed published papers related to miRNAs as non-invasive biomarkers in GC. Because of the overwhelming amount of published data, we focused solely on miRNAs as non-invasive diagnostic biomarkers in GC and excluded the data to functional alterations, prognostic and predictive role. In the first part, we will briefly review the specific and unique issues related to GC crucial for understanding the disease, provide the compiling data showing the current stand of the research and highlight the need for the future development and new directions in the field.
GC is a multifactorial heterogenic disease with unique cascade of genetic and epigenetic events leading to the cancer. There are multiple factors that, in more or less fashion, responsible for the clinical and biologically-relevant tumor heterogeneity, which may substantially impact an identification of potential diagnostic biomarkers. Those factors include geographical differences in prevalence of the risk factors, genetic background of the population, environmental factors and probably nutrition. For instance, the prevalence of GC in Asian countries and Russia is higher than in United States, Canada and northern Europe. Interestingly, geographical differences correlate with anatomical localization of primary gastric tumors. Tumors in corpus-distal subtype are predominant in Asian countries and junctional-proximal subtype in Europe. Among the most important etiological factors that may influence the tumor biology at least during the process of carcinogenesis is the Helicobacter pylori (H. pylori) infection, which is now acknowledged as an infectious disease with all the consequences of prevention and treatment[6]. For in depth review of the role of H. pylori in GC development, we refer to several recent publications[7,8]. Briefly, H. pylori is a chronic infectious diseases that causes almost always an active chronic inflammation of the gastric mucosa[6]. The persistent chronic inflammation of gastric mucosa causes different range of molecular alterations with increasing loss and accumulation of changes that leads to the phase of atrophic gastritis (AG) with intestinal metaplasie (IM) and dysplasia, which may further progress to GC and is known as Correa’s cascade[9]. AG and IM are well acknowledged preneoplastic stages of GC with an increased GC risk[10]. Close endoscopic follow up of patients with preneoplastic conditions and lesions is recommended[11]. The risk of H. pylori-related GC development may be associated with severity of mucosal inflammation and be partially dependent on bacterial virulence factors such as cytotoxin-associated gene A (CagA) and vacuolating cytotoxin A (VacA)[12]. We and others have recently shown that VacA is probably one of the most important bacterial factors that correlate with mucosa inflammation[13] and is strongly associated with anti-CagA-IgG production[14]. H. pylori eradication is suggested as the most effective way in GC reduction in high-prevalence regions[15]; however, its value is beneficial mainly for primary and secondary GC prevention without strong diagnostic value in GC screening.
Historically, GC is divided based on the Lauren’s classification into 2 histologic subtypes: intestinal and diffuse[16]. Lauren’s classification is of the remarkable value in treatment decisions and has prognostic and predictive value. With an advance of high throughput technologies, the value of Lauren’s classification has been questioned. The landmark work from The Cancer Genome Atlas (TCGA) has provided a new molecular classification subdividing GC in chromosomal instable (CIN), genomically stable (GS), microsatellite instable (MSI) and Epstein-Barr-Virus positive (EBV) tumor groups[17]. GS group shows relatively strong overlap with Lauren’s diffuse type tumors. Hence, one of the main advantages of TCGA molecular classification may be in further subdivision of intestinal subtype of GC tumors in CIN, MSI and EBV. Those tumor subtypes carry not only unique molecular patterns relevant for understanding the etiology but have potential predictive value for implementation of individualized novel therapeutic strategies.
To date, different molecules have been analyzed as potential biomarkers in patients with GC; however, as of 2018, there are no single blood-based biomarker that have sufficient sensitivity or specificity for implementation in GC screening routinely[8]. Several well-known antigens including carcinoembryonic antigen (CEA), cancer antigen 19-9 (CA19-9) and cancer antigen 72-4 (CA72-4) have been investigated in relation to GC[18]. Although the concentration of those antigens may be increased in some GC patients, the overall sensitivity of individual or combined CEA, CA19-9 and CA72-4 levels remains of insufficient discriminative power necessary for GC screening[18]. Besides typical “tumor-based” biomarkers, an effort has been made to establish a “functional” test for gastric mucosa also called “serological biopsy”[15]. Pepsinogen I and II (PGI and PGII) concentration correlate with AG, which is the preneoplastic condition of intestinal type of GC is intestinal type of GC, and is frequently used in Asian countries. In Europe the PGI and PGII panels is expanded by the use of Gastrin-17 (G17). G17 is produced by the G-cells and stimulates the hydrochloric acid and pepsinogen production and therefore is part of the same physiological cascade as PGI. Unfortunately, the use of G17 is hampered by the stability of the peptide and the net benefit in addition to PGI and II is a matter of ongoing research[19]. According to the recent systemic reviews and meta-analysis the sensitivity for AG identification is up to 70%[20,21]. Thus, PGI and PGII may be helpful for serological identification of patients with probability of AG in regions with limited resources and availability of endoscopy. However, upper GI endoscopy with careful assessment of the gastric mucosa and targeted biopsies remains the gold-standard for GC screening and identification of patients at risk. One’s upper GI endoscopy is performed, there is no additional diagnostic benefit of PGI/PGII/G17 for instance in GC screening or risk assessment[22].
MiRNAs have gathered a lot of scientific attention during the past 10 years. As we have mentioned above, miRNAs are unique subgroup of ncRNAs with crucial role in multiple biological processes. MiRNAs are involved in regulation of different molecular pathways including cell differentiation, cell cycle progression or apoptosis through post-transcriptional regulation of gene expression[23]. Deregulation of miRNAs can influence carcinogenesis through mRNA targets encoding tumor suppressor genes or oncogenes[24]. Due to its unique biogenesis, miRNAs have several features that make them an attractive group of molecules in biomarker research field. MiRNAs are very stable and are easily and reproducibly retrieved from different biological material including tissues, blood, feces, saliva, ascites and even paraffin embedded blocks[25-32]. Due to those properties, miRNAs carry a huge potential as biomarkers and have been recently extensively explored in GC. For detailed information we kindly refer to our recent reviews[4,5].
Multiple studies have demonstrated the differential expression of miRNAs in GC tissues and its functional role in GC has been suggested[4]. It is believed that miRNA alterations appear early in the cascade of the preneoplastic events. For instance, differential expression of miRNAs is identified in subjects with H. pylori infection and expression of miR-155 and miR-223 showed gradual increase in correlation to Correa’s cascade both in antrum and corpus mucosa[25]. TCGA group has shown that different molecular subtypes of GC have unique miRNA expression profiles[17], which is in support of several other profiling studies[26,33]. Understanding of the mechanisms of miRNA expression such as CpG island promoter methylation, provides valuable information for biomarker research. For example, miR-137 was implicated in GI cancers showing differential methylation in colorectal cancer (CRC) and GC patients, although the magnitude of changes was superior in CRC[34]. MiR-29c expression is reduced early in gastric carcinogenesis and has been suggested as a diagnostic and therapeutic biomarker for patients with GC[35]. The interplay of the two transcription factors HNF4γ and NR2F2 and their coordinated regulation by miR-30 and miR-194, respectively, may represent a miRNA related network responsible for expression regulation of intestinal transcripts in the development of intestinal metaplasia[36].
Another growing field of miRNA research in GC is related to the analysis of single nucleotide polymorphisms (SNPs) in miRNA genes. Variation in miRNA-sequences may lead to the expression differences and modify regulatory function of miRNAs[37,38]. To date, both gene sequences encoding precursor miRNAs[39,40] as well as variations in miRNA binding regions of target genes[41] have been extensively explored in cancer studies. The most investigated SNPs in GC are related to miR-27a, miR-146a, miR-421, miR-449a; miR-196a-2, miR-492 and miR-608[42]. Although some associations between the risk of GC development and miRNA-related SNPs have been suggested, none of the identified associations is ready to be applied in clinical settings in particular in GC screening.
Having shown the miRNA changes in tumor tissues, multiple research groups simultaneously evaluated the potential of miRNAs as non-invasive biomarkers in various specimens. Three publications related to large B-cell lymphoma, prostate and lung cancer appeared in 2008 strongly suggesting miRNAs as potential biomarkers for cancer[43-45]. This knowledge has been further extended to systematic analysis in feces in CRC[28], pancreatic cancer[29], and other diseases and specimens. Recently, it has been shown that miRNAs can be reproducibly measured in peritoneal fluid and ascites from cancer patients with peritoneal carcinomatosis and ascites[31,46]. It seems that basically any kind of body fluids (breast milk, urine, synovial fluids etc.) have measurable expression of miRNAs, which may potentially reflect a normal condition or be associated with pathophysiological alterations and therefore used as a biomarkers[5]. With existence of the overwhelming data to miRNAs in GC, we provide in the next chapter the most comprehensive summary of the exiting published data to miRNAs as non-invasive diagnostic biomarkers in GC.
To identify all available papers, we performed a systematic search with following steps: (1) Identify papers in MEDLINE/PUBMED using following criteria: gastric cancer, stomach cancer, microRNA, miRNA, biomarkers, plasma, serum, blood (until 30th November 2017); (2) We further screened all available abstracts manually one by one and excluded following papers: reviews, duplicates, retracted work, paper primarily related to miRNAs in tissues, in vitro or as prognostic or predictive biomarkers, papers without confirmed GC at the time point of the analysis or missing control group for direct comparison; (3) in January 2018 we updated the list including the papers published in December 2017 by applying more stringed criteria: diagnostic biomarkers, GC, plasma, serum. Overall, we obtained 75 original papers analyzing the expression of miRNAs in blood/serum/plasma between GC patients and controls. Among those, 18 papers refer to profiling of circulating miRNAs in GC compared to controls. Seventy-four full text papers and 1 abstract were systematically reviewed and the data were entered into the database. GraphPad Prism 7 (La Jolla, CA, United States) was used to create the figures.
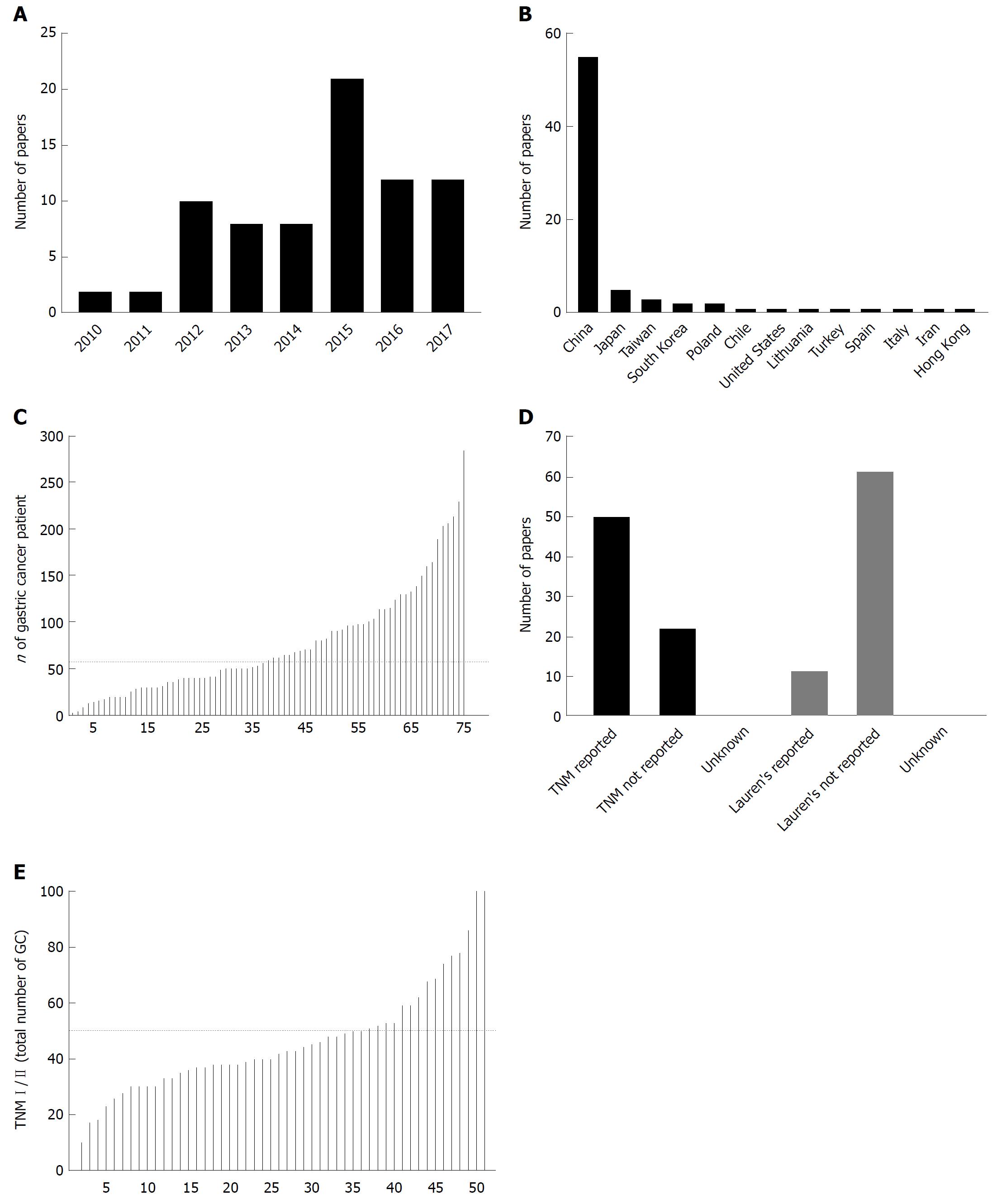
The first data to potential of miRNAs as diagnostic biomarkers in GC appeared in 2010 only two years after the first reports to detection of circulating miRNAs. We identified 75 publications from 2010 to 2017 where miRNA expression was studied in blood of GC patients and controls independently to the primary aim of the study. Over the period of 8 years, as shown in the Figure 1, we observed an increasing number of publication per year with maximum of 21 papers published in 2015 suggesting an increasing interest to the topic during the last years. To access the regional differences, we evaluated the origin of the used specimens as a surrogate. Majority of GC specimens and according published papers originate from China (55/75 or 73.3%) followed by Japan, Taiwan, Korea and others, further suggesting the regional difference in priority of the research topic and potential clinical relevance (Figure 1B).
As next, we aimed to systematically assess the reporting quality and translational potential of the published work. For this, we created 4 internal quality measures: (1) how many samples from GC patients was used; (2) is TNM-Stage reported; (3) is Lauren’s classification reported; and (4) proportion of patients with TNM I/II in total amount of GC tumors. Figure 1C shows the number of GC samples used among the 75 published papers. The median number included in the study was 57 (range 3-285) and total 5699 specimens were analyzed. Among those, 5 reports included over 200 samples each with highest number of included samples published by Qiu et al[47].
As we have shown in the introduction part, the Lauren’s classification is among the most valuable tools to assess the histological subtype, which further correlates with molecular subtype[17]. Unfortunately, the data to Lauren’s classification were available only for 16% (12/74) of the studies. TNM staging was used as another reporting quality surrogate as it correlated with prognosis and potential biomarkers need to be able to identify early cancers. Interestingly, TNM staging was reported only in 69% (51/74) of studies, which may substantially limit the quality assessment of the published work (Figure 1D). Among the studies with reported TNM staging, the proportion of GC with relatively early stages (TNM I/II) was quite heterogenic between the studies (Figure 1E). Only two studies focused solely on samples from GC patients with TNM stage I and II, while 68.8% (35/51) of studies had more than 50% of samples from patients with metastatic GC (lymph node or distant metastases).
Among the identified papers, 38 papers studied the expression of miRNAs in plasma, 32 studies in serum, 3 studies in blood and only 2 reports for peripheral blood mononuclear cell (PBMC) (Figure 2). For miRNA extraction, the mirVana, Trizol and miRNeasy were the most frequently used kits. There were substantial differences between the methods for detection/analysis. SYBR Green-based method was applied in 57% (42/74) more frequently used as TaqMan-based method (Figure 2C).
As next, we focused on the data to internal normalization of circulating miRNAs (Figure 2D). In several previous publications, RNU6b has been clearly criticized for unsuitability for normalization of blood samples as it shows different biogenesis, stability and may not reflect biogenesis of miRNAs[5]. Nevertheless, almost 60% of studies used the RNU6b-method for internal normalization of blood specimens. In similar fashion, 15% of papers applied miR-16-based method (alone or in combination with other methods), although an increasing evidence suggests that miR-16-based method may not be the best way for normalization of circulating miRNAs. Spiked-in-based method (most frequently cel-miR-39) is still considered as the most appropriate currently available methods for miRNAs normalization in blood and was used in up to 26% of studies, even though, it may probably not be the perfect long-term solution for miRNA normalization.
Challenges with normalization of circulating miRNAs may probably be the most disappointing issue related to this topic. Several papers deal with the normalization issues and compare several methods, most frequently miR-16 and RNU6b. For instance, Song et al[48] studied the expression of several miRNAs in blood and showed that there is quite a large degree of variation among miRNAs. In particular, the expression of miR-21 was higher in stage IV GC patients; however, only if the samples were normalized to miR-16, miR-93 or miR-16 and miR-93 together. This was not the case if normalization was done based on the volume[48]. Interestingly, both miR-16 and miR-93 showed substantial variation and lower expression in healthy controls arguing against it usefulness as a normalizer. Peng et al[49] studied the expression of miR-191 and miR-425 in serum of 57 GC patients. No difference was found if the authors used RNU6b, while miR-16-based normalization led to significantly higher values of miR-191. In another study, Shiotani et al[50] also compared different normalization methods in GC samples from GC patients after endoscopic submucosal dissection. The authors conclude that only normalization to miR-16, but not RNU6b, led to the higher expression of miR-106b and let-7. According to our search (Table 1)[26,47,48,51-120], there are 3 reports to differential expression of miR-16 in GC with 399 studies samples[51-53]. Two reports show up- and one report downregulation of miR-16 in plasma and sera samples, further questioning the usefulness of miR-16 for normalization of circulating miRNAs.
| microRNA | Changes inGC | Changes inGC tissue | Controls (n) | GC(n) | TNM reported | ROC | Validation | Source (Pl/Bl/Se) | qPCR | Normalization(qPCR) | Ref. |
| let-7a | ↓ | ↓ | 30 | 69 | Yes | Yes | Plasma | TaqMan | RNU6b | [56] | |
| ↓ | ↓ | 45 | 80 | Yes | Serum | SYBR | RNU6b | [57] | |||
| ↔ | 30 | 30 | Yes | Plasma | TaqMan | RNU6b | [58] | ||||
| let-7c | ↔ | 202 | 214 | No | Serum | SYBR | standard curve | [59] | |||
| let-7e | ↑ | 82 | 82 | No | 0.7 | Yes | Serum | TaqMan | cel-miR-39 | [60] | |
| let-7f | ↔ | 30 | 30 | Yes | Plasma | TaqMan | RNU6b | [58] | |||
| ↑ | 202 | 214 | No | Serum | SYBR | standard curve | [59] | ||||
| let-7g | ↔ | 30 | 30 | No | Serum | SYBR | miR-16 | [61] | |||
| let-7i | ↑ | 202 | 214 | No | Serum | SYBR | standard curve | [59] | |||
| miR-1 | ↑ | 127 | 164 | Yes | Serum | TaqMan | volume | [62] | |||
| ↔ | 30 | 30 | No | Plasma | SYBR | cel-miR-39 | [63] | ||||
| miR-100 | ↑ | 47 | 50 | Yes | 0.71 | Serum | TaqMan | RNU6b | [52] | ||
| miR-103 | ↑ | 14 | 17 | No | Plasma | SYBR | Sp6 | [64] | |||
| ↔ | 50 | 50 | No | 0.548 | Plasma | SYBR | cel-miR-39 | [65] | |||
| miR-106a | ↑ | 30 | 69 | Yes | Yes | Plasma | TaqMan | RNU6b | [56] | ||
| ↑ | 27 | 41 | No | 0.684 | PBMC | SYBR | RNU6b | [66] | |||
| ↔ | 30 | 30 | No | Plasma | SYBR | cel-miR-39 | [63] | ||||
| ↑ | ↑ | 22 | 48 | NA | Serum | NA | NA | [67] | |||
| ↑ | ↑ | 20 | 20 | Yes | Yes | Serum | TaqMan | cel-miR-39 miR-191-5p | [68] | ||
| ↑ | 130 | 130 | No | 0.786 | Yes | Serum + Exosomes | TaqMan | cel-miR-39 | [69] | ||
| miR-106b | ↑ | ↑ | 30 | 69 | Yes | 0.721 | Yes | Plasma | TaqMan | RNU6b | [56] |
| ↑ | 90 | 90 | Yes | 0.773 | Yes | Plasma | SYBR | cel-miR-39 | [63] | ||
| ↑ | 15 | 31 | Yes | Serum | TaqMan | cel-miR-39 | [70] | ||||
| ↑ | ↑ | 20 | 20 | Yes | Plasma | TaqMan | cel-miR-39 RNU6b | [71] | |||
| ↓ | 36 | 40 | Yes | 0.856 | Serum | SYBR | QuantoEC | [72] | |||
| ↑ | 65 | 65 | Yes | 0.898 | Plasma | TaqMan | RNU6b | [73] | |||
| miR-107 | ↑ | ↑ | 36 | 36 | No | 0.63 | Serum | SYBR | 5srRNA | [74] | |
| ↑ | 14 | 14 | Yes | Serum | TaqMan | RNU6b | [75] | ||||
| ↔ | 50 | 50 | No | 0.563 | Plasma | SYBR | cel-miR-39 | [65] | |||
| miR-10b-5p | ↑ | ↑ | 167 | 203 | Yes | 0.627 | Yes | Serum + exosomes | SYBR | cel-miR-39 miR-16 | [76] |
| miR-122 | ↔ | 36 | 96 | Yes | Plasma | SYBR | ath-miR-159a | [77] | |||
| miR-1233 | ↓ | 3 | 3 | No | Plasma | SYBR | RNU6b | [78] | |||
| miR-130a | ↑ | ↑ | 41 | 41 | No | 0.905 | Serum | SYBR | RNU6B | [79] | |
| ↑ | ↑ | 20 | 20 | Yes | Yes | Serum | TaqMan | cel-miR-39 miR-191-5p | [68] | ||
| miR-132-3p | ↑ | ↑ | 167 | 203 | Yes | 0.652 | Yes | Serum + exosomes | SYBR | cel-miR-39 miR-16 | [76] |
| miR-139 | ↓ | ↑ | 18 | 25 | No | 0.940 | Plasma | SYBR | RNU6b | [80] | |
| miR-140-5p | ↑ | 50 | 50 | Yes | Plasma | SYBR | RNU6B | [81] | |||
| miR-141 | ↓ | 3 | 3 | No | Plasma | SYBR | RNU6b | [78] | |||
| ↓ | 14 | 17 | No | Plasma | SYBR | Sp6 | [64] | ||||
| miR-142-3p | ↓ | 285 | 285 | Yes | 0.839 | Yes | Plasma | TaqMan | cel-miR-39 | [47] | |
| miR-143-3p | ↔ | 73 | 206 | Yes | Yes | Serum | SYBR | RNU6b | [82] | ||
| miR-144 | ↓ | ↓ | 40 | 96 | Yes | 0.821 | Serum | SYBR | RNU6b | [83] | |
| miR-146a | ↔ | 15 | 31 | Yes | Serum | TaqMan | cel-miR-39 | [70] | |||
| ↔ | 73 | 206 | Yes | Yes | Serum | SYBR | RNU6b | [82] | |||
| ↑ | ↔ | 20 | 20 | Yes | Yes | Serum | TaqMan | cel-miR-39 miR-191-5p | [68] | ||
| miR-146b | ↑ | ↔ | 20 | 20 | Yes | Yes | Serum | TaqMan | cel-miR-39 miR-191-5p | [68] | |
| miR-148a | ↔ | 15 | 31 | Yes | Serum | TaqMan | cel-miR-39 | [70] | |||
| ↓ | 285 | 285 | Yes | 0.842 | Yes | Plasma | TaqMan | cel-miR-39 | [47] | ||
| ↓ | ↓ | 39 | 38 | Yes | 0.349 | Plasma | TaqMan | miR-16-5p | [26] | ||
| miR-151-5p | ↑ | 130 | 230 | Yes | 0.625 | Yes | Plasma | TaqMan | RNU6b | [58] | |
| miR-155 | ↑ | 20 | 30 | No | Yes | Plasma | TaqMan | RNU6b | [84] | ||
| ↔ | 15 | 15 | No | Plasma | TaqMan | RNU6b | [85] | ||||
| miR-15b-5p | ↑ | ↑ | 100 | 100 | No | Plasma | SYBR | RNU6b | [86] | ||
| miR-16 | ↑ | 106 | 160 | Yes | 0.768-0.925 | Yes | Plasma | TaqMan | cel-miR-39 | [51] | |
| ↑ | 47 | 50 | Yes | 0.90 | Serum | TaqMan | RNU6b | [52] | |||
| ↓ | 129 | 189 | Yes | 0.772 | Yes | Plasma | TaqMan | cel-miR-39 miR-16-5p | [53] | ||
| miR-17 | ↑ | 27 | 41 | No | 0.743 | PBMC | SYBR | RNU6b | [66] | ||
| ↓ | 36 | 40 | Yes | 0.879 | Serum | SYBR | QuantoEC | [72] | |||
| ↑ | 30 | 69 | Yes | Yes | Plasma | TaqMan | RNU6b | [56] | |||
| ↔ | 30 | 30 | No | Plasma | SYBR | cel-miR-39 | [63] | ||||
| ↔ | 20 | 20 | No | Serum + exosomes | TaqMan | cel-miR-39 | [69] | ||||
| miR-181a | ↑ | ↑ | 18 | 25 | No | 0.882 | Plasma | SYBR | RNU6b | [80] | |
| miR-181b | ↓ | 89 | 92 | Yes | Yes | Serum | SYBR | miR-16 | [61] | ||
| miR-181c | ↑ | ↑ | 60 | 30 | No | Plasma | SYBR | RNU6b | [87] | ||
| miR-185 | ↑ | ↑ | 109 | 133 | Yes | 0.65 | Yes | Plasma | SYBR | cel-miR-39 RNU6b | [88] |
| ↑ | ↔ | 167 | 203 | Yes | 0.637 | Yes | Serum + exosomes | SYBR | cel-miR-39 miR-16 | [76] | |
| miR-187-3p | ↑ | 61 | 61 | Yes | Yes | Serum | SYBR | RNU6b | [89] | ||
| miR-18a | ↑ | 50 | 50 | Yes | Plasma | SYBR | RNU6B | [81] | |||
| ↑ | ↑ | 65 | 104 | Yes | 0.805 | Plasma | TaqMan | RNU6b | [90] | ||
| miR-191 | ↑ | 82 | 82 | No | 0.63 | Yes | Serum | TaqMan | cel-miR-39 | [60] | |
| ↑ | 58 | 57 | Yes | 0.849 | Serum | TaqMan | miR-16 | [49] | |||
| miR-192 | ↔ | 36 | 96 | Yes | Plasma | SYBR | ath-miR-159a | [77] | |||
| miR-194 | ↑ | 3 | 3 | No | Plasma | SYBR | RNU6b | [78] | |||
| ↔ | 50 | 50 | No | 0.512 | Plasma | SYBR | cel-miR-39 | [65] | |||
| miR-195 | ↓ | 285 | 285 | Yes | 0.765 | Yes | Plasma | TaqMan | cel-miR-39 | [47] | |
| ↓ | ↓ | 36 | 62 | Yes | Yes | Serum | SYBR | RNU6b | [91] | ||
| ↓ | 190 | 20 | No | Plasma | (TaqMan) | global mean | [92] | ||||
| ↑ | ↔ | 167 | 203 | Yes | 0.683 | Yes | Serum + exosomes | SYBR | cel-miR-39 miR-16 | [76] | |
| mir-196a | ↓ | ↑ | 14 | 17 | No | Plasma | SYBR | Sp6 | [64] | ||
| ↑ | ↑ | 126 | 98 | Yes | 0.864 | Plasma | SYBR | miR-16 | [93] | ||
| miR-196b | ↑ | ↑ | 126 | 98 | Yes | 0.811 | Plasma | SYBR | miR-16 | [93] | |
| miR-198 | ↔ | 30 | 30 | Yes | Plasma | TaqMan | RNU6b | [58] | |||
| miR-199a-3p | ↑ | 70 | 80 | Yes | 0.818 | Yes | Plasma | TaqMan | RNU6b | [94] | |
| ↑ | 130 | 230 | Yes | 0.837 | Yes | Plasma | TaqMan | RNU6b | [58] | ||
| ↑ | 50 | 50 | Yes | Plasma | SYBR | RNU6B | [81] | ||||
| miR-19a | ↑ | ↑ | 20 | 20 | Yes | Yes | Serum | TaqMan | cel-miR-39 miR-191-5p | [68] | |
| miR-19b | ↑ | ↔ | 20 | 20 | Yes | Yes | Serum | TaqMan | cel-miR-39 miR-191-5p | [68] | |
| ↑ | 3 | 3 | No | Plasma | SYBR | RNU6b | [78] | ||||
| ↓ | 129 | 189 | Yes | 0.749 | Yes | Plasma | TaqMan | cel-miR-39, miR-16-5p | [53] | ||
| ↑ | 130 | 130 | No | 0.769 | Yes | Serum + exosomes | TaqMan | cel-miR-39 | [69] | ||
| miR-200b | ↓ | 14 | 17 | No | Plasma | SYBR | Sp6 | [64] | |||
| miR-200c | ↑ | 15 | 52 | Yes | 0.715 | blood | SYBR | RNU6b 5SrRNA | [95] | ||
| ↑ | 100 | 98 | Yes | Serum | SYBR | RNU6b | [96] | ||||
| miR-203 | ↓ | 89 | 92 | Yes | Yes | Serum | SYBR | miR-16 | [61] | ||
| ↓ | 22 | 130 | Yes | 0.707 | Yes | Serum | TaqMan | cel-miR-39 | [97] | ||
| miR-204 | ↓ | 40 | 115 | Yes | Yes | Serum | SYBR | RNU6b | [98] | ||
| miR-206 | ↓ | 150 | 150 | Yes | 0.89 | Serum | TaqMan | cel-miR-39 | [99] | ||
| miR-20a | ↑ | 127 | 164 | Yes | Serum | TaqMan | volume | [62] | |||
| ↑ | 90 | 90 | Yes | 0.859 | Yes | Plasma | SYBR | cel-miR-39 | [63] | ||
| ↑ | ↑ | 30 | 30 | No | Plasma | SYBR | RNU6b | [100] | |||
| ↑ | ↑ | 109 | 133 | Yes | 0.67 | Yes | Plasma | SYBR | cel-miR-39 RNU6b | [88] | |
| ↑ | ↑ | 28 | 28 | No | Plasma | SYBR | RNU6b | [101] | |||
| ↑ | ↔ | 167 | 203 | Yes | 0.637 | Yes | Serum + exosomes | SYBR | cel-miR-39 miR-16 | [76] | |
| ↑ | 12 | 12 | No | Serum | SYBR | cel-miR-39 | [102] | ||||
| miR-21 | ↑ | 30 | 69 | Yes | Yes | Plasma | TaqMan | RNU6b | [56] | ||
| ↑ | 20 | 53 | Yes | 0.853 | Blood | SYBR | RNU6b | [103] | |||
| ↑ | 70 | 70 | Yes | 0.794 | Yes | Plasma | TaqMan | cel-miR-39 | [104] | ||
| ↑ | 39 | 30 | Yes | 0.81 | Serum | SYBR | miR-16 | [105] | |||
| ↑ | 20 | 40 | Yes | Serum | All-in-one | Volume miR-16, miR-93 | [48] | ||||
| ↔ | 90 | 90 | Yes | Plasma | SYBR | cel-miR-39 | [63] | ||||
| ↔ | 15 | 31 | Yes | Serum | TaqMan | cel-miR-39 | [70] | ||||
| ↔ | 15 | 15 | No | Plasma | TaqMan | RNU6b | [85] | ||||
| ↑ | 50 | 50 | Yes | 0.912 | Serum | SYBR | RNU6b | [106] | |||
| ↑ | 50 | 50 | Yes | 0.898 | PBMC | SYBR | RNU6b | [106] | |||
| ↑ | ↑ | 14 | 17 | No | Plasma | SYBR | Sp6 | [64] | |||
| ↑ | 89 | 92 | Yes | Yes | Serum | SYBR | miR-16 | [61] | |||
| ↑ | ↑ | 20 | 20 | Yes | Yes | Serum | TaqMan | cel-miR-39 miR-191-5p | [68] | ||
| miR-210 | ↑ | ↑ | 109 | 133 | Yes | 0.75 | Yes | Plasma | SYBR | cel-miR-39 RNU6b | [88] |
| miR-212 | ↑ | ↔ | 20 | 20 | Yes | Yes | Serum | TaqMan | cel-miR-39 miR-191-5p | [68] | |
| miR-218 | ↓ | 70 | 70 | Yes | 0.743 | Yes | Plasma | TaqMan | cel-miR-39 | [104] | |
| ↓ | 56 | 68 | Yes | Serum | SYBR | cel-miR-39 | [107] | ||||
| miR-220 | ↑ | ↔ | 20 | 20 | Yes | Yes | Serum | TaqMan | cel-miR-39 miR-191-5p | [68] | |
| miR-221 | ↑ | 82 | 82 | No | 0.7 | Yes | Serum | TaqMan | cel-miR-39 | [60] | |
| ↑ | 90 | 90 | Yes | 0.796 | Yes | Plasma | SYBR | cel-miR-39 | [63] | ||
| ↑ | 14 | 17 | No | Plasma | SYBR | Sp6 | [64] | ||||
| miR-222 | ↑ | 82 | 82 | No | 0.65 | Yes | Serum | TaqMan | cel-miR-39 | [60] | |
| ↑ | 56 | 114 | Yes | 0.85 | Plasma | TaqMan | RNU6b | [108] | |||
| miR-223 | ↑ | 70 | 70 | Yes | 0.91 | Yes | Plasma | TaqMan | cel-miR-39 | [104] | |
| ↑ | 15 | 31 | Yes | Serum | TaqMan | cel-miR-39 | [70] | ||||
| ↑ | 47 | 50 | Yes | 0.85 | Serum | TaqMan | RNU6b | [52] | |||
| ↑ | ↑ | 50 | 50 | Yes | 0.81 | Plasma | SYBR | RNU6b | [109] | ||
| ↑ | 3 | 3 | No | Plasma | SYBR | RNU6b | [78] | ||||
| ↑ | ↑ | 20 | 20 | Yes | Yes | Serum | TaqMan | cel-miR-39 miR-191-5p | [68] | ||
| ↑ | ↑ | 20 | 15 | Yes | Plasma | NCode | cel-miR-39/-54/-238; miR-16 | [110] | |||
| ↑ | ↑ | 39 | 38 | Yes | 0.671 | Plasma | TaqMan | miR-16-5p | [26] | ||
| miR-23a | ↑ | 14 | 17 | No | Plasma | SYBR | Sp6 | [64] | |||
| miR-23b | ↑ | 50 | 138 | Yes | 0.80 | Plasma | SYBR | RNU6b | [111] | ||
| miR-25 | ↔ | 10 | 10 | No | Plasma | TaqMan | cel-miR-39 | [104] | |||
| ↑ | 106 | 160 | Yes | 0.694-0.925 | Yes | Plasma | TaqMan | cel-miR-39 | [51] | ||
| ↑ | ↑ | 20 | 20 | Yes | Plasma | TaqMan | cel-miR-39; RNU6b | [71] | |||
| ↑ | ↑ | 109 | 133 | Yes | 0.65 | Yes | Plasma | SYBR | cel-miR-39; RNU6b | [88] | |
| ↔ | ↑ | 70 | 70 | Yes | Plasma | TaqMan | RNU6B | [112] | |||
| ↑ | 14 | 14 | Yes | Serum | TaqMan | RNU6b | [75] | ||||
| ↑ | 65 | 65 | Yes | 0.817 | Plasma | TaqMan | RNU6b | [73] | |||
| miR-26a | ↓ | 285 | 285 | Yes | 0.882 | Yes | Plasma | TaqMan | cel-miR-39 | [47] | |
| miR-26b | ↔ | 30 | 30 | Yes | Plasma | TaqMan | RNU6b | [58] | |||
| miR-27a | ↑ | 127 | 164 | Yes | Serum | TaqMan | volume | [62] | |||
| ↑ | 82 | 82 | No | 0.67 | Yes | Serum | TaqMan | cel-miR-39 | [60] | ||
| ↔ | 30 | 30 | No | Plasma | SYBR | cel-miR-39 | [63] | ||||
| ↔ | 15 | 31 | Yes | Serum | TaqMan | cel-miR-39 | [70] | ||||
| ↑ | ↑ | 35 | 35 | Yes | 0.70 | Yes | Plasma | TaqMan | RNU6b | [85] | |
| miR-27b | ↑ | 82 | 82 | No | 0.66 | Yes | Serum | TaqMan | cel-miR-39 | [60] | |
| miR-296 | ↑ | ↔ | 20 | 20 | Yes | Yes | Serum | TaqMan | cel-miR-39; miR-191-5p | [68] | |
| ↑ | ↑ | 167 | 203 | Yes | 0.652 | Yes | Serum + exosomes | SYBR | cel-miR-39 miR-16 | [76] | |
| miR-30a-5p | ↔ | 20 | 20 | No | Serum + exosomes | TaqMan | cel-miR-39 | [69] | |||
| miR-30c | ↑ | ↔ | 20 | 20 | Yes | Yes | Serum | TaqMan | cel-miR-39; miR-191-5p | [68] | |
| miR-31 | ↓ | 89 | 92 | Yes | Yes | Serum | SYBR | miR-16 | [61] | ||
| miR-32 | ↑ | ↑ | 40 | 40 | No | Plasma | SYBR | RNU6b | [113] | ||
| miR-323-3p | ↑ | ↔ | 20 | 20 | Yes | Yes | Serum | TaqMan | cel-miR-39; miR-191-5p | [68] | |
| miR-331 | ↑ | ↑ | 20 | 20 | Yes | Yes | Serum | TaqMan | cel-miR-39; miR-191-5p | [68] | |
| miR-335 | ↓ | ↓ | 7 | 4 | No | Plasma | TaqMan | RNU6b | [114] | ||
| miR-34 | ↔ | 30 | 30 | No | Plasma | SYBR | cel-miR-39 | [63] | |||
| miR-346 | ↓ | 14 | 17 | No | Plasma | SYBR | Sp6 | [64] | |||
| miR-34a | ↑ | 127 | 164 | Yes | Serum | TaqMan | volume | [62] | |||
| miR-365 | ↑ | ↔ | 20 | 20 | Yes | Yes | Serum | TaqMan | cel-miR-39; miR-191-5p | [68] | |
| miR-370 | ↑ | ↑ | 12 | 40 | Yes | 0.79 | Plasma | TaqMan | miR-16 | [115] | |
| miR-371-5p | ↑ | 61 | 61 | Yes | Yes | Serum | SYBR | RNU6b | [89] | ||
| miR-374 | ↑ | ↑ | 20 | 20 | Yes | Yes | Serum | TaqMan | cel-miR-39; miR-191-5p | [68] | |
| miR-375 | ↓ | ↓ | 20 | 20 | No | 0.835 | Serum | TaqMan | RNU6b | [116] | |
| ↓ | ↓ | 39 | 38 | Yes | 0.32 | Plasma | TaqMan | miR-16-5p | [26] | ||
| miR-376a | ↔ | ↑ | 108 | 65 | Yes | Plasma | TaqMan | RNU6B | [117] | ||
| miR-376c | ↑ | 82 | 82 | No | 0.71 | Yes | Serum | TaqMan | cel-miR-39 | [60] | |
| ↔ | 30 | 30 | No | Plasma | SYBR | cel-miR-39 | [63] | ||||
| ↑ | ↑ | 108 | 65 | Yes | 0.77 | Plasma | TaqMan | RNU6B | [117] | ||
| miR-378 | ↑ | ↓ | 61 | 61 | Yes | 0.861 | Yes | Serum | SYBR | RNU6b | [89] |
| ↔ | 30 | 30 | No | Plasma | SYBR | cel-miR-39 | [63] | ||||
| ↓ | 14 | 17 | No | Plasma | SYBR | Sp6 | [64] | ||||
| miR-421 | ↑ | 17 | 40 | Yes | 0.773 | PBMC | SYBR | RNU6b | [118] | ||
| ↑ | 50 | 50 | No | Yes | Serum (?) | SYBR | RNU6b | [119] | |||
| ↑ | 90 | 90 | No | 0.779 | Serum | SYBR | RNU6b | [120] | |||
| ↑ | 90 | 90 | No | 0.821 | PBMC | SYBR | RNU6b | [120] | |||
| miR-423-5p | ↑ | 127 | 164 | Yes | Serum | TaqMan | volume | [62] | |||
| ↔ | 30 | 30 | No | Plasma | SYBR | cel-miR-39 | [63] | ||||
| miR-425 | ↔ | 58 | 57 | Yes | Serum | TaqMan | miR-16 | [49] | |||
| miR-433 | ↔ | 15 | 31 | Yes | Serum | TaqMan | cel-miR-39 | [70] | |||
| ↑ | ↔ | 20 | 20 | Yes | Yes | Serum | TaqMan | cel-miR-39; miR-191-5p | [68] | ||
| miR-451 | ↑ | ↓ | 30 | 56 | Yes | 0.96 | Yes | Plasma | TaqMan | RNU6b | [54] |
| ↔ | 90 | 90 | Yes | Plasma | SYBR | cel-miR-39 | [63] | ||||
| ↑ | ↔ | 20 | 20 | Yes | Yes | Serum | TaqMan | cel-miR-39; miR-191-5p | [68] | ||
| ↑ | 106 | 160 | Yes | 0.790-0.850 | Yes | Plasma | TaqMan | cel-miR-39 | [51] | ||
| ↔ | 73 | 206 | Yes | Yes | Serum | SYBR | RNU6b | [82] | |||
| miR-484 | ↓ | ↓ | 14 | 17 | No | Plasma | SYBR | Sp6 | [64] | ||
| miR-486 | ↑ | ↓ | 30 | 56 | Yes | 0.92 | Yes | Plasma | TaqMan | RNU6b | [54] |
| ↔ | 30 | 30 | No | Plasma | SYBR | cel-miR-39 | [63] | ||||
| ↑ | 106 | 160 | Yes | 0.779-0.863 | Yes | Plasma | TaqMan | cel-miR-39 | [51] | ||
| ↓ | 14 | 17 | No | Plasma | SYBR | Sp6 | [64] | ||||
| miR-501-3p | ↔ | 73 | 206 | Yes | Yes | Serum | SYBR | RNU6b | [82] | ||
| miR-518d | ↑ | ↔ | 20 | 20 | Yes | Yes | Serum | TaqMan | cel-miR-39; miR-191-5p | [68] | |
| miR-518f | ↑ | ↔ | 20 | 20 | Yes | Yes | Serum | TaqMan | cel-miR-39; miR-191-5p | [68] | |
| miR-627 | ↑ | ↑ | 111 | 123 | Yes | 0.937 | Yes | Plasma | SYBR | RNU6B | [81] |
| miR-629 | ↑ | ↑ | 111 | 123 | Yes | 0.912 | Yes | Plasma | SYBR | RNU6B | [81] |
| miR-652 | ↑ | ↑ | 111 | 123 | Yes | 0.918 | Yes | Plasma | SYBR | RNU6B | [81] |
| miR-720 | ↔ | 30 | 30 | Yes | Plasma | TaqMan | RNU6b | [58] | |||
| miR-744 | ↑ | 82 | 82 | No | 0.74 | Yes | Serum | TaqMan | cel-miR-39 | [60] | |
| ↔ | 30 | 30 | No | Plasma | SYBR | cel-miR-39 | [63] | ||||
| miR-92a | ↑ | 106 | 160 | Yes | 0.732-0.913 | Yes | Plasma | TaqMan | cel-miR-39 | [51] | |
| ↓ | 89 | 92 | Yes | Yes | Serum | SYBR | miR-16 | [61] | |||
| miR-92b | ↑ | ↑ | 109 | 133 | Yes | 0.69 | Yes | Plasma | SYBR | cel-miR-39; RNU6b | [88] |
| miR-93 | ↑ | ↑ | 20 | 20 | Yes | Plasma | TaqMan | cel-miR-39; RNU6b | [71] | ||
| ↑ | 65 | 65 | Yes | 0.756 | Plasma | TaqMan | RNU6b | [73] | |||
| miR-940 | ↓ | ↓ | 105 | 115 | Yes | 0.96 | Yes | Plasma | SYBR | miR-16 | [55] |
The approach of miR-16-based normalization may be also called as a “proportional normalization” as rather the proportion between certain miRNAs (in comparison to miR-16) and not the absolute value of studied miRNAs is used. This has been implemented by multiple studies and has been shown of potential diagnostic benefit independently to its scientific objectivity and validity. For instance, we analyzed miRNAs expression in ascites and showed that miR-21 was upregulated in patients with peritoneal carcinomatosis compared to control group[31]. However, also patients with peritonitis demonstrated similar increase as patients with peritoneal carcinomatosis. To overcome this limitation, we used the proportion of miR-21 (cancer-associated) and miR-223 (inflammation-associated) to differentiate the groups[31]. For miR-16, the very high values of miR-16 in erythrocytes strongly suggest that other factors such as tumor anemia or hemolysis may have an additional impact on the results. Nevertheless, this does not necessarily mean that miR-16 is not suitable, but rather that we need to know the influential factors and to know exact biogenesis of miR-16 in circulation. Further studies are needed to provide the comprehensive view on patients-related factors.
An alternative normalization way has been proposed where multiple miRNAs can be used simultaneously. For instance, miRCURY LNA Universal RT microRNA PCR System offers internal standard including miR-103a-3p, miR-191-5p, miR-423-3p and -5p and miR-451. However, we strongly doubt the usefulness of this method, as every single of those selected miRNAs have been reported as deregulated in cancer and in particular GC (Table 1). Furthermore, miR-451 is highly dependent on hemolysis and may provide some unexpected bias in analysis. Thus, in similar way as addressed for miR-16, additional studies are needed to confirm the usefulness, biological suitability and stability of the methods.
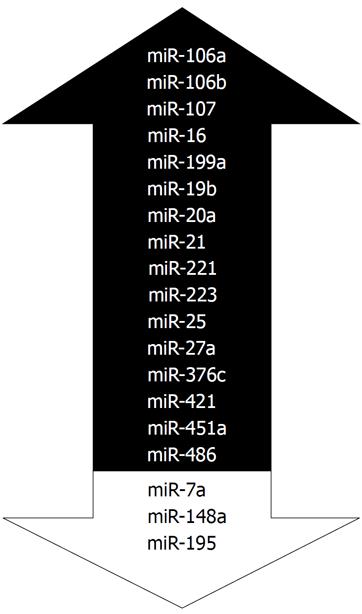
In the Table 1, we have summarized the miRNAs that have been analyzed for the differential expression between GC patients and controls. According to our search, we identified 106 miRNAs that were studied in different studies. Among those, 13 miRNAs such as let-7c, let-7g, miR-143-3p, miR-122, miR-192, miR-198 etc. showed no significant changes. As shown in in Table 1, multiple miRNAs have conflicting results showing differential expression in one cohort while no changes found in another. In the Figure 3 we summarize the most frequently deregulated miRNAs. Those were selected using two criteria: (1) at least 3 publications and (2) at least two with reproducible report on increased or decreased expression. Among the most consistent miRNAs are the miR-20a, miR-223 and miR-421. Among the most studied is the miR-21 with total 13 reports. Summarizing the data to miR-21 expression, three reports showed no differences while 10 reports show an upregulation of miR-21 both in sera and in plasma of GC patients. As shown in the Table 1, multiple groups used samples from independent cohorts to confirm the results, which substantially contributes to the quality of the reports.
To estimate the diagnostic potential of miRNAs as biomarkers in GC, multiple studies provided receiver operator curves (ROC) values. For instance, Konishi et al[54] reported the diagnostic accuracy for miR-451 reaching 0.96 with calculated sensitivity of 96% and specificity of 100%. Liu et al[55] studied miR-940 and reported the ROC value of 0.96 although with slightly lower sensitivity 81.2% and specificity 98.6%. Although those results are striking, we need to keep in mind that validation of the results from independent groups or cohorts in prospective studies are still to come. We believe that the data from our summary table will be helpful for the future search, validation and discussion of the results.
It is well known that the most promising way to identify the potential biomarkers is the unbiased profiling of the samples. This approach was applied in the pivotal work by Chen et al[45] in 2008 in lung cancers, where the authors used multiple pooled samples for profiling and performed independent validation using qPCR. As shown in the Table 2[47,51,53-55,58,60,62,64,68,76,78,81,82,88,92,121], this approach was very intensively used for miRNA-profiling in GC patients. Among the total eighteen miRNA-profiling studies, 50% (9/18) reported the use of the pooled samples. Majority of studies used single technical pooled profiling from 5-40 samples for GC and controls. Although, this may be an appropriate way for proof-of-principle studies, it has several limitations where inappropriate changes of miRNAs in few subjects may create very strong deviation and bias and therefore independent validation is mandatory. Among the 18 profiling studies (Table 2), 11 reports studied miRNA in plasma, 6 in sera and 1 in blood samples. From the technical perspective there was substantial variation in use of extraction kits and profiling platforms used for analysis. Similar to the overall data in total cohort (Table 1), researches from China with 12 profiling studies provided the most overwhelming data. Remaining work comes from Poland, Japan, Turkey, Hong-Kong and United States. The first studies tend to use relatively small number of samples while more recent studies provide increasing number of samples and include an impressive number of samples for validation analysis as well[68,76].
| Ref. | Year | Source | Extraction Kit | Platform | Sample Origin | GC(number of technical runs) | Controls (number of technical runs) | pooled samples | GC(number ofpooled samples) | Controls (number ofpooled samples) |
| Liu et al[62] | 2011 | Serum | phen/chlor | Solexa | China | 1 | 1 | Yes | 20 | 20 |
| Song et al[60] | 2012 | Serum | miRNeasy | qPCR-array | China | 1 | 1 | Yes | 14 | 14 |
| Liu et al[89] | 2012 | Serum | mirVana | Agilent | China | 7 | 10 | No | ||
| Konishi et al[54] | 2012 | Plasma | mirVana | microarray | Japan | 3 | 3 | No | ||
| Gorur et al[92] | 2013 | Plasma | high-pure miRNA Isolation | TaqMan Array | Turkey | 1 | 1 | Yes | 20 | 190 |
| Shah et al[121] | 2013 | Blood | miRNeasy | Geniom Biochip | Poland | 1 | 1 | Yes | 8 | 19 |
| Li et al[58] | 2013 | Plasma | mirVana | Agilent | China | 20 | 20 | No | ||
| Zhu et al[51] | 2014 | Plasma | miRNeasy | TaqMan Array | China | 1 | 1 | Yes | 40 | 40 |
| Zhang et al[53] | 2015 | Plasma | mirVana | Agilent | China | 16 | 18 | No | ||
| Zhou et al[88] | 2015 | Plasma | mirVana | Exiqon | China | 3 | 1 | Yes | 30 | 10 |
| Shin et al[81] | 2015 | Plasma | Trizol miRNeasy | miRCURY LNA | China | 5 | 5 | No | ||
| Zhang et al[78] | 2015 | Plasma | mirVana | Agilent | China | 3 | 3 | No | ||
| Liu et al[55] | 2016 | Plasma | miRNeasy | miRCURY LNA | Hong-Kong | 5 | 5 | No | ||
| Qiu et al[47] | 2016 | Plasma | miRNeasy | Agilent | China | 1 | 1 | Yes | 5 | 5 |
| Treece et al[64] | 2016 | Plasma | miRCURY | GastroGenus miR Panel | United States | 17 | 14 | No | ||
| Jiang et al[82] | 2017 | Serum | unknown | miSeq | China | 2 | 1 | Yes | 20 | 10 |
| Huang et al[76] | 2017 | Serum | mirVana | Exiqon | China | 3 | 1 | Yes | 30 | 10 |
| Sierzega et al[68] | 2017 | Serum | miRNA ABC Purification | TaqMan Array | Poland | 20 | 20 | No |
In this review, we systematically summarized and analyzed the existing data to miRNAs as non-invasive biomarkers for GC. There is no doubt that further studies with an improved study design will follow and it is a matter of time until the miRNA-based biomarkers will enter the clinical studies either in oncologic patients or patients with other diseases. However, following our critical review of the existing papers we would like to provide several cautionary notes for improvement.
The era of proof-of-principle studies in GC has passed and with 75 published data it is time to improve the quality of the work. There are several ways that may be considered: (1) description of patient’s cohort including Lauren’s Classification, TNM staging and precise number of patients is necessary; (2) reporting of the methodological steps (extraction kits, measurements, reproducibility, validation etc.) need to be carefully reviewed and reported; (3) for primary research an independent cohort of samples need to be included; and (4) reporting of ROC/AUC values, sensitivity and specificity are currently used in quite biased way in proportion of GC patients to controls mostly in 1:1; however, in real-life settings the proportion will be at least 100-1000:1. Unless the prospective study is done it is clear that the real-life settings are not possible to achieve and additional effort is needed to increase the number of control sample.
In the present review of the studies, there is a substantial technical heterogeneity among the studies: various extraction kits, qPCR methods and most importantly normalization. We have just recently published our data showing the differences between various extraction kits and miRNA expression in ascites[31]. Those results need to be taking to account and an independent validation of the primary samples will be needed. As we have recently reviewed[5], use of serum and plasma will probably have an independent effect on recovery and stability of miRNAs. Besides, contamination with various parts of circulating cells (erythrocytes, thrombocytes) may also provide an additional bias and caution in interpreting of miRNA data is necessary. As reviewed above, one of the larges differences among the studies is related to the choice of reference genes for normalization of miRNAs. Here we would like to caution the use of RNU6b for normalization of miRNAs and also probably reduce the use of miR-16 for normalization of circulating miRNAs unless additional studies provide the evidence for the objectivity of measurements.
Taking to account the unique biology of GC, further focus should be made to identify the high-risk patients with the highest risk of GC development. For instance, patients with moderate to severe AG or IM are at increased risk for development of intestinal type of GC. Here, much more effort needs to be done to characterize the miRNA changes not only in GC but also in patients with preneoplastic conditions or lesions. It would be extremely interesting to know the cascade of changes in miRNA expression in subjects with hereditary diffuse GC.
In currently available studies, all GC subtypes are pooled together and most effort is made to identify “all fits one” biomarkers for GC. This is definitely the most desirable way, where all GC patients could be diagnosed with the same biomarkers; however, we need also keep in mind that GC is a heterogenic disease with unique molecular alterations[17]. Since GC shows subtype-unique miRNA alterations, it may be very likely that circulating miRNAs, at least partly, may behave differently between intestinal and diffuse types of GC. With the knowledge of TCGA molecular classification, it would also very exciting to see if different molecular subtypes have characteristic circulating miRNA expression pattern.
High throughput genomic data sequencing showed that conventional miRNAs may differ both in length of the molecule and its’ sequence[122,123]. These variant miRNA-sequences are now being referred to as isoforms of miRNAs or isomiR’s. Up to date, the role of these molecules in GC remains largely unexplored. Nevertheless, increasing number of studies point out important deregulation patterns of isomiR’s in cancer. For instance, in a recent study, we identified 219 deregulated isomiR’s between gastrointestinal stromal tissue and tumor adjacent tissues[124]. To our knowledge, the only study that investigated isomiR’s in GC found that certain isomiR-types preferentially occur in normal gastric tissue but other types prefer GC tissue[122]. Although these data may be available, no published data is available to isomiR’s changes in blood samples of GC patients and may be focus of future studies.
As discussed in above, multiple studies have shown that circulating miRNAs may serve as diagnostic biomarkers for GC; however, the origin of these molecules is not fully clear. Using profiling data from sera and tumor tissues, Sierzega et al[68] identified differences in expression pattern in GC patients; however, miRNA expression data analysis did not support the conclusion that circulating miRNA originate primarily from the tumor tissue. Hence, it remain open if exosomal miRNAs or even certain blood-cells-related miRNAs may be more promising to study[27]. Furthermore, flow-cytometry or microfluidic-based single-cell or cell-type specific sequencing analysis may provide more specific transcription patterns with higher diagnostic and prognostic value[125]. Therefore, additional studies employing single-cell based analysis from blood samples of GC patients are needed in order to identify more sensitive and specific biomarkers.
During the past 7 years, large amount of data has been gathered to support the need for intensive research on miRNA-based biomarkers in GC. In this review, we systematically summarized the data to miRNAs as non-invasive diagnostic biomarkers in GC. Meticulous analysis of the published work revealed relatively high level of heterogeneity not only in methodological and technical aspects, but also in reporting quality of the studies. At present, no miRNA-based biomarkers are ready to be implemented for GC-screening other than in research studies; however, the provided data clearly highlight the potential of miRNAs as diagnostic biomarkers and also highlight the need for the further improvement. We hope that this first of its kind comprehensive review with critical points may lead to a “next wave” of “second generation” comprehensive studies that take into account not only technical aspects of miRNA research but also unique aspects of GC biology.
Manuscript source: Unsolicited manuscript
Specialty type: Gastroenterology and hepatology
Country of origin: Germany
Peer-review report classification
Grade A (Excellent): A, A, A
Grade B (Very good): B
Grade C (Good): C
Grade D (Fair): 0
Grade E (Poor): 0
P- Reviewer: Aurello P, Gurkan A, Kim GH, Lin JY, Yoshiyama H S- Editor: Gong ZM L- Editor: A E- Editor: Yin SY
| 1. | Chen W, Zheng R, Baade PD, Zhang S, Zeng H, Bray F, Jemal A, Yu XQ, He J. Cancer statistics in China, 2015. CA Cancer J Clin. 2016;66:115-132. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 11444] [Cited by in RCA: 13210] [Article Influence: 1467.8] [Reference Citation Analysis (3)] |
| 2. | Siegel RL, Miller KD, Jemal A. Cancer statistics, 2018. CA Cancer J Clin. 2018;68:7-30. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 11573] [Cited by in RCA: 13158] [Article Influence: 1879.7] [Reference Citation Analysis (4)] |
| 3. | Lu J, Getz G, Miska EA, Alvarez-Saavedra E, Lamb J, Peck D, Sweet-Cordero A, Ebert BL, Mak RH, Ferrando AA. MicroRNA expression profiles classify human cancers. Nature. 2005;435:834-838. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 7723] [Cited by in RCA: 7369] [Article Influence: 368.5] [Reference Citation Analysis (0)] |
| 4. | Link A, Kupcinskas J, Wex T, Malfertheiner P. Macro-role of microRNA in gastric cancer. Dig Dis. 2012;30:255-267. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 76] [Cited by in RCA: 78] [Article Influence: 6.0] [Reference Citation Analysis (0)] |
| 5. | Link A, Goel A. MicroRNA in gastrointestinal cancer: a step closer to reality. Adv Clin Chem. 2013;62:221-268. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 23] [Cited by in RCA: 28] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 6. | Sugano K, Tack J, Kuipers EJ, Graham DY, El-Omar EM, Miura S, Haruma K, Asaka M, Uemura N, Malfertheiner P; faculty members of Kyoto Global Consensus Conference. Kyoto global consensus report on Helicobacter pylori gastritis. Gut. 2015;64:1353-1367. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1322] [Cited by in RCA: 1181] [Article Influence: 118.1] [Reference Citation Analysis (0)] |
| 7. | Malfertheiner P, Link A, Selgrad M. Helicobacter pylori: perspectives and time trends. Nat Rev Gastroenterol Hepatol. 2014;11:628-638. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 133] [Cited by in RCA: 120] [Article Influence: 10.9] [Reference Citation Analysis (0)] |
| 8. | Bornschein J, Leja M, Kupcinskas J, Link A, Weaver J, Rugge M, Malfertheiner P. Molecular diagnostics in gastric cancer. Front Biosci (Landmark Ed). 2014;19:312-338. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 24] [Cited by in RCA: 22] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 9. | Chiba T, Marusawa H, Ushijima T. Inflammation-associated cancer development in digestive organs: mechanisms and roles for genetic and epigenetic modulation. Gastroenterology. 2012;143:550-563. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 268] [Cited by in RCA: 297] [Article Influence: 22.8] [Reference Citation Analysis (0)] |
| 10. | Song H, Ekheden IG, Zheng Z, Ericsson J, Nyrén O, Ye W. Incidence of gastric cancer among patients with gastric precancerous lesions: observational cohort study in a low risk Western population. BMJ. 2015;351:h3867. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 182] [Cited by in RCA: 205] [Article Influence: 20.5] [Reference Citation Analysis (0)] |
| 11. | Beardwell I, Holden L, Claydon T. Human resource management: a contemporary approach [Internet]. 2004;. |
| 12. | Atherton JC. The pathogenesis of Helicobacter pylori-induced gastro-duodenal diseases. Annu Rev Pathol. 2006;1:63-96. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 394] [Cited by in RCA: 410] [Article Influence: 22.8] [Reference Citation Analysis (0)] |
| 13. | Basso D, Zambon CF, Letley DP, Stranges A, Marchet A, Rhead JL, Schiavon S, Guariso G, Ceroti M, Nitti D. Clinical relevance of Helicobacter pylori cagA and vacA gene polymorphisms. Gastroenterology. 2008;135:91-99. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 283] [Cited by in RCA: 294] [Article Influence: 17.3] [Reference Citation Analysis (0)] |
| 14. | Link A, Langner C, Schirrmeister W, Habendorf W, Weigt J, Venerito M, Tammer I, Schlüter D, Schlaermann P, Meyer TF. Helicobacter pylori vacA genotype is a predominant determinant of immune response to Helicobacter pylori CagA. World J Gastroenterol. 2017;23:4712-4723. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 29] [Cited by in RCA: 26] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 15. | Malfertheiner P, Megraud F, O’Morain CA, Gisbert JP, Kuipers EJ, Axon AT, Bazzoli F, Gasbarrini A, Atherton J, Graham DY. Management of Helicobacter pylori infection-the Maastricht V/Florence Consensus Report. Gut. 2017;66:6-30. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2220] [Cited by in RCA: 1983] [Article Influence: 247.9] [Reference Citation Analysis (1)] |
| 16. | Lauren P. The two histological main types of gastric carcinoma: diffuse and so-called intestinal-type carcinoma. An attempt at a histo-clinical classification. Acta Pathol Microbiol Scand. 1965;64:31-49. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4011] [Cited by in RCA: 4322] [Article Influence: 149.0] [Reference Citation Analysis (0)] |
| 17. | Cancer Genome Atlas Research Network. Comprehensive molecular characterization of gastric adenocarcinoma. Nature. 2014;513:202-209. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 5015] [Cited by in RCA: 4848] [Article Influence: 440.7] [Reference Citation Analysis (2)] |
| 18. | Shimada H, Noie T, Ohashi M, Oba K, Takahashi Y. Clinical significance of serum tumor markers for gastric cancer: a systematic review of literature by the Task Force of the Japanese Gastric Cancer Association. Gastric Cancer. 2014;17:26-33. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 264] [Cited by in RCA: 367] [Article Influence: 33.4] [Reference Citation Analysis (0)] |
| 19. | Goni E, Venerito M, Schulz C, Weigt J, Langner C, Link A, Malfertheiner P. Influence of laboratory-related and endoscopy-related factors on the assessment of serum pepsinogens and gastrin-17. Eur J Gastroenterol Hepatol. 2017;29:1340-1345. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 7] [Cited by in RCA: 6] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 20. | Zagari RM, Rabitti S, Greenwood DC, Eusebi LH, Vestito A, Bazzoli F. Systematic review with meta-analysis: diagnostic performance of the combination of pepsinogen, gastrin-17 and anti-Helicobacter pylori antibodies serum assays for the diagnosis of atrophic gastritis. Aliment Pharmacol Ther. 2017;46:657-667. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 84] [Cited by in RCA: 133] [Article Influence: 16.6] [Reference Citation Analysis (0)] |
| 21. | Huang YK, Yu JC, Kang WM, Ma ZQ, Ye X, Tian SB, Yan C. Significance of Serum Pepsinogens as a Biomarker for Gastric Cancer and Atrophic Gastritis Screening: A Systematic Review and Meta-Analysis. PLoS One. 2015;10:e0142080. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 82] [Cited by in RCA: 125] [Article Influence: 12.5] [Reference Citation Analysis (0)] |
| 22. | Bornschein J, Selgrad M, Wex T, Kuester D, Malfertheiner P. Serological assessment of gastric mucosal atrophy in gastric cancer. BMC Gastroenterol. 2012;12:10. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 50] [Cited by in RCA: 61] [Article Influence: 4.7] [Reference Citation Analysis (0)] |
| 23. | Lujambio A, Lowe SW. The microcosmos of cancer. Nature. 2012;482:347-355. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 802] [Cited by in RCA: 911] [Article Influence: 70.1] [Reference Citation Analysis (0)] |
| 24. | McLean MH, El-Omar EM. Genetics of gastric cancer. Nat Rev Gastroenterol Hepatol. 2014;11:664-674. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 250] [Cited by in RCA: 296] [Article Influence: 26.9] [Reference Citation Analysis (0)] |
| 25. | Link A, Schirrmeister W, Langner C, Varbanova M, Bornschein J, Wex T, Malfertheiner P. Differential expression of microRNAs in preneoplastic gastric mucosa. Sci Rep. 2015;5:8270. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 40] [Cited by in RCA: 42] [Article Influence: 4.2] [Reference Citation Analysis (0)] |
| 26. | Juzėnas S, Saltenienė V, Kupcinskas J, Link A, Kiudelis G, Jonaitis L, Jarmalaite S, Kupcinskas L, Malfertheiner P, Skieceviciene J. Analysis of Deregulated microRNAs and Their Target Genes in Gastric Cancer. PLoS One. 2015;10:e0132327. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 29] [Cited by in RCA: 38] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 27. | Juzenas S, Venkatesh G, Hübenthal M, Hoeppner MP, Du ZG, Paulsen M, Rosenstiel P, Senger P, Hofmann-Apitius M, Keller A. A comprehensive, cell specific microRNA catalogue of human peripheral blood. Nucleic Acids Res. 2017;45:9290-9301. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 129] [Cited by in RCA: 136] [Article Influence: 17.0] [Reference Citation Analysis (0)] |
| 28. | Link A, Balaguer F, Shen Y, Nagasaka T, Lozano JJ, Boland CR, Goel A. Fecal MicroRNAs as novel biomarkers for colon cancer screening. Cancer Epidemiol Biomarkers Prev. 2010;19:1766-1774. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 240] [Cited by in RCA: 251] [Article Influence: 16.7] [Reference Citation Analysis (0)] |
| 29. | Link A, Becker V, Goel A, Wex T, Malfertheiner P. Feasibility of fecal microRNAs as novel biomarkers for pancreatic cancer. PLoS One. 2012;7:e42933. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 66] [Cited by in RCA: 68] [Article Influence: 5.2] [Reference Citation Analysis (0)] |
| 30. | Yu L, Todd NW, Xing L, Xie Y, Zhang H, Liu Z, Fang H, Zhang J, Katz RL, Jiang F. Early detection of lung adenocarcinoma in sputum by a panel of microRNA markers. Int J Cancer. 2010;127:2870-2878. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 260] [Cited by in RCA: 287] [Article Influence: 20.5] [Reference Citation Analysis (0)] |
| 31. | Schindler P, Kupcinskas J, Juzenas S, Skieceviciene J, Salteniene V, Schulz C, Weigt J, Malfertheiner P, Link A. Expression of microRNAs in the ascites of patients with peritoneal carcinomatosis and peritonitis. Cancer Cytopathol. 2018;126:353-363. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 17] [Cited by in RCA: 16] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 32. | Weber JA, Baxter DH, Zhang S, Huang DY, Huang KH, Lee MJ, Galas DJ, Wang K. The microRNA spectrum in 12 body fluids. Clin Chem. 2010;56:1733-1741. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1810] [Cited by in RCA: 2084] [Article Influence: 138.9] [Reference Citation Analysis (0)] |
| 33. | Ueda T, Volinia S, Okumura H, Shimizu M, Taccioli C, Rossi S, Alder H, Liu CG, Oue N, Yasui W. Relation between microRNA expression and progression and prognosis of gastric cancer: a microRNA expression analysis. Lancet Oncol. 2010;11:136-146. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 644] [Cited by in RCA: 676] [Article Influence: 45.1] [Reference Citation Analysis (0)] |
| 34. | Steponaitiene R, Kupcinskas J, Langner C, Balaguer F, Venclauskas L, Pauzas H, Tamelis A, Skieceviciene J, Kupcinskas L, Malfertheiner P. Epigenetic silencing of miR-137 is a frequent event in gastric carcinogenesis. Mol Carcinog. 2016;55:376-386. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 45] [Cited by in RCA: 52] [Article Influence: 5.8] [Reference Citation Analysis (0)] |
| 35. | Han TS, Hur K, Xu G, Choi B, Okugawa Y, Toiyama Y, Oshima H, Oshima M, Lee HJ, Kim VN. MicroRNA-29c mediates initiation of gastric carcinogenesis by directly targeting ITGB1. Gut. 2015;64:203-214. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 102] [Cited by in RCA: 116] [Article Influence: 11.6] [Reference Citation Analysis (0)] |
| 36. | Sousa JF, Nam KT, Petersen CP, Lee HJ, Yang HK, Kim WH, Goldenring JR. miR-30-HNF4γ and miR-194-NR2F2 regulatory networks contribute to the upregulation of metaplasia markers in the stomach. Gut. 2016;65:914-924. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 39] [Cited by in RCA: 47] [Article Influence: 5.2] [Reference Citation Analysis (0)] |
| 37. | Mishra PJ, Bertino JR. MicroRNA polymorphisms: the future of pharmacogenomics, molecular epidemiology and individualized medicine. Pharmacogenomics. 2009;10:399-416. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 207] [Cited by in RCA: 202] [Article Influence: 12.6] [Reference Citation Analysis (0)] |
| 38. | Liu C, Rennie WA, Carmack CS, Kanoria S, Cheng J, Lu J, Ding Y. Effects of genetic variations on microRNA: target interactions. Nucleic Acids Res. 2014;42:9543-9552. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 36] [Cited by in RCA: 42] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 39. | Kupcinskas J, Bruzaite I, Juzenas S, Gyvyte U, Jonaitis L, Kiudelis G, Skieceviciene J, Leja M, Pauzas H, Tamelis A. Lack of association between miR-27a, miR-146a, miR-196a-2, miR-492 and miR-608 gene polymorphisms and colorectal cancer. Sci Rep. 2014;4:5993. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 53] [Cited by in RCA: 54] [Article Influence: 4.9] [Reference Citation Analysis (0)] |
| 40. | Kupcinskas J, Wex T, Link A, Leja M, Bruzaite I, Steponaitiene R, Juzenas S, Gyvyte U, Ivanauskas A, Ancans G. Gene polymorphisms of micrornas in Helicobacter pylori-induced high risk atrophic gastritis and gastric cancer. PLoS One. 2014;9:e87467. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 57] [Cited by in RCA: 58] [Article Influence: 5.3] [Reference Citation Analysis (0)] |
| 41. | Petkevicius V, Salteniene V, Juzenas S, Wex T, Link A, Leja M, Steponaitiene R, Skieceviciene J, Kupcinskas L, Jonaitis L. Polymorphisms of microRNA target genes IL12B, INSR, CCND1 and IL10 in gastric cancer. World J Gastroenterol. 2017;23:3480-3487. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 18] [Cited by in RCA: 18] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 42. | Fu B, Song P, Lu M, Wang B, Zhao Q. The association between miR-146a gene rs2910164 polymorphism and gastric cancer risk: a meta-analysis. Biomed Pharmacother. 2014;68:923-928. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 12] [Cited by in RCA: 14] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 43. | Lawrie CH, Gal S, Dunlop HM, Pushkaran B, Liggins AP, Pulford K, Banham AH, Pezzella F, Boultwood J, Wainscoat JS. Detection of elevated levels of tumour-associated microRNAs in serum of patients with diffuse large B-cell lymphoma. Br J Haematol. 2008;141:672-675. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1258] [Cited by in RCA: 1333] [Article Influence: 78.4] [Reference Citation Analysis (0)] |
| 44. | Mitchell PS, Parkin RK, Kroh EM, Fritz BR, Wyman SK, Pogosova-Agadjanyan EL, Peterson A, Noteboom J, O’Briant KC, Allen A. Circulating microRNAs as stable blood-based markers for cancer detection. Proc Natl Acad Sci U S A. 2008;105:10513-10518. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 5636] [Cited by in RCA: 6311] [Article Influence: 371.2] [Reference Citation Analysis (0)] |
| 45. | Chen X, Ba Y, Ma L, Cai X, Yin Y, Wang K, Guo J, Zhang Y, Chen J, Guo X. Characterization of microRNAs in serum: a novel class of biomarkers for diagnosis of cancer and other diseases. Cell Res. 2008;18:997-1006. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3218] [Cited by in RCA: 3553] [Article Influence: 209.0] [Reference Citation Analysis (0)] |
| 46. | Tokuhisa M, Ichikawa Y, Kosaka N, Ochiya T, Yashiro M, Hirakawa K, Kosaka T, Makino H, Akiyama H, Kunisaki C. Exosomal miRNAs from Peritoneum Lavage Fluid as Potential Prognostic Biomarkers of Peritoneal Metastasis in Gastric Cancer. PLoS One. 2015;10:e0130472. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 115] [Cited by in RCA: 140] [Article Influence: 14.0] [Reference Citation Analysis (0)] |
| 47. | Qiu X, Zhang J, Shi W, Liu S, Kang M, Chu H, Wu D, Tong N, Gong W, Tao G. Circulating MicroRNA-26a in Plasma and Its Potential Diagnostic Value in Gastric Cancer. PLoS One. 2016;11:e0151345. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 25] [Cited by in RCA: 35] [Article Influence: 3.9] [Reference Citation Analysis (0)] |
| 48. | Song J, Bai Z, Han W, Zhang J, Meng H, Bi J, Ma X, Han S, Zhang Z. Identification of suitable reference genes for qPCR analysis of serum microRNA in gastric cancer patients. Dig Dis Sci. 2012;57:897-904. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 204] [Cited by in RCA: 223] [Article Influence: 17.2] [Reference Citation Analysis (0)] |
| 49. | Peng WZ, Ma R, Wang F, Yu J, Liu ZB. Role of miR-191/425 cluster in tumorigenesis and diagnosis of gastric cancer. Int J Mol Sci. 2014;15:4031-4048. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 42] [Cited by in RCA: 56] [Article Influence: 5.1] [Reference Citation Analysis (0)] |
| 50. | Shiotani A, Murao T, Kimura Y, Matsumoto H, Kamada T, Kusunoki H, Inoue K, Uedo N, Iishi H, Haruma K. Identification of serum miRNAs as novel non-invasive biomarkers for detection of high risk for early gastric cancer. Br J Cancer. 2013;109:2323-2330. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 69] [Cited by in RCA: 78] [Article Influence: 6.5] [Reference Citation Analysis (0)] |
| 51. | Zhu C, Ren C, Han J, Ding Y, Du J, Dai N, Dai J, Ma H, Hu Z, Shen H. A five-microRNA panel in plasma was identified as potential biomarker for early detection of gastric cancer. Br J Cancer. 2014;110:2291-2299. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 159] [Cited by in RCA: 187] [Article Influence: 17.0] [Reference Citation Analysis (0)] |
| 52. | Wang H, Wang L, Wu Z, Sun R, Jin H, Ma J, Liu L, Ling R, Yi J, Wang L. Three dysregulated microRNAs in serum as novel biomarkers for gastric cancer screening. Med Oncol. 2014;31:298. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 48] [Cited by in RCA: 56] [Article Influence: 5.1] [Reference Citation Analysis (0)] |
| 53. | Zhang J, Song Y, Zhang C, Zhi X, Fu H, Ma Y, Chen Y, Pan F, Wang K, Ni J. Circulating MiR-16-5p and MiR-19b-3p as Two Novel Potential Biomarkers to Indicate Progression of Gastric Cancer. Theranostics. 2015;5:733-745. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 106] [Cited by in RCA: 122] [Article Influence: 12.2] [Reference Citation Analysis (0)] |
| 54. | Konishi H, Ichikawa D, Komatsu S, Shiozaki A, Tsujiura M, Takeshita H, Morimura R, Nagata H, Arita T, Kawaguchi T. Detection of gastric cancer-associated microRNAs on microRNA microarray comparing pre- and post-operative plasma. Br J Cancer. 2012;106:740-747. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 136] [Cited by in RCA: 160] [Article Influence: 12.3] [Reference Citation Analysis (0)] |
| 55. | Liu X, Kwong A, Sihoe A, Chu KM. Plasma miR-940 may serve as a novel biomarker for gastric cancer. Tumour Biol. 2016;37:3589-3597. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 24] [Cited by in RCA: 31] [Article Influence: 3.1] [Reference Citation Analysis (0)] |
| 56. | Tsujiura M, Ichikawa D, Komatsu S, Shiozaki A, Takeshita H, Kosuga T, Konishi H, Morimura R, Deguchi K, Fujiwara H. Circulating microRNAs in plasma of patients with gastric cancers. Br J Cancer. 2010;102:1174-1179. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 449] [Cited by in RCA: 509] [Article Influence: 33.9] [Reference Citation Analysis (0)] |
| 57. | Wang WN, Chen Y, Zhang YD, Hu TH. The regulatory mechanism of CCR7 gene expression and its involvement in the metastasis and progression of gastric cancer. Tumour Biol. 2013;34:1865-1871. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 25] [Cited by in RCA: 28] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 58. | Li C, Li JF, Cai Q, Qiu QQ, Yan M, Liu BY, Zhu ZG. miRNA-199a-3p in plasma as a potential diagnostic biomarker for gastric cancer. Ann Surg Oncol. 2013;20 Suppl 3:S397-S405. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 39] [Cited by in RCA: 51] [Article Influence: 3.9] [Reference Citation Analysis (0)] |
| 59. | Liu WJ, Xu Q, Sun LP, Dong QG, He CY, Yuan Y. Expression of serum let-7c, let-7i, and let-7f microRNA with its target gene, pepsinogen C, in gastric cancer and precancerous disease. Tumour Biol. 2015;36:3337-3343. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 24] [Cited by in RCA: 27] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 60. | Song MY, Pan KF, Su HJ, Zhang L, Ma JL, Li JY, Yuasa Y, Kang D, Kim YS, You WC. Identification of serum microRNAs as novel non-invasive biomarkers for early detection of gastric cancer. PLoS One. 2012;7:e33608. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 141] [Cited by in RCA: 159] [Article Influence: 12.2] [Reference Citation Analysis (0)] |
| 61. | Huang S, Wang J, Li J, Luo Q, Zhao M, Zheng L, Dong X, Chen C, Che Y, Liu P. Serum microRNA expression profile as a diagnostic panel for gastric cancer. Jpn J Clin Oncol. 2016;46:811-818. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 11] [Cited by in RCA: 18] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 62. | Liu R, Zhang C, Hu Z, Li G, Wang C, Yang C, Huang D, Chen X, Zhang H, Zhuang R. A five-microRNA signature identified from genome-wide serum microRNA expression profiling serves as a fingerprint for gastric cancer diagnosis. Eur J Cancer. 2011;47:784-791. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 308] [Cited by in RCA: 328] [Article Influence: 21.9] [Reference Citation Analysis (0)] |
| 63. | Cai H, Yuan Y, Hao YF, Guo TK, Wei X, Zhang YM. Plasma microRNAs serve as novel potential biomarkers for early detection of gastric cancer. Med Oncol. 2013;30:452. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 66] [Cited by in RCA: 79] [Article Influence: 6.6] [Reference Citation Analysis (0)] |
| 64. | Treece AL, Duncan DL, Tang W, Elmore S, Morgan DR, Dominguez RL, Speck O, Meyers MO, Gulley ML. Gastric adenocarcinoma microRNA profiles in fixed tissue and in plasma reveal cancer-associated and Epstein-Barr virus-related expression patterns. Lab Invest. 2016;96:661-671. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 37] [Cited by in RCA: 40] [Article Influence: 4.4] [Reference Citation Analysis (0)] |
| 65. | Oze I, Shimada S, Nagasaki H, Akiyama Y, Watanabe M, Yatabe Y, Matsuo K, Yuasa Y. Plasma microRNA-103, microRNA-107, and microRNA-194 levels are not biomarkers for human diffuse gastric cancer. J Cancer Res Clin Oncol. 2017;143:551-554. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 8] [Cited by in RCA: 9] [Article Influence: 1.1] [Reference Citation Analysis (0)] |
| 66. | Zhou H, Guo JM, Lou YR, Zhang XJ, Zhong FD, Jiang Z, Cheng J, Xiao BX. Detection of circulating tumor cells in peripheral blood from patients with gastric cancer using microRNA as a marker. J Mol Med (Berl). 2010;88:709-717. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 109] [Cited by in RCA: 115] [Article Influence: 7.7] [Reference Citation Analysis (0)] |
| 67. | Yuan R, Wang G, Xu Z, Zhao H, Chen H, Han Y, Wang B, Zhou J, Hu H, Guo Z. Up-regulated Circulating miR-106a by DNA Methylation Promised a Potential Diagnostic and Prognostic Marker for Gastric Cancer. Anticancer Agents Med Chem. 2016;16:1093-1100. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 24] [Cited by in RCA: 27] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 68. | Sierzega M, Kaczor M, Kolodziejczyk P, Kulig J, Sanak M, Richter P. Evaluation of serum microRNA biomarkers for gastric cancer based on blood and tissue pools profiling: the importance of miR-21 and miR-331. Br J Cancer. 2017;117:266-273. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 64] [Cited by in RCA: 81] [Article Influence: 10.1] [Reference Citation Analysis (0)] |
| 69. | Wang N, Wang L, Yang Y, Gong L, Xiao B, Liu X. A serum exosomal microRNA panel as a potential biomarker test for gastric cancer. Biochem Biophys Res Commun. 2017;493:1322-1328. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 70] [Cited by in RCA: 101] [Article Influence: 12.6] [Reference Citation Analysis (0)] |
| 70. | Kim SY, Jeon TY, Choi CI, Kim DH, Kim DH, Kim GH, Ryu DY, Lee BE, Kim HH. Validation of circulating miRNA biomarkers for predicting lymph node metastasis in gastric cancer. J Mol Diagn. 2013;15:661-669. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 67] [Cited by in RCA: 66] [Article Influence: 5.5] [Reference Citation Analysis (0)] |
| 71. | Zhang R, Wang W, Li F, Zhang H, Liu J. MicroRNA-106b~25 expressions in tumor tissues and plasma of patients with gastric cancers. Med Oncol. 2014;31:243. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 33] [Cited by in RCA: 38] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 72. | Zeng Q, Jin C, Chen W, Xia F, Wang Q, Fan F, Du J, Guo Y, Lin C, Yang K. Downregulation of serum miR-17 and miR-106b levels in gastric cancer and benign gastric diseases. Chin J Cancer Res. 2014;26:711-716. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 19] [Reference Citation Analysis (0)] |
| 73. | Li F, Guo Y, Liu J, Zhang R. The significance of elevated plasma expression of microRNA 106b~25 clusters in gastric cancer. PLoS One. 2017;12:e0178427. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 14] [Cited by in RCA: 19] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 74. | Ayremlou N, Mozdarani H, Mowla SJ, Delavari A. Increased levels of serum and tissue miR-107 in human gastric cancer: Correlation with tumor hypoxia. Cancer Biomark. 2015;15:851-860. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 23] [Cited by in RCA: 28] [Article Influence: 3.1] [Reference Citation Analysis (0)] |
| 75. | Zhang M, Wang X, Li W, Cui Y. miR-107 and miR-25 simultaneously target LATS2 and regulate proliferation and invasion of gastric adenocarcinoma (GAC) cells. Biochem Biophys Res Commun. 2015;460:806-812. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 30] [Cited by in RCA: 40] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 76. | Huang Z, Zhu D, Wu L, He M, Zhou X, Zhang L, Zhang H, Wang W, Zhu J, Cheng W. Six Serum-Based miRNAs as Potential Diagnostic Biomarkers for Gastric Cancer. Cancer Epidemiol Biomarkers Prev. 2017;26:188-196. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 112] [Cited by in RCA: 110] [Article Influence: 13.8] [Reference Citation Analysis (0)] |
| 77. | Chen Q, Ge X, Zhang Y, Xia H, Yuan D, Tang Q, Chen L, Pang X, Leng W, Bi F. Plasma miR-122 and miR-192 as potential novel biomarkers for the early detection of distant metastasis of gastric cancer. Oncol Rep. 2014;31:1863-1870. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 72] [Cited by in RCA: 78] [Article Influence: 7.1] [Reference Citation Analysis (0)] |
| 78. | Zhang ZZ, Wang CJ, Niu L, Xu J, Wang M, Cao H, Hu B. Analysis of plasma MicroRNAs to identifying early diagnostic molecule for gastric cancer. Int J Clin Exp Med. 2015;8:3700-3706. [PubMed] |
| 79. | Jiang H, Yu WW, Wang LL, Peng Y. miR-130a acts as a potential diagnostic biomarker and promotes gastric cancer migration, invasion and proliferation by targeting RUNX3. Oncol Rep. 2015;34:1153-1161. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 57] [Cited by in RCA: 53] [Article Influence: 5.3] [Reference Citation Analysis (0)] |
| 80. | Chen S, Zhu J, Yu F, Tian Y, Ma S, Liu X. Combination of miRNA and RNA functions as potential biomarkers for gastric cancer. Tumour Biol. 2015;36:9909-9918. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 14] [Cited by in RCA: 19] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 81. | Shin VY, Ng EK, Chan VW, Kwong A, Chu KM. A three-miRNA signature as promising non-invasive diagnostic marker for gastric cancer. Mol Cancer. 2015;14:202. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 65] [Cited by in RCA: 87] [Article Influence: 8.7] [Reference Citation Analysis (0)] |
| 82. | Jiang X, Wang W, Yang Y, Du L, Yang X, Wang L, Zheng G, Duan W, Wang R, Zhang X. Identification of circulating microRNA signatures as potential noninvasive biomarkers for prediction and prognosis of lymph node metastasis in gastric cancer. Oncotarget. 2017;65132-65142. [RCA] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 20] [Cited by in RCA: 22] [Article Influence: 2.8] [Reference Citation Analysis (1)] |
| 83. | Liu S, Suo J, Wang C, Sun X, Wang D, He L, Zhang Y, Li W. Prognostic significance of low miR-144 expression in gastric cancer. Cancer Biomark. 2017;20:547-552. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 24] [Cited by in RCA: 30] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 84. | Tang G, Shen X, Lv K, Wu Y, Bi J, Shen Q. Different normalization strategies might cause inconsistent variation in circulating microRNAs in patients with hepatocellular carcinoma. Med Sci Monit. 2015;21:617-624. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 39] [Cited by in RCA: 42] [Article Influence: 4.2] [Reference Citation Analysis (0)] |
| 85. | Park JL, Kim M, Song KS, Kim SY, Kim YS. Cell-Free miR-27a, a Potential Diagnostic and Prognostic Biomarker for Gastric Cancer. Genomics Inform. 2015;13:70-75. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 15] [Cited by in RCA: 23] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 86. | Zhao C, Li Y, Chen G, Wang F, Shen Z, Zhou R. Overexpression of miR-15b-5p promotes gastric cancer metastasis by regulating PAQR3. Oncol Rep. 2017;38:352-358. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 34] [Cited by in RCA: 41] [Article Influence: 5.1] [Reference Citation Analysis (0)] |
| 87. | Cui MH, Hou XL, Lei XY, Mu FH, Yang GB, Yue L, Fu Y, Yi GX. Upregulation of microRNA 181c expression in gastric cancer tissues and plasma. Asian Pac J Cancer Prev. 2013;14:3063-3066. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 17] [Cited by in RCA: 16] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 88. | Zhou X, Zhu W, Li H, Wen W, Cheng W, Wang F, Wu Y, Qi L, Fan Y, Chen Y. Diagnostic value of a plasma microRNA signature in gastric cancer: a microRNA expression analysis. Sci Rep. 2015;5:11251. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 97] [Cited by in RCA: 110] [Article Influence: 11.0] [Reference Citation Analysis (0)] |
| 89. | Liu H, Zhu L, Liu B, Yang L, Meng X, Zhang W, Ma Y, Xiao H. Genome-wide microRNA profiles identify miR-378 as a serum biomarker for early detection of gastric cancer. Cancer Lett. 2012;316:196-203. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 185] [Cited by in RCA: 208] [Article Influence: 14.9] [Reference Citation Analysis (0)] |
| 90. | Tsujiura M, Komatsu S, Ichikawa D, Shiozaki A, Konishi H, Takeshita H, Moriumura R, Nagata H, Kawaguchi T, Hirajima S. Circulating miR-18a in plasma contributes to cancer detection and monitoring in patients with gastric cancer. Gastric Cancer. 2015;18:271-279. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 57] [Cited by in RCA: 70] [Article Influence: 7.0] [Reference Citation Analysis (0)] |
| 91. | Shen YH, Xie ZB, Yue AM, Wei QD, Zhao HF, Yin HD, Mai W, Zhong XG, Huang SR. Expression level of microRNA-195 in the serum of patients with gastric cancer and its relationship with the clinicopathological staging of the cancer. Eur Rev Med Pharmacol Sci. 2016;20:1283-1287. [PubMed] |
| 92. | Gorur A, Balci Fidanci S, Dogruer Unal N, Ayaz L, Akbayir S, Yildirim Yaroglu H, Dirlik M, Serin MS, Tamer L. Determination of plasma microRNA for early detection of gastric cancer. Mol Biol Rep. 2013;40:2091-2096. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 49] [Cited by in RCA: 60] [Article Influence: 4.6] [Reference Citation Analysis (0)] |
| 93. | Tsai MM, Wang CS, Tsai CY, Huang CG, Lee KF, Huang HW, Lin YH, Chi HC, Kuo LM, Lu PH. Circulating microRNA-196a/b are novel biomarkers associated with metastatic gastric cancer. Eur J Cancer. 2016;64:137-148. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 39] [Cited by in RCA: 51] [Article Influence: 5.7] [Reference Citation Analysis (1)] |
| 94. | Li C, Li JF, Cai Q, Qiu QQ, Yan M, Liu BY, Zhu ZG. MiRNA-199a-3p: A potential circulating diagnostic biomarker for early gastric cancer. J Surg Oncol. 2013;108:89-92. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 59] [Cited by in RCA: 70] [Article Influence: 5.8] [Reference Citation Analysis (0)] |
| 95. | Valladares-Ayerbes M, Reboredo M, Medina-Villaamil V, Iglesias-Díaz P, Lorenzo-Patiño MJ, Haz M, Santamarina I, Blanco M, Fernández-Tajes J, Quindós M. Circulating miR-200c as a diagnostic and prognostic biomarker for gastric cancer. J Transl Med. 2012;10:186. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 104] [Cited by in RCA: 123] [Article Influence: 9.5] [Reference Citation Analysis (0)] |
| 96. | Zhang HP, Sun FB, Li SJ. Serum miR-200c expression level as a prognostic biomarker for gastric cancer. Genet Mol Res. 2015;14:15913-15920. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 16] [Cited by in RCA: 18] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 97. | Imaoka H, Toiyama Y, Okigami M, Yasuda H, Saigusa S, Ohi M, Tanaka K, Inoue Y, Mohri Y, Kusunoki M. Circulating microRNA-203 predicts metastases, early recurrence, and poor prognosis in human gastric cancer. Gastric Cancer. 2016;19:744-753. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 57] [Cited by in RCA: 69] [Article Influence: 7.7] [Reference Citation Analysis (0)] |
| 98. | Chen X, Liu XS, Liu HY, Lu YY, Li Y. Reduced expression of serum miR-204 predicts poor prognosis of gastric cancer. Genet Mol Res. 2016;15. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 6] [Cited by in RCA: 17] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 99. | Hou CG, Luo XY, Li G. Diagnostic and Prognostic Value of Serum MicroRNA-206 in Patients with Gastric Cancer. Cell Physiol Biochem. 2016;39:1512-1520. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 22] [Cited by in RCA: 29] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 100. | Du Y, Zhu M, Zhou X, Huang Z, Zhu J, Xu J, Cheng G, Shu Y, Liu P, Zhu W. miR-20a enhances cisplatin resistance of human gastric cancer cell line by targeting NFKBIB. Tumour Biol. 2016;37:1261-1269. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 34] [Cited by in RCA: 42] [Article Influence: 4.2] [Reference Citation Analysis (0)] |
| 101. | Zhu M, Zhou X, Du Y, Huang Z, Zhu J, Xu J, Cheng G, Shu Y, Liu P, Zhu W. miR-20a induces cisplatin resistance of a human gastric cancer cell line via targeting CYLD. Mol Med Rep. 2016;14:1742-1750. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 40] [Cited by in RCA: 53] [Article Influence: 5.9] [Reference Citation Analysis (0)] |
| 102. | Yang R, Fu Y, Zeng Y, Xiang M, Yin Y, Li L, Xu H, Zhong J, Zeng X. Serum miR-20a is a promising biomarker for gastric cancer. Biomed Rep. 2017;6:429-434. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 35] [Cited by in RCA: 46] [Article Influence: 5.8] [Reference Citation Analysis (0)] |
| 103. | Zheng Y, Cui L, Sun W, Zhou H, Yuan X, Huo M, Chen J, Lou Y, Guo J. MicroRNA-21 is a new marker of circulating tumor cells in gastric cancer patients. Cancer Biomark. 2011;10:71-77. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 51] [Cited by in RCA: 62] [Article Influence: 4.8] [Reference Citation Analysis (0)] |
| 104. | Li BS, Zhao YL, Guo G, Li W, Zhu ED, Luo X, Mao XH, Zou QM, Yu PW, Zuo QF. Plasma microRNAs, miR-223, miR-21 and miR-218, as novel potential biomarkers for gastric cancer detection. PLoS One. 2012;7:e41629. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 152] [Cited by in RCA: 188] [Article Influence: 14.5] [Reference Citation Analysis (0)] |
| 105. | Wang B, Zhang Q. The expression and clinical significance of circulating microRNA-21 in serum of five solid tumors. J Cancer Res Clin Oncol. 2012;138:1659-1666. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 173] [Cited by in RCA: 179] [Article Influence: 13.8] [Reference Citation Analysis (0)] |
| 106. | Wu J, Li G, Wang Z, Yao Y, Chen R, Pu X, Wang J. Circulating MicroRNA-21 Is a Potential Diagnostic Biomarker in Gastric Cancer. Dis Markers. 2015;2015:435656. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 46] [Cited by in RCA: 75] [Article Influence: 7.5] [Reference Citation Analysis (0)] |
| 107. | Xin SY, Feng XS, Zhou LQ, Sun JJ, Gao XL, Yao GL. Reduced expression of circulating microRNA-218 in gastric cancer and correlation with tumor invasion and prognosis. World J Gastroenterol. 2014;20:6906-6911. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 35] [Cited by in RCA: 37] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 108. | Fu Z, Qian F, Yang X, Jiang H, Chen Y, Liu S. Circulating miR-222 in plasma and its potential diagnostic and prognostic value in gastric cancer. Med Oncol. 2014;31:164. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 40] [Cited by in RCA: 53] [Article Influence: 4.8] [Reference Citation Analysis (0)] |
| 109. | Zhou X, Ji G, Chen H, Jin W, Yin C, Zhang G. Clinical role of circulating miR-223 as a novel biomarker in early diagnosis of cancer patients. Int J Clin Exp Med. 2015;8:16890-16898. [PubMed] |
| 110. | Fassan M, Saraggi D, Balsamo L, Realdon S, Scarpa M, Castoro C, Coati I, Salmaso R, Farinati F, Guzzardo V. Early miR-223 Upregulation in Gastroesophageal Carcinogenesis. Am J Clin Pathol. 2017;147:301-308. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 27] [Cited by in RCA: 28] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 111. | Zhuang K, Han K, Tang H, Yin X, Zhang J, Zhang X, Zhang L. Up-Regulation of Plasma miR-23b is Associated with Poor Prognosis of Gastric Cancer. Med Sci Monit. 2016;22:356-361. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 17] [Cited by in RCA: 28] [Article Influence: 3.1] [Reference Citation Analysis (0)] |
| 112. | Li BS, Zuo QF, Zhao YL, Xiao B, Zhuang Y, Mao XH, Wu C, Yang SM, Zeng H, Zou QM. MicroRNA-25 promotes gastric cancer migration, invasion and proliferation by directly targeting transducer of ERBB2, 1 and correlates with poor survival. Oncogene. 2015;34:2556-2565. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 93] [Cited by in RCA: 99] [Article Influence: 9.0] [Reference Citation Analysis (0)] |
| 113. | Yan C, Yu J, Liu Y, Kang W, Ma Z, Zhou L. MiR-32 promotes gastric carcinoma tumorigenesis by targeting Kruppel-like factor 4. Biochem Biophys Res Commun. 2015;467:913-920. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 27] [Cited by in RCA: 29] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 114. | Sandoval-Bórquez A, Polakovicova I, Carrasco-Véliz N, Lobos-González L, Riquelme I, Carrasco-Avino G, Bizama C, Norero E, Owen GI, Roa JC. MicroRNA-335-5p is a potential suppressor of metastasis and invasion in gastric cancer. Clin Epigenetics. 2017;9:114. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 47] [Cited by in RCA: 59] [Article Influence: 7.4] [Reference Citation Analysis (0)] |
| 115. | Lo SS, Hung PS, Chen JH, Tu HF, Fang WL, Chen CY, Chen WT, Gong NR, Wu CW. Overexpression of miR-370 and downregulation of its novel target TGFβ-RII contribute to the progression of gastric carcinoma. Oncogene. 2012;31:226-237. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 80] [Cited by in RCA: 93] [Article Influence: 6.6] [Reference Citation Analysis (0)] |
| 116. | Zhang WH, Gui JH, Wang CZ, Chang Q, Xu SP, Cai CH, Li YN, Tian YP, Yan L, Wu B. The identification of miR-375 as a potential biomarker in distal gastric adenocarcinoma. Oncol Res. 2012;20:139-147. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 33] [Cited by in RCA: 36] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 117. | Hung PS, Chen CY, Chen WT, Kuo CY, Fang WL, Huang KH, Chiu PC, Lo SS. miR-376c promotes carcinogenesis and serves as a plasma marker for gastric carcinoma. PLoS One. 2017;12:e0177346. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 24] [Cited by in RCA: 32] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 118. | Zhou H, Xiao B, Zhou F, Deng H, Zhang X, Lou Y, Gong Z, Du C, Guo J. MiR-421 is a functional marker of circulating tumor cells in gastric cancer patients. Biomarkers. 2012;17:104-110. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 43] [Cited by in RCA: 52] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 119. | Zhao G, Xu L, Hui L, Zhao J. Level of circulated microRNA-421 in gastric carcinoma and related mechanisms. Int J Clin Exp Pathol. 2015;8:14252-14256. [PubMed] |
| 120. | Wu J, Li G, Yao Y, Wang Z, Sun W, Wang J. MicroRNA-421 is a new potential diagnosis biomarker with higher sensitivity and specificity than carcinoembryonic antigen and cancer antigen 125 in gastric cancer. Biomarkers. 2015;20:58-63. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 34] [Cited by in RCA: 43] [Article Influence: 3.9] [Reference Citation Analysis (0)] |
| 121. | Shah AA, Leidinger P, Backes C, Keller A, Karpinski P, Sasiadek MM, Blin N, Meese E. A set of specific miRNAs is connected with murine and human gastric cancer. Genes Chromosomes Cancer. 2013;52:237-249. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 9] [Cited by in RCA: 10] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 122. | Li SC, Liao YL, Ho MR, Tsai KW, Lai CH, Lin WC. miRNA arm selection and isomiR distribution in gastric cancer. BMC Genomics. 2012;13 Suppl 1:S13. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 96] [Cited by in RCA: 107] [Article Influence: 8.2] [Reference Citation Analysis (0)] |
| 123. | Telonis AG, Rigoutsos I. Race Disparities in the Contribution of miRNA Isoforms and tRNA-Derived Fragments to Triple-Negative Breast Cancer. Cancer Res. 2018;78:1140-1154. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 65] [Cited by in RCA: 93] [Article Influence: 11.6] [Reference Citation Analysis (0)] |
| 124. | Gyvyte U, Juzenas S, Salteniene V, Kupcinskas J, Poskiene L, Kucinskas L, Jarmalaite S, Stuopelyte K, Steponaitiene R, Hemmrich-Stanisak G. MiRNA profiling of gastrointestinal stromal tumors by next-generation sequencing. Oncotarget. 2017;8:37225-37238. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 30] [Cited by in RCA: 33] [Article Influence: 4.1] [Reference Citation Analysis (0)] |
| 125. | Lawson DA, Bhakta NR, Kessenbrock K, Prummel KD, Yu Y, Takai K, Zhou A, Eyob H, Balakrishnan S, Wang CY. Single-cell analysis reveals a stem-cell program in human metastatic breast cancer cells. Nature. 2015;526:131-135. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 604] [Cited by in RCA: 696] [Article Influence: 69.6] [Reference Citation Analysis (0)] |