Published online Jan 14, 2018. doi: 10.3748/wjg.v24.i2.248
Peer-review started: October 17, 2017
First decision: November 14, 2017
Revised: November 15, 2017
Accepted: November 28, 2017
Article in press: November 28, 2017
Published online: January 14, 2018
To investigate possible effects of IRF5 polymorphisms in the 3’ UTR region of the IFR5 locus on susceptibility to hepatitis B virus (HBV) infection and progression of liver diseases among clinically classified Vietnamese patients.
Four IFR5 SNPs (rs13242262A/T, rs77416878C/T, rs10488630A/G, and rs2280714T/C) were genotyped in clinically classified HBV patients [chronic hepatitis B (CHB). n = 99; liver cirrhosis (LC), n = 131; hepatocellular carcinoma (HCC), n = 149] and in 242 healthy controls by direct sequencing and TaqMan real-time PCR assays.
Comparing patients and controls, no significant association was observed for the four IFR5 variants. However, the alleles rs13242262T and rs10488630G contributed to an increased risk of liver cirrhosis (LC vs CHB: OR = 1.5, 95%CI: 1.1-2.3, adjusted P = 0.04; LC vs CHB: OR = 1.7, 95%CI: 1.1-2.6, adjusted P = 0.019). Haplotype IRF5*TCGT constructed from 4 SNPs was observed frequently in LC compared to CHB patients (OR = 2.1, 95%CI: 1.2-3.3, adjusted P = 0.008). Haplotype IRF5*TCAT occurred rather among CHB patients than in the other HBV patient groups (LC vs CHB: OR = 0.4, 95%CI: 0.2-0.8, adjusted P = 0.03; HCC vs CHB: OR = 0.3, 95%CI: 0.15-0.7, adjusted P = 0.003). The IRF5*TCAT haplotype was also associated with increased levels of ALT, AST and bilirubin.
Our study shows that IFR5 variants may contribute as a host factor in determining the pathogenesis in chronic HBV infections.
Core tip:IFR5 is expressed in immune cells and mediates Toll-like receptor signal transduction, playing a vital role in the induction of antiviral and inflammatory response. So far, multiple IFR5 single nucleotide polymorphisms have been shown to be associated with autoimmune diseases. This study investigated the effects of four IFR5 variants on susceptibility to hepatitis B virus (HBV) infection and liver disease outcomes in HBV infected patients. Two IFR5 variants (rs13242262, rs10488630) and constructed haplotypes (TCGT, TCAT) were associated with clinical outcomes suggesting that IFR5 variants may contribute to determine the pathogenesis of HBV infection.
- Citation: Sy BT, Hoan NX, Tong HV, Meyer CG, Toan NL, Song LH, Bock CT, Velavan TP. Genetic variants of interferon regulatory factor 5 associated with chronic hepatitis B infection. World J Gastroenterol 2018; 24(2): 248-256
- URL: https://www.wjgnet.com/1007-9327/full/v24/i2/248.htm
- DOI: https://dx.doi.org/10.3748/wjg.v24.i2.248
Hepatitis B virus (HBV) infection is a major health concern affecting approximately two billion individuals worldwide. 350 million people are chronically infected, putting them at risk to develop liver cirrhosis (LC) and hepatocellular carcinoma (HCC)[1]. The clinical outcome of HBV infection is heterogeneous and a consequence of the complex interaction between viral and host factors. The host´s genetic background is crucial for the outcome of the disease. Evidence for a host genetic effects are based on a twin study[2] and genome wide association studies (GWASs)[3-5]. GWASs examine possible associations of large number of genetic variants across the entire human genome, taking into account distinct disease phenotypes of HBV infection[6]. Many important candidate genes have been shown to be significantly associated with susceptibility to HBV infection and the progression of HBV-related liver diseases[6-8].
HBV is a noncytopathic virus as observed in a number of asymptomatic HBV carriers who have minimal hepatocellular injury and liver necroinflammation despite high levels of HBV replication[9]. Thus, hepatocellular injury is strongly dependent on the host immune responses[10]. Induction of type I interferons (IFNs) by viruses is crucial for innate immunity, which is primarily controlled by several transcriptional factors, in particular by interferon regulatory factors (IRFs)[11]. The IRF family comprises of nine members (IRF1 to IRF9), which are characterized by two major domains, a highly conserved amino (N)-terminal DNA binding domain and a C-terminal IFR association domain (IAD)[12]. These regions are important in mediating the interaction with transcription co-activators[13]. IRF5, a member of the IRF family, is expressed in B cells and innate immune cells and mediates Toll-like receptor signal transduction, leading to production of several inflammatory cytokines such as interleukin 12 and IFN-α[14-16]. Therefore, IRF5 plays a vital role in the induction of antiviral and inflammatory response [17,18].
So far, multiple IRF5 single nucleotide polymorphisms (SNPs) have been shown to be associated with autoimmune diseases, including systemic lupus erythematosus and rheumatoid arthritis[19,20]. However, there are so far no data available on associations of IRF5 variants with susceptibility to HBV infection and the clinical course of HBV-related liver diseases. This study aims to investigate possible effects of IRF5 polymorphisms on susceptibility to HBV infection and progression of liver diseases among HBV patients in a Vietnamese population.
379 unrelated Vietnamese HBV-infected patients were randomly recruited in a case-control design at the 108 Military Central Hospital and the 103 Military Hospital of the Vietnam Military Medical University, Hanoi, Vietnam. Patients were assigned to subgroups of disease based on clinical manifestations and liver function tests. Subgroups included chronic hepatitis (CHB, n = 99), liver cirrhosis (LC, n = 131) and hepatocellular carcinoma (HCC, n = 149). The diagnostic criteria have previously been described[21]. Based on clinical manifestations and laboratory parameters, patients were assigned to the different clinical subgroups as previously described. Briefly, the CHB patients were characterized based upon clinical syndromes such as fatigue, anorexia, jaundice, hepatomegaly, hard density of the liver, splenomegaly, hyperbilirubinemia, elevated levels of AST and ALT, HBsAg positive for longer than 6 mo. The HBV-related LC patients were characterized as patients infected with HBV (HBsAg positive) showing the clinical manifestations such as anorexia, nausea, vomiting, malaise, weight loss, abdominal distress, jaundice, edema, cutaneous arterial ‘‘Spider’’ angiomas, palma erythema, ascites, shrunken liver, splenomegaly, hyperbilirubinemia, elevated levels of AST and ALT, prolonged serum prothrombin time, and decreased serum albumin. The HBV-related hepatocellular carcinoma patients were characterized as patients infected with chronically HBV (HBsAg positive), abdominal pain, an abdominal mass in the right upper quadrant, blood-tinged ascites, weight loss, anorexia, fatigue, jaundice, prolonged serum prothrombin time, hyperbilirubinemia, elevated levels of AST, ALT and serum a-fetoprotein (AFP), ultrasound showed tumor, liver biopsy and histopathology showing tumor cells. None of the patients were under any antivirals during sampling. None of the patients had a history of alcohol or drug abuse. All participants were confirmed to be negative for anti-HCV and anti-HIV antibodies by ELISA assays. Liver function tests including the assessment of alanine transaminase (ALT), aspartate transaminase (AST), total bilirubin and direct bilirubin, albumin and prothrombin levels were performed using an autoanalyser (AU640 Chemistry Analyzer, Beckman Coulter, CA, United States). 242 blood samples from healthy individuals (HC) were collected from blood banks as the control group. All 242 control individuals were negative for HBsAg, anti-HCV and anti-HIV antibodies. All specimens were frozen at -20 °C until use.
The four IRF5 SNPs rs13242262A/T, rs77416878C/T, rs10488630A/G, and rs2280714G/A located closely at the 3′ downstream regions of the IRF5 locus were selected for this study. Two SNPs (rs13242262, and rs2280714) have been shown to be associated with IRF5 mRNA expression and activation of the interferon α pathway in different world populations[22].
Genomic DNA was isolated from 200 μL of whole blood using a DNA purification kit (Qiagen, Hilden, Germany). The fragments containing the SNPs rs13242262A/T and rs77416878C/T were amplified by PCR using the primer pairs IRF5-F1: 5’-AGG CCT GTG CAG TTC TAC TCC C-3’ and IRF5-R1: 5’-CCT CAC ACT GGC CTG CCT TTA C-3’. PCR amplifications were carried out in 25 μL reaction volumes containing: 1 x PCR buffer, 0.2 mmol/L dNTPs, 1 mmol/L MgCl2, 0.15 mmol/L of each primer, 1 unit of Taq polymerase and 50 ng of genomic DNA. Cycling conditions: denaturation at 95 °C for 5 min, followed by 35 cycles of three-step cycling with denaturation at 94 °C for 40 s, annealing at 61 °C for 40 s, and extension at 72 °C for 45 s and a final extension at 72 °C for 7 min.
PCR products were purified using the Exo-SAP-IT PCR Product Cleanup Reagent (Affymetrix Santa Clara, mmol/L) 5 μL of purified PCR products were used as templates. Sequencing was performed using the BigDye terminator v.1.1 cycle sequencing kit (Applied Biosystems, Foster City, CA, United States) on an ABI 3130XL DNA sequencer according to the manufacturer’s instructions. The polymorphisms were identified by assembling DNA sequences with the reference sequence of the IFR5 gene obtained from the NCBI database (GenBank accession number: NC-00007). In addition, the two SNPs rs10488630A/G, and rs2280714G/A were genotyped using TaqMan® SNP genotyping assays according to the instruction of the manufacturer.
The data were analyzed using R version 3.1.2 (http://www.r-project.org). Permutation tests were used to compare groups for quantitative variables permuted for 1000 iterations. Genotype and haplotype frequencies were analyzed by gene counting and expectation-maximum (EM) algorithms and the significance of deviation from Hardy-Weinberg equilibrium was tested using the random-permutation procedure as implemented in the Arlequin v. 3.5.1.2 software (http://lgb.unige.ch/arlequin). We used a binary logistic regression model adjusted for age and gender to analyze associations of IRF5 variants and haplotypes with HBV-related liver diseases. The false discovery rate correction method was used for multiple comparisons[23] and adjusted P values are given. The level of significance was set at a value of P < 0.05 and all reported P values are two-sided.
The baseline characteristics of the 379 HBV-infected patients from the different subgroups with well-characterized clinical profiles and from the 242 healthy controls are described in Table 1. Most patients and controls were male (81% and 64%). The median age of patients increased according to progression of liver disease; healthy controls were younger than the patients. The levels of ALT, AST and bilirubin were significantly higher in patients with CHB compared to the other subgroups (P < 0.0001). As expected, the albumin and prothrombin levels as well as platelet counts were significantly lower in patients with LC compared to patients without LC (P < 0.001). Alpha-fetoprotein (AFP) levels were higher in HCC patients with or without LC compared to CHB and LC groups (P < 0.0001). Viral loads did not differ significantly between HBV virus subgroups (P > 0.05).
| Characteristics | HC (n = 242) | CHB (n = 99) | LC (n = 131) | HCC (n = 149) | P values |
| Age (yr) | 39 (18-79) | 41 (19-78) | 52 (17-78) | 53 (18-79) | < 0.0001 |
| Gender (Male/Female) | 156/86 | 82/17 | 105/26 | 119/30 | < 0.0001 |
| AST (IU/L) | NR | 219 (17-3732) | 74 (12-720) | 59 (16-513) | < 0.0001 |
| ALT (IU/L) | NR | 158 (12-4593) | 59 (9-1354) | 47 (13-471) | < 0.0001 |
| Total bilirubin (µmol/L) | NR | 46.6 (1.8-795) | 31 (1.2-722) | 17 (2-290) | < 0.0001 |
| Direct bilirubin (µmol/L) | NR | 29.9 (1-512) | 17 (1-450) | 7.1 (1.2-189) | < 0.0001 |
| Albumin (g/L) | NR | 42 (23-48) | 30 (20-47) | 39 (27-49) | < 0.0001 |
| Prothrombin (% of standard) | NR | 85 (50-120) | 47.5 (15-101) | 80 (31-115) | < 0.0001 |
| HBV-DNA (copies/mL) | NA | 1.8 × 105 (4 × 102-8.1 × 106) | 8.3 × 104 (2 × 102-4.1 × 106) | 9.4 × 104 (2.9 × 102-1.0 × 105) | NS |
| Alfa Feto Protein (IU/L) | NR | 4.3 (1.5-300) | 8.6 (1.2-400) | 196 (1.1- 438) | < 0.0001 |
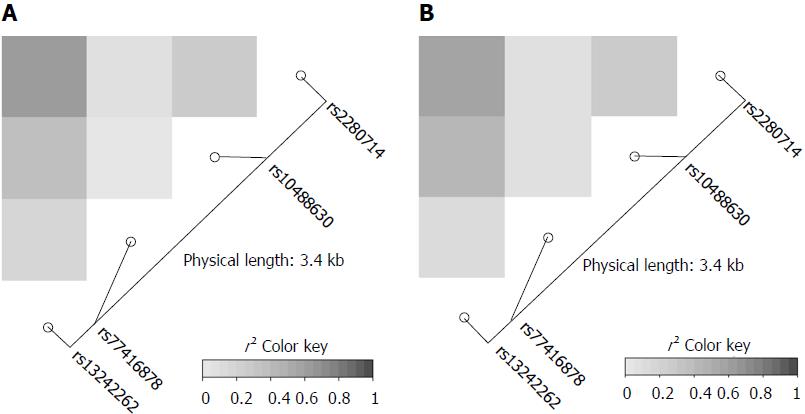
The genotype frequencies of the four IRF5 variants rs13242262A/T, rs10488630A/G, rs77416878C/T, rs2280714T/C in HBV patients and in HCs were in Hardy-Weinberg equilibrium (P > 0.05). Linkage disequilibrium analysis revealed strong allelic combinations between rs13242262 and rs2280714; rs13242262 and rs10488630; rs10488630 and rs2280714 for both HBV infected patients and HCs (Figure 1). Genotype and allele frequencies of the IRF5 SNPs in patients and HCs as well as the comparisons between different subgroups are given in Tables 2 and 3.
| IRF5 variants | HC, n = 242 | CHB, n = 99 | LC, n = 131 | HCC, n = 149 | LC vs CHB | |
| OR (95%CI) | P value | |||||
| rs13242262 A/T | ||||||
| AA | 67 (27.7) | 27 (27.3) | 32 (24.4) | 45 (30.2) | Reference | |
| AT | 119 (49.2) | 59 (59.6) | 65 (49.6) | 77 (51.7) | 1.0 (0.5-2.0) | NS |
| TT | 56 (23.1) | 13 (13.1) | 34 (26.0) | 27 (18.1) | 3.1 (1.2-7.8) | 0.014 |
| Allele | ||||||
| A | 253 (52.3) | 113 (57) | 129 (49.2) | 165 (55.3) | Reference | |
| T | 231 (47.7) | 85 (43) | 133 (50.8) | 133 (44.7) | 1.5 (1.1-2.3) | 0.040 |
| Dominant | ||||||
| AA | 67 (27.7) | 27 (27.3) | 32(24.4) | 45 (30.2) | Reference | |
| AT + TT | 175 (72.3) | 72 (72.7) | 99 (75.6) | 104 (69.8) | 1.3 (0.7-2.6) | NS |
| Recessive | ||||||
| AA + AT | 186 (76.9) | 86 (87) | 97 (74.0) | 122 (81.9) | Reference | |
| TT | 56 (23.1) | 13 (13) | 34 (26.0) | 27 (18.1) | 2.8 (1.3-5.9) | 0.0057 |
| rs10488630 A/G | ||||||
| AA | 115 (47.5) | 58 (58.6) | 59 (45.1) | 73 (49.0) | Reference | |
| AG | 104 (43.0) | 36 (36.4) | 56 (42.7) | 63 (42.3) | 1.6 (0.9-2.9) | 0.100 |
| GG | 23 (9.5) | 5 (5.0) | 16 (12.2) | 13 (8.7) | 3.0 (1.0-9.5) | 0.045 |
| Allele | ||||||
| A | 334 (69) | 152 (76.8) | 174 (66.4) | 209 (72.3) | Reference | |
| G | 150 (31) | 46 (23.2) | 88 (33.6) | 89 (27.7) | 1.7 (1.1-2.6) | 0.019 |
| Dominant | ||||||
| AA | 115 (47.5) | 58 (58.6) | 59 (45.0) | 73 (49) | Reference | |
| AG + GG | 127 (52.5) | 41 (41.4) | 72 (55.0) | 76 (51) | 1.8 (1.0-3.2) | 0.035 |
| Recessive | ||||||
| AA + AG | 219 (90.5) | 94 (94.9) | 115 (87.8) | 136 (91.3) | Reference | |
| GG | 23 (9.5) | 5 (5.1) | 16 (12.2) | 13 (7.8) | 2.4 (0.8-7.2) | 0.100 |
| IRF5 variants | HC,n = 242 | CHB,n = 99 | LC,n = 131 | HCC,n = 149 |
| rs77416878 | ||||
| CC | 192 (79.4) | 79 (79.8) | 98 (74.8) | 115 (77.2) |
| CT | 48 (19.8) | 19 (19.2) | 32 (24.4) | 31 (20.8) |
| TT | 2 (0.8) | 1 (1) | 1 (0.8) | 3 (2) |
| Allele | ||||
| C | 432 (89.3) | 177 (89.4) | 228 (87) | 261 (87.6) |
| T | 52 (10.7) | 21 (10.6) | 34 (13) | 37 (12.4) |
| Dominant | ||||
| CC | 192 (79.4) | 79 (79.8) | 98 (74.8) | 115 (77.2) |
| CT + TT | 50 (20.6) | 20 (20.2) | 33 (24.4) | 34 (22.8) |
| Recessive | ||||
| CC + CT | 240 (99.2) | 98 (99) | 130 (99.2) | 146 (98) |
| TT | 2 (0.8) | 1 (1) | 1 (0.8) | 3 (2) |
| rs2280714 | ||||
| TT | 84 (34.7) | 31 (31.3) | 47 (35.9) | 39 (26.2) |
| TC | 114 (47.1) | 52 (52.5) | 69 (52.7) | 87 (58.4) |
| CC | 44 (18.2) | 16 (16.2) | 15 (11.4) | 23 (15.4) |
| Allele | ||||
| T | 282 (58.3) | 114 (57.6) | 163 (62.2) | 165 (55.4) |
| C | 202 (41.7) | 84 (42.4) | 99 (37.8) | 133 (44.6) |
| Dominant | ||||
| TT | 84 (34.7) | 31 (31.3) | 47 (35.9) | 39 (26.2) |
| TC + TT | 158 (65.3) | 78 (68.7) | 84 (64.1) | 110 (73.8) |
| Recessive | ||||
| TT + TC | 198 (81.8) | 83 (83.8) | 116 (88.6) | 126 (84.6) |
| CC | 44 (18.2) | 16 (16.2) | 15 (11.4) | 23 (15.4) |
Genotype and allele frequencies of the four IRF5 SNPs did not differ between HBV patients or subgroups and controls, indicating that IRF5 SNPs are not associated with HBV infection per se. Among chronic HBV carriers, rs13242262TT and rs10488630GG genotype were significantly more frequent among LC patients compared to CHB patients (rs13242262TT: OR = 3.1, 95%CI: 1.2-7.8, adjusted P = 0.014; rs10488630GG, OR = 3.0, 95%CI: 1.0-9.5, adjusted P = 0.045, Table 2). A similar trend was observed for rs13242262T (OR = 1.5, 95%CI: 1.1-2.3, adjusted P = 0.04) and rs10488630G (OR = 1.7, 95%CI: 1.1-2.6, adjusted P = 0.019; Table 2). For SNPs rs77416878C/T, and rs2280714T/C all comparisons between patient subgroups using binary logistic model adjusted for age and gender did not indicate any significant difference (Table 3). These results show that, of the four SNPs genotyped, the two variants rs13242262 and rs10488630 are associated with liver disease progression.
Haplotypes were constructed based on the four SNPs. Among nine IRF5 haplotypes detected, the five common haplotypes rs13242262/rs77416878/rs10488630/rs2280714 ACAC, TCGT, TCAT, ACAT, and TTAT were observed in both HCs and HBV patients. Their frequencies are summarized in Table 4. We compared the haplotype frequencies between HCs and all HBV patients as well as the disease subgroups (HC vs all HBV; HC vs LC; HC vs CHB; HC vs HCC). The results did not indicate any significant difference (data not shown).
| Haplotypes | HC,n = 484 | CHB,n = 198 | LC,n = 298 | HCC,n = 262 | LC vs CHB | HCC vs CHB | HCC vs LC | |||
| OR (95% CI) | P value | OR (95% CI) | P value | OR (95%CI) | P value | |||||
| ACAC | 196 (40.6) | 83 (41.9) | 98 (37.4) | 132 (44.3) | Reference | Reference | Reference | |||
| TCGT | 141 (29.0) | 37 (18.7) | 86 (32.7) | 81 (27.2) | 2.1 (1.2-3.3) | 0.008 | NS | 0.7 (0.4-1.1) | 0.08 | |
| TCAT | 33 (6.8) | 27 (13.6) | 12 (4.6) | 13 (4.4) | 0.4 (0.2-0.8) | 0.037 | 0.3 (0.15-0.7) | 0.003 | NS | |
| ACAT | 54 (11.2) | 24 (12) | 30 (11.5) | 31 (10.4) | NS | NS | NS | |||
| TTAT | 49 (10.2) | 18 (9.1) | 34 (13) | 33 (11.1) | NS | NS | NS | |||
| ACGC | 2 (0.4) | 1 (0.5) | 0 (0) | 1 (0.3) | NS | NS | NS | |||
| ACGT | 5 (1) | 5 (2.5) | 1 (0.4) | 3 (1) | NS | NS | NS | |||
| TCGC | 2 (0.4) | 0 (0) | 1 (0.4) | 0 (0) | NS | NS | NS | |||
| TTGT | 2 (0.4) | 3 (1.5) | 0 (0) | 4 (1.3) | NS | NS | NS | |||
We further compared haplotype frequencies between the HBV subgroups. Haplotype TCGT was found more frequently among LC compared to CHB patients (LC vs CHB: OR = 2.1, 95%CI: 1.2-3.3, adjusted P = 0.008), indicating that this haplotype may contribute significantly to an increased risk of LC in HBV carriers. However, a contradictory finding was observed for the haplotype TCAT, which was observed significantly more frequent in CHB compared to LC and HCC patients (LC vs CHB: OR = 0.4, 95%CI: 0.2-0.8, adjusted P = 0.037 and HCC vs CHB: OR = 0.3, 95%CI: 0.15-0.7, adjusted P = 0.003). This haplotype appears to partly protect from the risk of the advanced clinical manifestations LC and HCC in chronic HBV carriers. There were no differences in comparisons of the TCGT and TCAT haplotype frequencies between LC and HCC patients. Also, no significant difference was observed when frequencies of other haplotypes were compared (Table 4).
To explore the possible impact of the four SNPs on disease outcomes, the SNP frequencies were correlated with several liver function tests, a cancer marker, and viral loads. No significant associations of IRF5 genotypes with the parameters ALT, AST, total and direct bilirubin, prothrombin, AFP and HBV-DNA loads were observed (adjusted P > 0.05).
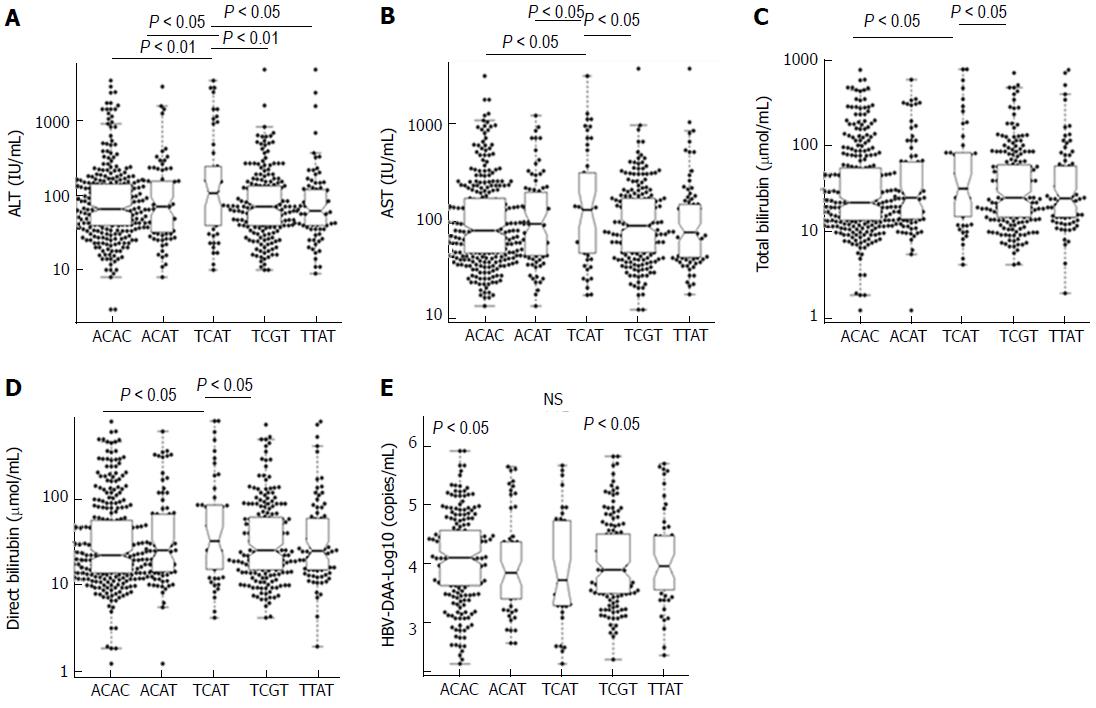
We further examined the association of five common IRF5 haplotypes with clinical outcomes of HBV infection. Patients with haplotype TCAT had higher levels of AST, ALT, total bilirubin and direct bilirubin compared to the other haplotypes (Figure 2). No significant differences of viral loads, prothrombin and AFP levels among five common haplotypes were observed.
IRF5 is a particularly interesting member of the IRF family, which are crucial in the innate immune response with a variety of activities like activation of type I IFN genes, inflammatory cytokines and tumor suppressors[24,25]. Therefore, IRF5 is involved in many conditions, including autoimmune diseases, viral infections and cancers[11,19,20]. In this study, we studied the role of IRF5 polymorphisms in HBV infected patients. IRF5 variants are associated with LC progression in patients with CHB while the constructed haplotypes are associated with LC and HCC progression in CHB patients. In addition, IRF5 variants and their constructed haplotypes are associated with clinical outcomes of HBV infection. For the first time we provide evidence of the functional role of IRF5 in immune response to the clinical outcome of HBV infection.
Host immune factors are crucial to the pathogenesis of HBV infection through genetic and epigenetic modifications and via the effects of cytokines[26]. Interferons are produced by the host in response to certain viral infections in order to inhibit viral replication. Induction of IFN is required for the defense against hepatitis viruses and further progression of related liver disease[27]. IRF5 is a transcriptional factor that can induce type I interferons and, therefore, appears to play an important role in the clinical course of HBV infection. To the best of our knowledge, this study is the first exploratory investigation of IRF5 polymorphisms addressing the clinical outcome of HBV-related liver diseases. Among four SNPs studied here, rs13242262A/T and rs10488630A/G appeared were with liver cirrhosis. In addition, the IRF5 haplotypes TCGT and TCAT are associated with liver cirrhosis in patients with chronic hepatitis B. SNP rs10488631 located in the same region was identified to be associated with primary biliary cirrhosis in populations of European descent[28]. However, this SNP was homogeneous in Asian populations and therefore excluded from analyses in this study. In addition, SNPs rs3807306 and rs4728142 in the IRF5 gene have been implicated as susceptibility loci for primary biliary cirrhosis[29].
The process of liver cirrhosis in HBV infection is a results of the interplay between viral factors and host immune responses through activation of inflammatory cytokines[30]. A recent study has shown that among several IRF5 SNPs, the variants rs13242262, rs2280714 and rs10488630 in the 3’UTR region are associated with increased IRF5 mRNA expression[22]. Studies have indicated that a variety of cytokines are dependent on IRF5[24,31]. Several IRF5-modulated genes (e.g., ISGs and STATs) involved in the type I IFN signaling pathway are significantly over-expressed in response to viral infection[18]. This supports the findings of our study, namely that these SNPs may contribute to progression of HBV-related liver diseases through regulating IRF5 expression and subsequent activation of genes in the type I IFN signaling pathway like ISG15 as seen in our study[7]. Furthermore, although all four studied SNPs were not associated with HCC, the haplotype TCAT contributes to a decreased risk of HCC development in patients with chronic hepatitis B. Data concerning the association between IRF5 and HCC are scarce. Nevertheless, methylation of IRF5 has been suggested to be associated with HCC in a Korean study[32]. The role of IRF5 in the development of HBV-related HCC needs to be explored further.
Although SNPs rs77416878C/T and rs2280714T/C are not associated with HBV-related liver disease and no significant association of all four SNPs studied with clinical parameters, constructed haplotypes are associated with clinical outcomes. Notably, the haplotype TCAT was observed significantly more frequent in CHB compared to LC and HCC patients, suggesting that this haplotype appears to partly protect from the risk of the advanced clinical manifestations LC and HCC in chronic HBV carriers. However, patients with the haplotype TCAT had higher levels of AST, ALT, total bilirubin and direct bilirubin compared to the other haplotypes. In fact, the clinical outcome or clinical progression of liver diseases in HBV infected patients are affected by several factors and are considered as a result of viral-host interaction. Therefore, we believe that haplotype TCAT is an important host factor in HBV infection but this haplotype only may not be a host factor in determining the overall clinical outcome of disease.
Until now, most studies have identified distinct IRF5 haplotypes to be associated with high serum IFN-α activity and with systemic lupus erythematosus[33-36]. In addition, the IRF5 risk haplotype TCC, which contains the risk alleles rs13242262, rs10488631 and rs2280714 are associated with increased IRF5, IFN-α, and IFN-inducible chemokine expression in healthy individuals[22]. However, our study did not assess the relationship of the IRF5 risk haplotypes ACAC, TCAT, TCGT with IRF5 expression, IFN-α activity and other related IFN-α gene. This is one of the study’s limitations; in fact the function of IRF5 in HBV infection needs further investigations. Nevertheless, we assume that the IRF5 risk haplotypes may affect the expression of multiple downstream genes in the IFN-α signaling pathway and certain inflammatory cytokines in HBV infection.
In conclusion, IRF5 variants rs13242262A/T and rs10488630A/G are associated with LC progression in patients with CHB. IRF5 haplotypes appear to influence the outcome of HBV infection. Further studies in this direction will provide insights into a role of IRF5 variants as prognostic markers of HBV-related liver diseases.
Hepatitis B virus (HBV) infection is a major health concern in Vietnam. Investigations were carried out to determine IRF5 polymorphisms in the 3’ UTR region of the IFR5 locus on susceptibility to HBV infection and progression of liver diseases among clinically classified Vietnamese patients.
IRF5 is a particularly interesting member of the IRF family, which are crucial in the innate immune response with a variety of activities like activation of type I IFN genes, inflammatory cytokines and tumor suppressors. There are so far no data available on associations of IRF5 variants with susceptibility to HBV infection and the clinical course of HBV-related liver diseases.
This study aims to investigate possible effects of IRF5 polymorphisms on susceptibility to HBV infection and progression of liver diseases among clinically classified Vietnamese patients.
The four IRF5 SNPs rs13242262A/T, rs77416878C/T, rs10488630A/G, and rs2280714G/A located closely at the 3′ downstream regions of the IRF5 locus were selected for this study. IRF5 variant genotyping was performed by direct sanger sequencing and by application of TaqMan® SNP genotyping assays.
Three hundred seventy-nine unrelated Vietnamese HBV-infected patients were randomly recruited in a case-control design. IRF5 variants are associated with LC progression in patients with CHB while the constructed haplotypes are associated with LC and HCC progression in CHB patients. In addition, IRF5 variants and their constructed haplotypes are associated with clinical outcomes of HBV infection.
Host immune factors are crucial to the pathogenesis of HBV infection. For the first time the authors provide evidence of the functional role of human IRF5 in immune response to the clinical outcome of HBV infection. IRF5 variants rs13242262A/T and rs10488630A/G are associated with LC progression in patients with CHB. IRF5 haplotypes appear to influence the outcome of HBV infection.
Further studies in this direction will provide insights into a role of IRF5 variants as prognostic markers of HBV-related liver diseases.
Manuscript source: Unsolicited manuscript
Specialty type: Gastroenterology and hepatology
Country of origin: Germany
Peer-review report classification
Grade A (Excellent): 0
Grade B (Very good): B
Grade C (Good): C, C
Grade D (Fair): 0
Grade E (Poor): 0
P- Reviewer: Silva LD, Sipos F, Waheed Y S- Editor: Ma YJ L- Editor: A E- Editor: Ma YJ
| 1. | Trépo C, Chan HL, Lok A. Hepatitis B virus infection. Lancet. 2014;384:2053-2063. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1004] [Cited by in F6Publishing: 1041] [Article Influence: 104.1] [Reference Citation Analysis (0)] |
| 2. | Lin TM, Chen CJ, Wu MM, Yang CS, Chen JS, Lin CC, Kwang TY, Hsu ST, Lin SY, Hsu LC. Hepatitis B virus markers in Chinese twins. Anticancer Res. 1989;9:737-741. [PubMed] [Cited in This Article: ] |
| 3. | Hu Z, Liu Y, Zhai X, Dai J, Jin G, Wang L, Zhu L, Yang Y, Liu J, Chu M. New loci associated with chronic hepatitis B virus infection in Han Chinese. Nat Genet. 2013;45:1499-1503. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 111] [Cited by in F6Publishing: 118] [Article Influence: 10.7] [Reference Citation Analysis (0)] |
| 4. | Jiang DK, Sun J, Cao G, Liu Y, Lin D, Gao YZ, Ren WH, Long XD, Zhang H, Ma XP. Genetic variants in STAT4 and HLA-DQ genes confer risk of hepatitis B virus-related hepatocellular carcinoma. Nat Genet. 2013;45:72-75. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 198] [Cited by in F6Publishing: 226] [Article Influence: 18.8] [Reference Citation Analysis (0)] |
| 5. | Li S, Qian J, Yang Y, Zhao W, Dai J, Bei JX, Foo JN, McLaren PJ, Li Z, Yang J. GWAS identifies novel susceptibility loci on 6p21.32 and 21q21.3 for hepatocellular carcinoma in chronic hepatitis B virus carriers. PLoS Genet. 2012;8:e1002791. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 143] [Cited by in F6Publishing: 151] [Article Influence: 12.6] [Reference Citation Analysis (0)] |
| 6. | Thursz M, Yee L, Khakoo S. Understanding the host genetics of chronic hepatitis B and C. Semin Liver Dis. 2011;31:115-127. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 67] [Cited by in F6Publishing: 72] [Article Influence: 5.5] [Reference Citation Analysis (0)] |
| 7. | Hoan NX, Van Tong H, Giang DP, Toan NL, Meyer CG, Bock CT, Kremsner PG, Song LH, Velavan TP. Interferon-stimulated gene 15 in hepatitis B-related liver diseases. Oncotarget. 2016;7:67777-67787. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 7] [Cited by in F6Publishing: 9] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 8. | Hu L, Zhai X, Liu J, Chu M, Pan S, Jiang J, Zhang Y, Wang H, Chen J, Shen H. Genetic variants in human leukocyte antigen/DP-DQ influence both hepatitis B virus clearance and hepatocellular carcinoma development. Hepatology. 2012;55:1426-1431. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 127] [Cited by in F6Publishing: 139] [Article Influence: 11.6] [Reference Citation Analysis (0)] |
| 9. | Chang JJ, Lewin SR. Immunopathogenesis of hepatitis B virus infection. Immunol Cell Biol. 2007;85:16-23. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 140] [Cited by in F6Publishing: 148] [Article Influence: 8.2] [Reference Citation Analysis (0)] |
| 10. | Rehermann B, Nascimbeni M. Immunology of hepatitis B virus and hepatitis C virus infection. Nat Rev Immunol. 2005;5:215-229. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1202] [Cited by in F6Publishing: 1174] [Article Influence: 61.8] [Reference Citation Analysis (0)] |
| 11. | Tamura T, Yanai H, Savitsky D, Taniguchi T. The IRF family transcription factors in immunity and oncogenesis. Annu Rev Immunol. 2008;26:535-584. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 876] [Cited by in F6Publishing: 924] [Article Influence: 57.8] [Reference Citation Analysis (0)] |
| 12. | Chen W, Royer WE Jr. Structural insights into interferon regulatory factor activation. Cell Signal. 2010;22:883-887. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 67] [Cited by in F6Publishing: 64] [Article Influence: 4.6] [Reference Citation Analysis (0)] |
| 13. | Chen W, Lam SS, Srinath H, Jiang Z, Correia JJ, Schiffer CA, Fitzgerald KA, Lin K, Royer WE Jr. Insights into interferon regulatory factor activation from the crystal structure of dimeric IRF5. Nat Struct Mol Biol. 2008;15:1213-1220. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 91] [Cited by in F6Publishing: 94] [Article Influence: 5.9] [Reference Citation Analysis (0)] |
| 14. | Chang Foreman HC, Van Scoy S, Cheng TF, Reich NC. Activation of interferon regulatory factor 5 by site specific phosphorylation. PLoS One. 2012;7:e33098. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 51] [Cited by in F6Publishing: 56] [Article Influence: 4.7] [Reference Citation Analysis (0)] |
| 15. | Lien C, Fang CM, Huso D, Livak F, Lu R, Pitha PM. Critical role of IRF-5 in regulation of B-cell differentiation. Proc Natl Acad Sci USA. 2010;107:4664-4668. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 71] [Cited by in F6Publishing: 70] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 16. | Savitsky DA, Yanai H, Tamura T, Taniguchi T, Honda K. Contribution of IRF5 in B cells to the development of murine SLE-like disease through its transcriptional control of the IgG2a locus. Proc Natl Acad Sci USA. 2010;107:10154-10159. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 70] [Cited by in F6Publishing: 70] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 17. | Fang CM, Roy S, Nielsen E, Paul M, Maul R, Paun A, Koentgen F, Raval FM, Szomolanyi-Tsuda E, Pitha PM. Unique contribution of IRF-5-Ikaros axis to the B-cell IgG2a response. Genes Immun. 2012;13:421-430. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 37] [Cited by in F6Publishing: 37] [Article Influence: 3.1] [Reference Citation Analysis (0)] |
| 18. | Barnes BJ, Richards J, Mancl M, Hanash S, Beretta L, Pitha PM. Global and distinct targets of IRF-5 and IRF-7 during innate response to viral infection. J Biol Chem. 2004;279:45194-45207. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 169] [Cited by in F6Publishing: 178] [Article Influence: 8.9] [Reference Citation Analysis (0)] |
| 19. | Tang L, Chen B, Ma B, Nie S. Association between IRF5 polymorphisms and autoimmune diseases: a meta-analysis. Genet Mol Res. 2014;13:4473-4485. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 29] [Cited by in F6Publishing: 29] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 20. | Li Y, Chen S, Li P, Wu Z, Li J, Liu B, Zhang F, Li Y. Association of the IRF5 rs2070197 polymorphism with systemic lupus erythematosus: a meta-analysis. Clin Rheumatol. 2015;34:1495-1501. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 7] [Cited by in F6Publishing: 7] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 21. | Hoan NX, Tong HV, Hecht N, Sy BT, Marcinek P, Meyer CG, Song le H, Toan NL, Kurreck J, Kremsner PG. Hepatitis E Virus Superinfection and Clinical Progression in Hepatitis B Patients. EBioMedicine. 2015;2:2080-2086. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 36] [Cited by in F6Publishing: 42] [Article Influence: 4.7] [Reference Citation Analysis (0)] |
| 22. | Rullo OJ, Woo JM, Wu H, Hoftman AD, Maranian P, Brahn BA, McCurdy D, Cantor RM, Tsao BP. Association of IRF5 polymorphisms with activation of the interferon alpha pathway. Ann Rheum Dis. 2010;69:611-617. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 48] [Cited by in F6Publishing: 50] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 23. | Benjamini Y. Discovering the false discovery rate. J R Stat Soc. 2010;72:405-416. [DOI] [Cited in This Article: ] [Cited by in Crossref: 357] [Cited by in F6Publishing: 362] [Article Influence: 25.9] [Reference Citation Analysis (0)] |
| 24. | Yanai H, Chen HM, Inuzuka T, Kondo S, Mak TW, Takaoka A, Honda K, Taniguchi T. Role of IFN regulatory factor 5 transcription factor in antiviral immunity and tumor suppression. Proc Natl Acad Sci USA. 2007;104:3402-3407. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 160] [Cited by in F6Publishing: 167] [Article Influence: 9.8] [Reference Citation Analysis (0)] |
| 25. | Barnes BJ, Moore PA, Pitha PM. Virus-specific activation of a novel interferon regulatory factor, IRF-5, results in the induction of distinct interferon alpha genes. J Biol Chem. 2001;276:23382-23390. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 300] [Cited by in F6Publishing: 297] [Article Influence: 12.9] [Reference Citation Analysis (0)] |
| 26. | Li X, Liu X, Tian L, Chen Y. Cytokine-Mediated Immunopathogenesis of Hepatitis B Virus Infections. Clin Rev Allergy Immunol. 2016;50:41-54. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 45] [Cited by in F6Publishing: 61] [Article Influence: 7.6] [Reference Citation Analysis (0)] |
| 27. | Rijckborst V, Janssen HL. The Role of Interferon in Hepatitis B Therapy. Curr Hepat Rep. 2010;9:231-238. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 34] [Cited by in F6Publishing: 28] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 28. | Hirschfield GM, Liu X, Han Y, Gorlov IP, Lu Y, Xu C, Lu Y, Chen W, Juran BD, Coltescu C. Variants at IRF5-TNPO3, 17q12-21 and MMEL1 are associated with primary biliary cirrhosis. Nat Genet. 2010;42:655-657. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 167] [Cited by in F6Publishing: 177] [Article Influence: 12.6] [Reference Citation Analysis (0)] |
| 29. | Juran BD, Hirschfield GM, Invernizzi P, Atkinson EJ, Li Y, Xie G, Kosoy R, Ransom M, Sun Y, Bianchi I. Immunochip analyses identify a novel risk locus for primary biliary cirrhosis at 13q14, multiple independent associations at four established risk loci and epistasis between 1p31 and 7q32 risk variants. Hum Mol Genet. 2012;21:5209-5221. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 118] [Cited by in F6Publishing: 124] [Article Influence: 10.3] [Reference Citation Analysis (0)] |
| 30. | Zhou WC, Zhang QB, Qiao L. Pathogenesis of liver cirrhosis. World J Gastroenterol. 2014;20:7312-7324. [PubMed] [DOI] [Cited in This Article: ] [Cited by in CrossRef: 335] [Cited by in F6Publishing: 335] [Article Influence: 33.5] [Reference Citation Analysis (8)] |
| 31. | Takaoka A, Yanai H, Kondo S, Duncan G, Negishi H, Mizutani T, Kano S, Honda K, Ohba Y, Mak TW. Integral role of IRF-5 in the gene induction programme activated by Toll-like receptors. Nature. 2005;434:243-249. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 738] [Cited by in F6Publishing: 761] [Article Influence: 40.1] [Reference Citation Analysis (0)] |
| 32. | Shin SH, Kim BH, Jang JJ, Suh KS, Kang GH. Identification of novel methylation markers in hepatocellular carcinoma using a methylation array. J Korean Med Sci. 2010;25:1152-1159. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 39] [Cited by in F6Publishing: 38] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 33. | Graham RR, Kozyrev SV, Baechler EC, Reddy MV, Plenge RM, Bauer JW, Ortmann WA, Koeuth T, González Escribano MF; Argentine and Spanish Collaborative Groups, Pons-Estel B, Petri M, Daly M, Gregersen PK, Martín J, Altshuler D, Behrens TW, Alarcón-Riquelme ME. A common haplotype of interferon regulatory factor 5 (IRF5) regulates splicing and expression and is associated with increased risk of systemic lupus erythematosus. Nat Genet. 2006;38:550-555. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 494] [Cited by in F6Publishing: 493] [Article Influence: 27.4] [Reference Citation Analysis (0)] |
| 34. | Graham RR, Kyogoku C, Sigurdsson S, Vlasova IA, Davies LR, Baechler EC, Plenge RM, Koeuth T, Ortmann WA, Hom G. Three functional variants of IFN regulatory factor 5 (IRF5) define risk and protective haplotypes for human lupus. Proc Natl Acad Sci USA. 2007;104:6758-6763. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 341] [Cited by in F6Publishing: 364] [Article Influence: 21.4] [Reference Citation Analysis (0)] |
| 35. | Niewold TB, Kelly JA, Flesch MH, Espinoza LR, Harley JB, Crow MK. Association of the IRF5 risk haplotype with high serum interferon-alpha activity in systemic lupus erythematosus patients. Arthritis Rheum. 2008;58:2481-2487. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 208] [Cited by in F6Publishing: 222] [Article Influence: 13.9] [Reference Citation Analysis (0)] |
| 36. | Niewold TB, Kelly JA, Kariuki SN, Franek BS, Kumar AA, Kaufman KM, Thomas K, Walker D, Kamp S, Frost JM. IRF5 haplotypes demonstrate diverse serological associations which predict serum interferon alpha activity and explain the majority of the genetic association with systemic lupus erythematosus. Ann Rheum Dis. 2012;71:463-468. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 107] [Cited by in F6Publishing: 109] [Article Influence: 8.4] [Reference Citation Analysis (0)] |