Copyright
©The Author(s) 2017.
World J Gastroenterol. Oct 7, 2017; 23(37): 6817-6832
Published online Oct 7, 2017. doi: 10.3748/wjg.v23.i37.6817
Published online Oct 7, 2017. doi: 10.3748/wjg.v23.i37.6817
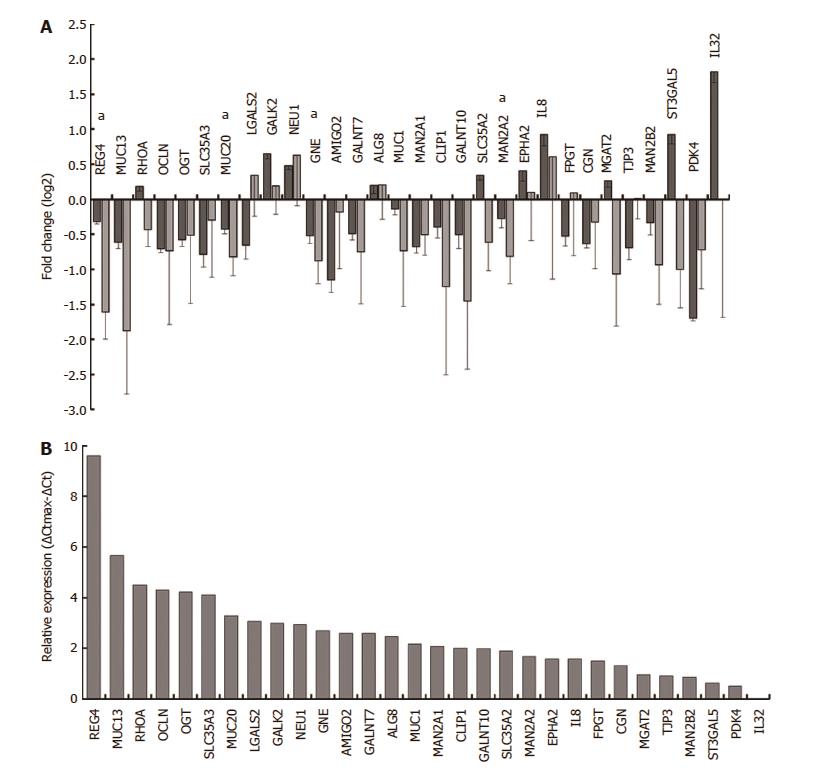
Figure 3 qRT-PCR validation of selected genes.
Differential expression of selected genes upon Helicobacter pylori (H. pylori) infection of E12 cells was compared between microarray (dark grey) and qRT-PCR (light grey) analyses. qRT-PCR validation was carried out on RNAs from samples collected independently of the microarray samples. A: Microarray data (n = 3) was normalised across samples with GeneSpring. qRT-PCR data (n = 3) were normalised using geNorm and a panel of three housekeeping genes. Error bars show standard deviations determined using propagation of error rules; B: Relative expression of each gene in uninfected E12 cells was estimated by qRT-PCR using ΔCt determinations. ΔCtmax was the ΔCt for the lowest expressing gene (IL32) after correcting Ct values across samples with the housekeeping gene GAPDH. Further details are given in Materials and Methods. All genes were significantly differentially expressed in microarray analysis and genes marked with asterisks (a) were confirmed significant by qRT-PCR.
- Citation: Cairns MT, Gupta A, Naughton JA, Kane M, Clyne M, Joshi L. Glycosylation-related gene expression in HT29-MTX-E12 cells upon infection by Helicobacter pylori. World J Gastroenterol 2017; 23(37): 6817-6832
- URL: https://www.wjgnet.com/1007-9327/full/v23/i37/6817.htm
- DOI: https://dx.doi.org/10.3748/wjg.v23.i37.6817