Published online May 28, 2017. doi: 10.3748/wjg.v23.i20.3713
Peer-review started: January 22, 2017
First decision: February 21, 2017
Revised: March 7, 2017
Accepted: March 21, 2017
Article in press: March 21, 2017
Published online: May 28, 2017
To analyze the incidence of hepatocellular carcinoma (HCC) in a population that underwent health checkups and had high serum miR-106b levels.
A total of 335 subjects who underwent checkups in the Digestive and Liver Disease Department of our hospital were randomly selected. RT-PCR was used to detect the level of miR-106b in serum samples. Laboratory and imaging examinations were carried out to confirm the HCC diagnosis in patients who had a > 2-fold change in miR-106b levels. Ultrasound-guided biopsy was also used for HCC diagnosis when necessary. On this basis, the clinical data of these subjects, including history of hepatitis virus infection, obesity, long-term history of alcohol use and stage of HCC, were collected. Then, the impact of these factors on the level of miR-106b in serum was analyzed. Furthermore, receiver operating characteristic (ROC) curve was drawn to evaluate the diagnostic efficacy of miR-106b for HCC.
A total of 35 subjects had abnormal serum miR-106b levels, of which 20 subjects were diagnosed with HCC. t-test revealed that the difference in serum miR-106b level in terms of sex, age, history of hepatitis virus infection, obesity and long-term history of alcohol use was not statistically significant. However, serum miR-106b levels in patients with advanced HCC (stage III/IV) was higher than in patients with early HCC (stage I/II), and the difference was statistically significant (P = 0.000). Moreover, the ROC curve revealed that the area under the curve value for miR-106b was 0.885, which shows that serum miR-106b level has a certain clinical value for HCC diagnosis.
The random sampling survey shows that serum miR-106b level is a valuable diagnostic marker for HCC. However, the diagnostic threshold value needs to be further researched.
Core tip: Primary hepatocellular carcinoma (HCC) is the cause of a large number of patient deaths each year, and presents a heavy financial burden to the community and family. MicroRNAs (miRNAs) have been proven to be involved in the development of various cancers, as well as during the development process. Among these miRNAs, miR-106b has been shown to be a potential diagnostic marker for early HCC. We randomly selected the sera of medical examiners and detected their miR-106b level and further verified the diagnostic value of miR-106b, and provided more data to support the clinical application of miR-106b in evidence-based medicine.
- Citation: Shi BM, Lu W, Ji K, Wang YF, Xiao S, Wang XY. Study on the value of serum miR-106b for the early diagnosis of hepatocellular carcinoma. World J Gastroenterol 2017; 23(20): 3713-3720
- URL: https://www.wjgnet.com/1007-9327/full/v23/i20/3713.htm
- DOI: https://dx.doi.org/10.3748/wjg.v23.i20.3713
Primary hepatocellular carcinoma (HCC) is one of the most common malignancies in the world, is the cause of a large number of patient deaths each year and presents a heavy financial burden to the community and families[1,2]. The incidence of HCC in China has shown an increasing trend year by year, and the main risk factor of hepatitis B infection has gradually transformed into alcohol and other non-viral infectious factors[3-7]. For the occult onset of HCC, many patients with advanced HCC obtain their initial diagnosis after the best treatment window for HCC has closed[8-11]. Therefore, there is an urgent clinical need for effective diagnostic indicators for the early diagnosis of HCC.
At present, B ultrasound and serum alpha-fetoprotein (AFP) are mainly used by medical personnel for initial screening. However, these two methods have the shortcoming of poor sensitivity, and are insufficient for early warning in high-risk groups[12]. MicroRNAs (miRNAs) are a class of 18-24 nucleotide short-chain non-coding RNAs that have been proven to be involved in the development of various cancers, as well as during the normal organismal development process. Tumor cells may also be able to release miRNA into the bloodstream, and such circulating miRNAs have become a hot topic in recent years.
Serum miRNA has a high stability, is resistant to RNase hydrolysis, and its contents relatively remain at a constant level after repeated freezing and thawing. Among these miRNAs, miR-106b has been shown to be a potential diagnostic marker for early HCC[13-19]. However, such studies involved mostly targeted subjects and were not conducive for evaluating the true diagnostic efficacy of miRNA. Hence, there is a need to conduct a random sampling survey of the physical examination blood samples of patients with digestive and liver disease, in order to determine whether miR-106b can be used as an early warning indicator of HCC.
In this study, we randomly selected the sera of medical examinees and detected their miR-106b level. The level of miR-106b in serum of patients with HCC was further analyzed. Furthermore, the significance of miR-106b levels in the diagnosis of HCC was further analyzed. The results of this study further verified the diagnostic value of miR-106b, and provided more data to support the clinical application of miR-106b in evidence-based medicine.
Inclusion criteria: (1) randomly selected serum samples from outpatients who underwent physical examination in our hospital between January 2011 and August 2015; (2) clinical information is complete; and (3) time within 1 mo after the serum test and willing to accept further physical examination and follow-up. Exclusion criteria: (1) liver cancer patients; (2) incomplete information; and (3) refusal to accept further physical examination and follow-up.
A total of 335 serum samples were selected at different time intervals, and the mean age of the subjects was 55.4 ± 10.7 years. The relative content of miR-106b was significantly increased two-times or more in 35 subjects. Among these 35 subjects, 21 were male and 14 were female. This study was approved by the Ethics Committee, and all patients signed an informed consent form.
Clinical research methods: Real-time quantitative PCR detecting system (qPCR) was used to detect miRNA levels in serum samples. MiR-106b was amplified by selecting cel-miR-39 as an internal reference. The relative expression of miR-106b in the serum sample was calculated using the 2-ΔΔCt method. When the relative levels of miR-106b were significantly more than double, the notification to participate in the group was further reviewed. The patients were diagnosed with HCC after follow-up and were divided into two groups: non-HCC group and HCC group. Follow-up records included the patient’s age, sex, obesity status, history of hepatitis, and long-term drinking history. Patients diagnosed with HCC were actively treated.
Detection of serum miR-106b relative content: Serum total RNA extraction was performed. First, blood samples were collected using vacuum blood collection tubes, and were left at room temperature for 40 min. Then, the supernatants were collected by centrifugation (3000 rpm at 4 °C) and stored at -80 °C until use. The extraction of total RNA in serum was performed according to kit instructions (miRNeasy Serum/Plasma Kit; Qiagen, Hilden, Germany).
Serum samples were supplemented with 1000 μL of QIAzol (lysate). After standing for 5 min, 3.5 μL of miRNeasy Serum/Plasma Spike-In Control was added (concentration of 1.6 × 108 copies/L) as a control for RNA purification yield and amplification efficiency. This was followed by the addition of chloroform at room temperature for 3 min. The 600 μL supernatant was mixed well with 900 μL of absolute ethanol, and transferred to an RNeasy MinElute spin column and centrifuged at 12000 rpm at 4 °C. This was followed by RWT, RPE buffer and ethanol elution phenol, and other organic reagents, and finally with 14 μL of DEPC water-eluting RNA. The resultant products were stored at -80 °C until use.
The RT miRNA tailing method was performed next. The tailing method was used to reverse transcribe miRNA into cDNA, and the operation was carried out according to the instructions of the reverse transcription kit (MiScriptII RT Kit; Qiagen). Briefly, the RNA was extracted according to instructions, and the other solutions were prepared for use in the reverse transcription system. Then, the reaction system was placed on the PCR instrument for amplification. The resulting cDNA was stored at -80 °C until use.
Real-time fluorescence quantitative PCR was performed finally. The relative content of miR-106b was measured according to miRNA SYBR Green PCR detection kit instructions (Qiagen). Reactions were performed on an ABI 7500 Real Time PCR instrument. According to the relevant literature, cel-miR-39 was selected as the internal reference. MiR-106b was amplified. Then, the relative expression of miR-106b in serum samples was calculated using the 2-ΔΔCt method (ΔCt = Ct(miR-106b)-Ct(cel-miR-39)).
Retrospective evaluation of HCC patients with abnormal serum miR-106b levels: When the relative levels of miR-106b significantly more than doubled, the patient was followed-up and notified to further confirm the suspected population prevalence of HCC. Further diagnosis of suspected patients was conducted according to the Chinese Ministry of Health guidelines issued in 2011 for the diagnosis and treatment of primary liver cancer patients. Clinical symptoms, blood biochemical examination [aspartate aminotransferase (AST) or glutamic oxaloacetic transaminase], tumor marker detection (serum AFP and its heteroplasm), and imaging studies were applied on the risk assessment and diagnosis of suspected cancer patients. B-ultrasound and computed tomography imaging were used as basis for the early diagnosis of liver cancer. The imaging diagnosis process was completed by an ultrasound physician and associate professors, in order to ensure accuracy of the diagnostic results.
History of hepatitis, obesity, drinking and other factors were evaluated to determine whether these also caused any change in serum miR-106b. Furthermore, related medical history and clinical signs were collected and analyzed statistically.
SPSS 19.0 software was used to analyze the data. Measurement data were expressed as mean ± SD. Subjects were divided into two groups according to clinical examination results: early HCC group and non-HCC group. t-test was used to compare the basic clinical data between the two groups. On this basis, the curve derived from the patient (receiver operating characteristic (ROC) curve) was used to evaluate the diagnostic efficacy of miR-106b. P < 0.05 was considered statistically significant.
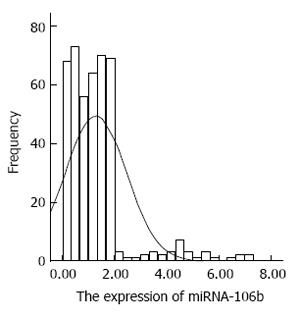
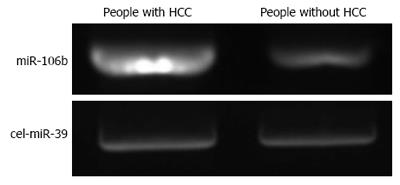
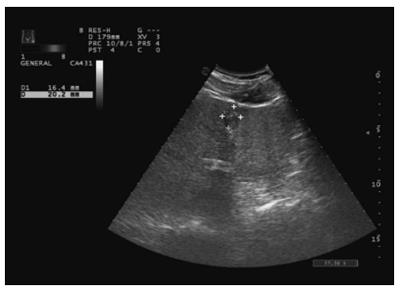
The distribution of miRNA-106b in serum samples obtained from patients is shown in Figure 1. The Kolmogorov-Smirnov test revealed normal distribution of the data (P = 0.000). The relative serum miRNA-106b expression level of all the subjects was 1.12 ± 0.89 times. MiRNA-106b increased more than two-times as an early warning indicator; a total of 35 subjects were suspected during the review, and results revealed that 25 of these subjects were diagnosed with HCC. The qPCR results of typical case samples are shown in Figure 2. The difference in cel-miR-39 in these two typical patients was not statistically significant, while the content of miRNA-106b in patients with hepatitis B was significantly higher than that in normal patients. Serum miR-106b increased in some of the suspected patients during the review as shown by ultrasound results (Figure 3). Early liver cancer lesions < 3 cm could be observed through ultrasound. There is a need for such to be carefully identified before they can be located.
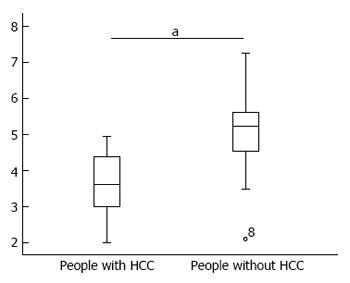
Patients were divided into two groups according to the review results, and the difference in serum miR-106b levels between these two groups is shown in Figure 4.
The t-test results revealed that patients with HCC had higher serum miR-106b levels than patients without HCC, and the difference was statistically significant (P = 0.000).
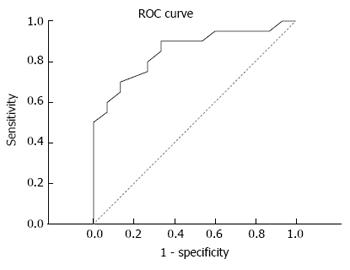
On this basis, the diagnostic efficacy of serum miRNA-106b was analyzed through the ROC curve. Results revealed that the area under the curve was 0.855, and sensitivity and specificity were 90.0% and 66.7%, respectively (Figure 5).
In clinical practice, obesity, viruses and alcohol cause liver damage, as well as changes in serum markers. Therefore, analyzing whether miR-106b is affected by these factors is of great significance for the diagnosis of HCC. The clinical data of the two groups of subjects are shown in Table 1. Age, sex, obesity, history of hepatitis and history of alcohol consumption had no significant effect on miRNA-106b expression (P > 0.05). However, serum miRNA-106b levels were significantly higher in patients with higher tumor stages (III/IV) than in patients in stage I/II (P = 0.000).
| Clinical parameter | n (%) | miRNA-106b level | t value | P value |
| History of hepatitis | ||||
| Have | 20 (57.14) | 4.62 ± 0.91 | 0.532 | 0.598 |
| None | 15 (42.86) | 4.45 ± 0.97 | ||
| Long-term drinking history | ||||
| Have | 24 (68.57) | 4.72 ± 0.92 | 1.378 | 0.177 |
| None | 11 (31.43) | 4.29 ± 0.69 | ||
| Obesity | ||||
| Have | 16 (45.71) | 4.62 ± 0.90 | 0.986 | 0.331 |
| None | 19 (54.29) | 4.35 ± 0.72 | ||
| Liver cancer stage | ||||
| I/II | 33 (94.26) | 4.64 ± 0.86 | -4.200 | 0.000 |
| III/IV | 2 (5.74) | 7.23 ± 0.02 |
HCC is one of the most common types of malignant tumors[11,20-22]. A survey has shown that HCC ranks second among deaths caused by diseases annually. Risk factors for liver cancer vary in different regions. In China, there is a high prevalence of liver cancer caused by hepatitis B.
Hence, liver cancer has become a major threat to national health. According to statistics, the number of patients with liver cancer in China has reached up to 50% of the cases worldwide[9,23-29]. At present, with tumor resection surgery, interventional techniques and the development of liver transplantation technology, the treatment of liver cancer has made remarkable progress.
However, even so, the survival rate of patients with liver cancer was not significantly improved. Furthermore, according to relevant studies on liver cancer patients, the overall 5-year survival rate is approximately 5%-9%[30-35]. The main reason for this startling statistic is that liver cells have good compensatory function, so that no obvious imaging and serological changes are found in early liver cancer patients during routine physical examination. Obvious clinical signs often manifest in the advance stage.
Therefore, clinically, there is an urgent need to develop high sensitivity and specificity diagnostic indicators for patients who may be suffering from liver cancer, in order to provide early warning of the disease in this population. At present, medical examiners mainly rely on B-ultrasound and serum AFP levels during the physical examination of high-risk groups for early warning. However, the accuracy of B-mode ultrasonography is directly related to the patient’s experience. Furthermore, the sensitivity and specificity of serum AFP diagnosis is low, which limits the diagnosis of early liver cancer and early treatment, and affects the prognosis of patients.
MiRNA is a non-coding small RNA with a length of approximately 22 nucleotides. MiRNAs can adjust the level of physiological functions in the cell after transcription, including that of cell proliferation, differentiation and apoptosis. Research has shown that the growth of tumor cells is subject to the corresponding miRNA regulation, such as miR-26a/b and miR-146b-5p, which affects the growth of tumor cell cycle regulation. MiR-7 inhibits tumor growth by regulating the PI3K/AKT/mTOR pathway during the migration process. Furthermore, miR-1826 can affect angiogenesis by down-regulating VEGFC.
More studies have found that tumor cells can release miRNA into the circulatory system. Furthermore, miRNA induces changes in blood, leading to tumor occurrence; hence, this development has important relevance[36-43]. In patients with liver cancer, miR-21, miR-122, miR-1, miR-25, miR-92a, miR-206, miR-106b and let-7f have been found to have potential as diagnostic indicators of liver cancer, with diagnostic sensitivity of ≥ 80%[13]. However, since many studies have involved targeted selected subjects, this resulted in the existence of certain bias in the selection process, which is not conducive to determining the true diagnosis of the evaluation of miRNA efficacy.
Therefore, in this study, we selected miR-106b as the research focus due to its high diagnostic value. Through the random screening of blood samples conducted by the medical staff, the efficacy of serum miR-106b in the diagnosis of liver cancer was analyzed, with a view of making some preliminary experiments for the entry of miRNA into routine laboratory diagnosis.
The miR-106b gene is located on human chromosome 7, as well as in glioma, prostate cancer and other tumors[44]. In HCC tissues, a study found that miR-106b can inhibit the APC gene and thereby promote cancer cell growth; and it can also significantly increase the serum miRNA of patients with liver cancer[44]. There are some controversies regarding the diagnostic limits of miR-106b in different studies. However, the relative serum miR-106b expression of patients with HCC was more than double in most studies[44-46]. Therefore, in order to include as many potential HCC patients as possible, in this study, relative serum miR-106b expression elevated above two-times was selected as the indicator. Furthermore, in this study, among the 35 subjects suspected to have miR-106b abnormalities, 20 were diagnosed with late stage HCC. The relative miR-106b expression level in subjects in the HCC group was significantly higher than that in the non-HCC group. These results suggest that miR-106b may serve as a potential biomarker for HCC.
In each study, the serum miRNA diagnosis threshold differs greatly depending on the subjects studied. In the study conducted by Choo et al[47], the sensitivity and specificity of serum miRNA for the diagnosis of HCC was 81% and 97%, respectively. In the present study, the ROC curve revealed that the area under the curve of miR-106b was 0.855, and sensitivity and specificity were 90.0% and 66.7%, respectively. This suggests that it has a certain diagnostic value. However, there may be a need to diagnose with other diagnostic indicators, in order to improve the specificity of liver cancer diagnosis. Although the present study was carried out with a large-scale sample survey, only 20 people were diagnosed with liver cancer among the 35 suspected subjects (accuracy rate: 57.14%). Therefore, the limits of the diagnostic value of miR-106b needs to be further studied with an expanded sample size.
In the study of the accuracy of AFP as a routine physical examination index, results revealed that serum miRNA significantly increased in these patients, and the difference in AFP was not significant. Furthermore, if 400 ng/mL were used as a diagnostic threshold for conventional diagnostic criteria, this would result in the missed opportunity to detect 6 patients with liver cancer. For early screening examinations, B-ultrasound is a routine diagnostic. However, in the absence of other high-risk diagnostic indicator warnings, physicians in the actual inspection process can easy misdiagnose the location of hidden and small ranges of early liver cancer.
If the presence of diagnostic markers predicts the risk of HCC, higher-resolution B-mode ultrasonography can be used to increase the detection rate in patients with early HCC. Taking into account that blood sample collection is a routine physical examination, as well as the high stability of miRNA in serum samples and other characteristics, miR-106b is suitable for use with AFP and other serum markers associated with early warning indicators for liver cancer. However, further studies are needed to determine the warning limits for miR-106b.
Liver cancer is an occult onset process. Furthermore, it is not easy to distinguish patients with early liver cancer from with patients with chronic liver disease through routine blood biochemistry and laboratory tests. In addition, obesity-induced fatty liver, virus-induced hepatitis and alcohol can induce liver damage, and at the same time can cause serological AFP, AST and other related enzymatic indicators to change[48-52]. Therefore, analyzing whether miR-106b is affected by these factors is of great significance in the diagnosis of HCC.
By comparing the miR-106b level of the suspected subjects, we found that in addition to tumor stage, hepatitis, obesity and long-term drinking history would not cause significant differences in miR-106b level. However, if liver cancer patients had a higher tumor stage, this significantly increased the serum miR-106b level; this finding is consistent with relevant literature reports, suggesting that in the future appropriate research should be conducted on the relationship between miRNA and tumor staging, in order to further analyze the value of serum miRNA for evaluating the condition of patients.
At the same time, this subject matter also has certain limitations. First of all, it is limited by objective conditions. In this study, only one miRNA was included in the assessment, and a single miRNA study does not accurately assess the value of the entire miRNA family for diagnosis. Hence, more representative miRNAs should be included in the study. At the same time, since the truncation values for miR-106b remain controversial, referral indicators used in this study may not be the optimal diagnostic threshold. This would induce the detection rate in subjects with suspected liver cancer to decline. Hence, these optimal diagnostic thresholds also need to be determined in larger clinical studies.
In summary, in this study, miR-106b was used as the research object, and the value of miRNAs in the diagnosis of HCC was evaluated by random sampling. MiR-106b has the potential to become an early serologic diagnostic marker for HCC.
Primary hepatocellular carcinoma (HCC) is the cause of a large number of patient deaths each year, and presents heavy financial burden to the community and family. MicroRNAs (miRNAs) have been proven to be involved in the development of various cancers, as well as during the normal organismal development process. Among these miRNAs, miR-106b has been shown to be a potential diagnostic marker for early HCC.
Although the treatment of liver cancer has made remarkable progress, the survival rate of patients with liver cancer has not significantly improved. MiRNA is a non-coding small RNA with a length of approximately 22 nucleotides. Moreover, studies have shown that tumor cells can release miRNA into the circulatory system, which means that they have the potential to forecast the occurrence of HCC.
In this study, miR-106b was chosen as the research object due to its high diagnostic value. The authors randomly selected the sera of medical examinees and detected their miR-106b level and further verified the diagnostic value of miR-106b, and provided more data to support the clinical application of miR-106b in evidence-based medicine.
This survey shows that serum miR-106b level is a valuable diagnostic marker for HCC. However, the diagnostic threshold value needs to be further researched.
Due to its high diagnostic value, serum miR-106b was chosen as the focus of this research. This study further showed that serum miR-106b level is a valuable diagnostic marker for HCC. However, more research should be carried to find out the diagnostic threshold value.
Manuscript source: Unsolicited manuscript
Specialty type: Gastroenterology and hepatology
Country of origin: China
Peer-review report classification
Grade A (Excellent): 0
Grade B (Very good): 0
Grade C (Good): C
Grade D (Fair): 0
Grade E (Poor): 0
P- Reviewer: Kantsevoy S S- Editor: Qi Y L- Editor: Filipodia E- Editor: Zhang FF
| 1. | Iwamoto T, Imai Y, Kogita S, Igura T, Sawai Y, Fukuda K, Yamaguchi Y, Matsumoto Y, Nakahara M, Morimoto O. Comparison of Contrast-Enhanced Ultrasound and Gadolinium-Ethoxybenzyl-Diethylenetriamine Pentaacetic Acid-Enhanced MRI for the Diagnosis of Macroscopic Type of Hepatocellular Carcinoma. Dig Dis. 2016;34:679-686. [PubMed] [DOI] [Cited in This Article: ] |
| 2. | Kawasaki T, Hata KY, Kinoshita D, Takayama M, Okuda H, Mizuno S, Kudo M. Radiofrequency Ablation Guided by Contrast-Enhanced Sonography versus B-Mode Sonography for Hepatocellular Carcinoma after Transcatheter Arterial Chemoembolization. Dig Dis. 2016;34:692-695. [PubMed] [DOI] [Cited in This Article: ] |
| 3. | Cheung OK, Cheng AS. Gender Differences in Adipocyte Metabolism and Liver Cancer Progression. Front Genet. 2016;7:168. [PubMed] [DOI] [Cited in This Article: ] |
| 4. | Gao C. Molecular pathological epidemiology in diabetes mellitus and risk of hepatocellular carcinoma. World J Hepatol. 2016;8:1119-1127. [PubMed] [DOI] [Cited in This Article: ] |
| 5. | Shi YX, Huang CJ, Yang ZG. Impact of hepatitis B virus infection on hepatic metabolic signaling pathway. World J Gastroenterol. 2016;22:8161-8167. [PubMed] [DOI] [Cited in This Article: ] |
| 6. | Yang N, Li S, Li G, Zhang S, Tang X, Ni S, Jian X, Xu C, Zhu J, Lu M. The role of extracellular vesicles in mediating progression, metastasis and potential treatment of hepatocellular carcinoma. Oncotarget. 2017;8:3683-3695. [PubMed] [DOI] [Cited in This Article: ] |
| 7. | Zhu Y, Liu H, Zhang M, Guo GL. Fatty liver diseases, bile acids, and FXR. Acta Pharm Sin B. 2016;6:409-412. [PubMed] [DOI] [Cited in This Article: ] |
| 8. | Lin J, Wu L, Bai X, Xie Y, Wang A, Zhang H, Yang X, Wan X, Lu X, Sang X. Combination treatment including targeted therapy for advanced hepatocellular carcinoma. Oncotarget. 2016;7:71036-71051. [PubMed] [DOI] [Cited in This Article: ] |
| 9. | Schütte K, Balbisi F, Malfertheiner P. Prevention of Hepatocellular Carcinoma. Gastrointest Tumors. 2016;3:37-43. [PubMed] [DOI] [Cited in This Article: ] |
| 10. | Vitale A, Peck-Radosavljevic M, Giannini EG, Vibert E, Sieghart W, Van Poucke S, Pawlik TM. Personalized treatment of patients with very early hepatocellular carcinoma. J Hepatol. 2017;66:412-423. [PubMed] [DOI] [Cited in This Article: ] |
| 11. | Zhu ZX, Huang JW, Liao MH, Zeng Y. Treatment strategy for hepatocellular carcinoma in China: radiofrequency ablation versus liver resection. Jpn J Clin Oncol. 2016;46:1075-1080. [PubMed] [DOI] [Cited in This Article: ] |
| 12. | AlSalloom AA. An update of biochemical markers of hepatocellular carcinoma. Int J Health Sci (Qassim). 2016;10:121-136. [PubMed] [Cited in This Article: ] |
| 13. | Afonso MB, Rodrigues PM, Simão AL, Castro RE. Circulating microRNAs as Potential Biomarkers in Non-Alcoholic Fatty Liver Disease and Hepatocellular Carcinoma. J Clin Med. 2016;5. [PubMed] [DOI] [Cited in This Article: ] |
| 14. | Chen WQ, Hu L, Chen GX, Deng HX. Role of microRNA-7 in digestive system malignancy. World J Gastrointest Oncol. 2016;8:121-127. [PubMed] [DOI] [Cited in This Article: ] |
| 15. | Fiorino S, Bacchi-Reggiani ML, Visani M, Acquaviva G, Fornelli A, Masetti M, Tura A, Grizzi F, Zanello M, Mastrangelo L. MicroRNAs as possible biomarkers for diagnosis and prognosis of hepatitis B- and C-related-hepatocellular-carcinoma. World J Gastroenterol. 2016;22:3907-3936. [PubMed] [DOI] [Cited in This Article: ] |
| 16. | Hayes CN, Chayama K. MicroRNAs as Biomarkers for Liver Disease and Hepatocellular Carcinoma. Int J Mol Sci. 2016;17:280. [PubMed] [DOI] [Cited in This Article: ] |
| 17. | Li H, Jiang JD, Peng ZG. MicroRNA-mediated interactions between host and hepatitis C virus. World J Gastroenterol. 2016;22:1487-1496. [PubMed] [DOI] [Cited in This Article: ] |
| 18. | Murakami Y, Kawada N. MicroRNAs in hepatic pathophysiology. Hepatol Res. 2017;47:60-69. [PubMed] [DOI] [Cited in This Article: ] |
| 19. | Hamou C, Callaghan MJ, Thangarajah H, Chang E, Chang EI, Grogan RH, Paterno J, Vial IN, Jazayeri L, Gurtner GC. Mesenchymal stem cells can participate in ischemic neovascularization. Plast Reconstr Surg. 2009;123:45S-55S. [PubMed] [DOI] [Cited in This Article: ] |
| 20. | Brunot A, Le Sourd S, Pracht M, Edeline J. Hepatocellular carcinoma in elderly patients: challenges and solutions. J Hepatocell Carcinoma. 2016;3:9-18. [PubMed] [DOI] [Cited in This Article: ] |
| 21. | Gallicchio R, Nardelli A, Mainenti P, Nappi A, Capacchione D, Simeon V, Sirignano C, Abbruzzi F, Barbato F, Landriscina M. Therapeutic Strategies in HCC: Radiation Modalities. Biomed Res Int. 2016;2016:1295329. [PubMed] [DOI] [Cited in This Article: ] |
| 22. | Bandiera S, Billie Bian C, Hoshida Y, Baumert TF, Zeisel MB. Chronic hepatitis C virus infection and pathogenesis of hepatocellular carcinoma. Curr Opin Virol. 2016;20:99-105. [PubMed] [DOI] [Cited in This Article: ] |
| 23. | Hemming AW, Berumen J, Mekeel K. Hepatitis B and Hepatocellular Carcinoma. Clin Liver Dis. 2016;20:703-720. [PubMed] [DOI] [Cited in This Article: ] |
| 24. | Konerman MA, Lok AS. Interferon Treatment for Hepatitis B. Clin Liver Dis. 2016;20:645-665. [PubMed] [DOI] [Cited in This Article: ] |
| 25. | Lee HW, Ahn SH. Prediction models of hepatocellular carcinoma development in chronic hepatitis B patients. World J Gastroenterol. 2016;22:8314-8321. [PubMed] [DOI] [Cited in This Article: ] |
| 26. | Luvisa BK, Hassanein TI. Hepatitis B Virus Infection and Liver Decompensation. Clin Liver Dis. 2016;20:681-692. [PubMed] [DOI] [Cited in This Article: ] |
| 27. | Takeda H, Takai A, Inuzuka T, Marusawa H. Genetic basis of hepatitis virus-associated hepatocellular carcinoma: linkage between infection, inflammation, and tumorigenesis. J Gastroenterol. 2017;52:26-38. [PubMed] [DOI] [Cited in This Article: ] |
| 28. | Van Hees S, Michielsen P, Vanwolleghem T. Circulating predictive and diagnostic biomarkers for hepatitis B virus-associated hepatocellular carcinoma. World J Gastroenterol. 2016;22:8271-8282. [PubMed] [DOI] [Cited in This Article: ] |
| 29. | Dong Y, Wu G. Analysis of short and long term therapeutic effects of radiofrequency hyperthermia combined with conformal radiotherapy in hepatocellular carcinoma. J BUON. 2016;21:407-411. [PubMed] [Cited in This Article: ] |
| 30. | Jiang SS, Tang Y, Zhang YJ, Weng DS, Zhou ZG, Pan K, Pan QZ, Wang QJ, Liu Q, He J. A phase I clinical trial utilizing autologous tumor-infiltrating lymphocytes in patients with primary hepatocellular carcinoma. Oncotarget. 2015;6:41339-41349. [PubMed] [DOI] [Cited in This Article: ] |
| 31. | Liu H, Wang ZG, Fu SY, Li AJ, Pan ZY, Zhou WP, Lau WY, Wu MC. Randomized clinical trial of chemoembolization plus radiofrequency ablation versus partial hepatectomy for hepatocellular carcinoma within the Milan criteria. Br J Surg. 2016;103:348-356. [PubMed] [DOI] [Cited in This Article: ] |
| 32. | Liu PH, Hsu CY, Lee YH, Hsia CY, Huang YH, Su CW, Chiou YY, Lin HC, Huo TI. When to Perform Surgical Resection or Radiofrequency Ablation for Early Hepatocellular Carcinoma?: A Nomogram-guided Treatment Strategy. Medicine (Baltimore). 2015;94:e1808. [PubMed] [DOI] [Cited in This Article: ] |
| 33. | Nagamatsu H, Sumie S, Niizeki T, Tajiri N, Iwamoto H, Aino H, Nakano M, Shimose S, Satani M, Okamura S. Hepatic arterial infusion chemoembolization therapy for advanced hepatocellular carcinoma: multicenter phase II study. Cancer Chemother Pharmacol. 2016;77:243-250. [PubMed] [DOI] [Cited in This Article: ] |
| 34. | Zhang Y, Fan W, Wang Y, Lu L, Fu S, Yang J, Huang Y, Yao W, Li J. Sorafenib With and Without Transarterial Chemoembolization for Advanced Hepatocellular Carcinoma With Main Portal Vein Tumor Thrombosis: A Retrospective Analysis. Oncologist. 2015;20:1417-1424. [PubMed] [DOI] [Cited in This Article: ] |
| 35. | Fukumoto I, Kikkawa N, Matsushita R, Kato M, Kurozumi A, Nishikawa R, Goto Y, Koshizuka K, Hanazawa T, Enokida H. Tumor-suppressive microRNAs (miR-26a/b, miR-29a/b/c and miR-218) concertedly suppressed metastasis-promoting LOXL2 in head and neck squamous cell carcinoma. J Hum Genet. 2016;61:109-118. [PubMed] [DOI] [Cited in This Article: ] |
| 36. | Ge S, Zhang L, Xie J, Liu F, He J, He J, Wang X, Xiang T. MicroRNA-146b regulates hepatic stellate cell activation via targeting of KLF4. Ann Hepatol. 2016;15:918-928. [PubMed] [DOI] [Cited in This Article: ] |
| 37. | Hirata H, Hinoda Y, Ueno K, Nakajima K, Ishii N, Dahiya R. MicroRNA-1826 directly targets beta-catenin (CTNNB1) and MEK1 (MAP2K1) in VHL-inactivated renal cancer. Carcinogenesis. 2012;33:501-508. [PubMed] [DOI] [Cited in This Article: ] |
| 38. | Hu Y, Yi B, He S, Gao L, Liu P, Wang L, Yu J, Wan D, Zhou J, Zhu X. Clinical Significance of miR-1826 as a Novel Prognostic Biomarker in Colorectal Cancer. Anticancer Agents Med Chem. 2016;16:1109-1116. [PubMed] [Cited in This Article: ] |
| 39. | Tan S, Ding K, Li R, Zhang W, Li G, Kong X, Qian P, Lobie PE, Zhu T. Identification of miR-26 as a key mediator of estrogen stimulated cell proliferation by targeting CHD1, GREB1 and KPNA2. Breast Cancer Res. 2014;16:R40. [PubMed] [DOI] [Cited in This Article: ] |
| 40. | Trompeter HI, Dreesen J, Hermann E, Iwaniuk KM, Hafner M, Renwick N, Tuschl T, Wernet P. MicroRNAs miR-26a, miR-26b, and miR-29b accelerate osteogenic differentiation of unrestricted somatic stem cells from human cord blood. BMC Genomics. 2013;14:111. [PubMed] [DOI] [Cited in This Article: ] |
| 41. | Wang S, Chen Y, Bai Y. p21 participates in the regulation of anaplastic thyroid cancer cell proliferation by miR-146b. Oncol Lett. 2016;12:2018-2022. [PubMed] [DOI] [Cited in This Article: ] |
| 42. | Zhu Y, Lu Y, Zhang Q, Liu JJ, Li TJ, Yang JR, Zeng C, Zhuang SM. MicroRNA-26a/b and their host genes cooperate to inhibit the G1/S transition by activating the pRb protein. Nucleic Acids Res. 2012;40:4615-4625. [PubMed] [DOI] [Cited in This Article: ] |
| 43. | Yen CS, Su ZR, Lee YP, Liu IT, Yen CJ. miR-106b promotes cancer progression in hepatitis B virus-associated hepatocellular carcinoma. World J Gastroenterol. 2016;22:5183-5192. [PubMed] [DOI] [Cited in This Article: ] |
| 44. | Shen G, Jia H, Chen D, Zhang J. [Effects of miR-106b expression on the proliferation of human hepatocellular carcinoma cells]. Zhonghua Zhongliu Zazhi. 2014;36:489-495. [PubMed] [Cited in This Article: ] |
| 45. | Tan W, Li Y, Lim SG, Tan TM. miR-106b-25/miR-17-92 clusters: polycistrons with oncogenic roles in hepatocellular carcinoma. World J Gastroenterol. 2014;20:5962-5972. [PubMed] [DOI] [Cited in This Article: ] |
| 46. | Chen G, Ni Y, Nagata N, Xu L, Ota T. Micronutrient Antioxidants and Nonalcoholic Fatty Liver Disease. Int J Mol Sci. 2016;17:pii: E1379. [PubMed] [DOI] [Cited in This Article: ] |
| 47. | Choo SP, Tan WL, Goh BK, Tai WM, Zhu AX. Comparison of hepatocellular carcinoma in Eastern versus Western populations. Cancer. 2016; Epub ahead of print. [PubMed] [DOI] [Cited in This Article: ] |
| 48. | Colagrande S, Inghilesi AL, Aburas S, Taliani GG, Nardi C, Marra F. Challenges of advanced hepatocellular carcinoma. World J Gastroenterol. 2016;22:7645-7659. [PubMed] [DOI] [Cited in This Article: ] |
| 49. | Fu S, Zhou RR, Li N, Huang Y, Fan XG. Hepatitis B virus X protein in liver tumor microenvironment. Tumour Biol. 2016; Epub ahead of print. [PubMed] [DOI] [Cited in This Article: ] |
| 50. | Kubo N, Araki K, Kuwano H, Shirabe K. Cancer-associated fibroblasts in hepatocellular carcinoma. World J Gastroenterol. 2016;22:6841-6850. [PubMed] [DOI] [Cited in This Article: ] |
| 51. | Mathew S, Ali A, Abdel-Hafiz H, Fatima K, Suhail M, Archunan G, Begum N, Jahangir S, Ilyas M, Chaudhary AG. Biomarkers for virus-induced hepatocellular carcinoma (HCC). Infect Genet Evol. 2014;26:327-339. [PubMed] [DOI] [Cited in This Article: ] |
| 52. | Zhu M, Guo J, Li W, Xia H, Lu Y, Dong X, Chen Y, Xie X, Fu S, Li M. HBx induced AFP receptor expressed to activate PI3K/AKT signal to promote expression of Src in liver cells and hepatoma cells. BMC Cancer. 2015;15:362. [PubMed] [DOI] [Cited in This Article: ] |