Copyright
©The Author(s) 2016.
World J Gastroenterol. Aug 28, 2016; 22(32): 7275-7288
Published online Aug 28, 2016. doi: 10.3748/wjg.v22.i32.7275
Published online Aug 28, 2016. doi: 10.3748/wjg.v22.i32.7275
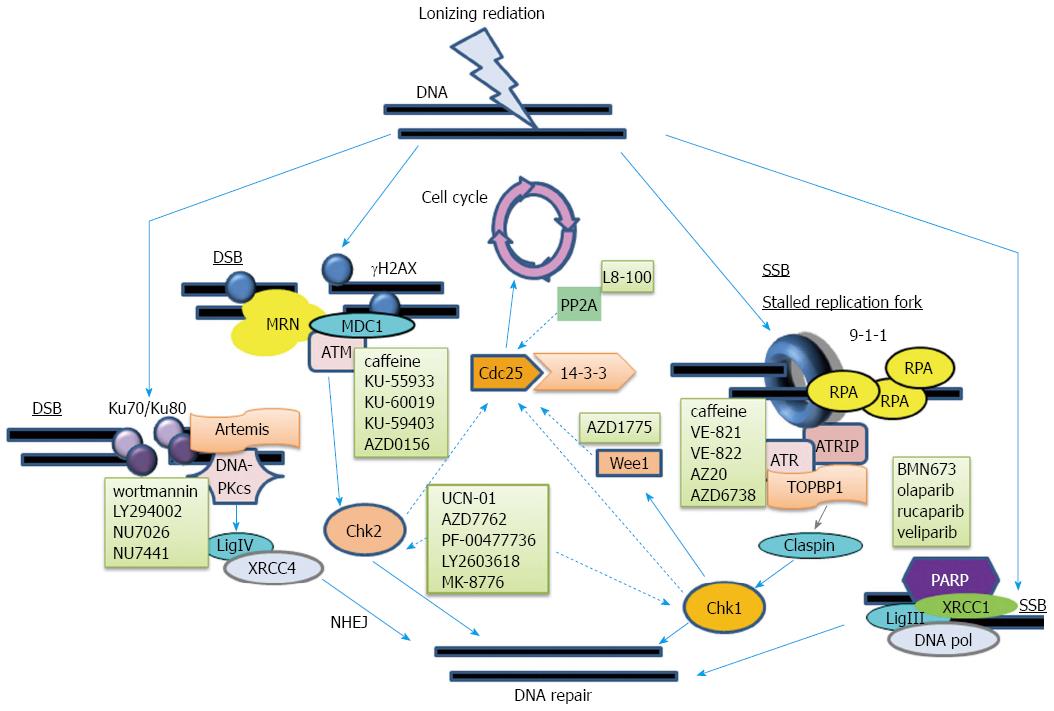
Figure 1 Major DNA repair pathways and molecular agents that inhibit DNA repair.
Here we demonstrate the main DNA repair pathways that are activated by IR-induced DNA damage. DNA repair machinery, with high affinity for ADP-ribose polymers, is recruited to DNA nicks after PARP binds SSB lesions; this executes DNA repair. RPA binds to ssDNA and recruits ATR and ATRIP. This is followed by recruitment of the 9-1-1 complex, TOPBP1, and claspin. Finally, Chk1 activates downstream repair machinery and phosphorylates Cdc25 proteins, which are inhibited by 14-3-3 proteins; this arrests the cell cycle. Wee1 and PP2A are also able to modulate the activity of Cdc25. DSB repair requires H2AX phosphorylation and recruitment of the MRN complex, MDC1, and ATM. Then Chk2 is phosphorylated, activated, and mediates repair and cell cycle arrest. NHEJ is the main IR-induced DSB repair method. It starts with Ku70/Ku80 heterodimer recruitment, which is followed by DNA-PKcs to form the active DNA-PK. Then DNA ligase IV and XRCC4 mediate DNA ligation. The respective molecular inhibitors for PARP, ATR, Chk1/Chk2, ATM, and DNA-PK are indicated. IR: Ionizing radiation; PARP: Poly ADP ribose polymerase; SSB: Single-strand break; DSB: Double-strand break; RPA: replication protein A; ATR: Ataxia-telangiectasia and Rad3 related; ATRIP: ATR-interacting protein; TOPBP1: Topoisomerase-binding protein-1; MRN: MRE11-RAD50-NBS1; NHEJ: Non-homologous end joining; ATM: Ataxia-telangiectasia mutated.
- Citation: Yang SH, Kuo TC, Wu H, Guo JC, Hsu C, Hsu CH, Tien YW, Yeh KH, Cheng AL, Kuo SH. Perspectives on the combination of radiotherapy and targeted therapy with DNA repair inhibitors in the treatment of pancreatic cancer. World J Gastroenterol 2016; 22(32): 7275-7288
- URL: https://www.wjgnet.com/1007-9327/full/v22/i32/7275.htm
- DOI: https://dx.doi.org/10.3748/wjg.v22.i32.7275