Published online Feb 28, 2015. doi: 10.3748/wjg.v21.i8.2343
Peer-review started: July 14, 2014
First decision: August 15, 2014
Revised: September 3, 2014
Accepted: November 19, 2014
Article in press: November 19, 2014
Published online: February 28, 2015
AIM: To find potential mutable sites by detecting mutations of the candidate gene in a kindred with polycystic liver disease (PCLD).
METHODS: First, we chose a kindred with PCLD and obtained five venous blood samples of this kindred after the family members signed the informed consent form. In the kindred two cases were diagnosed with PCLD, and the left three cases were normal individuals. All the blood samples were preserved at -85 °C. Second, we extracted the genomic DNA from the venous blood samples of the kindred using a QIAamp DNA Mini Kit and then performed long-range polymerase chain reaction (PCR) with different primers. The exons of PKD1 were all sequenced with the forward and reverse primers to ensure the accuracy of the results. Next, we purified the PCR products and directly sequenced them using Big Dye Terminator Chemistry version 3.1. The sequencing reaction was conducted with BiomekFX (Beckman). Finally, we analyzed the results.
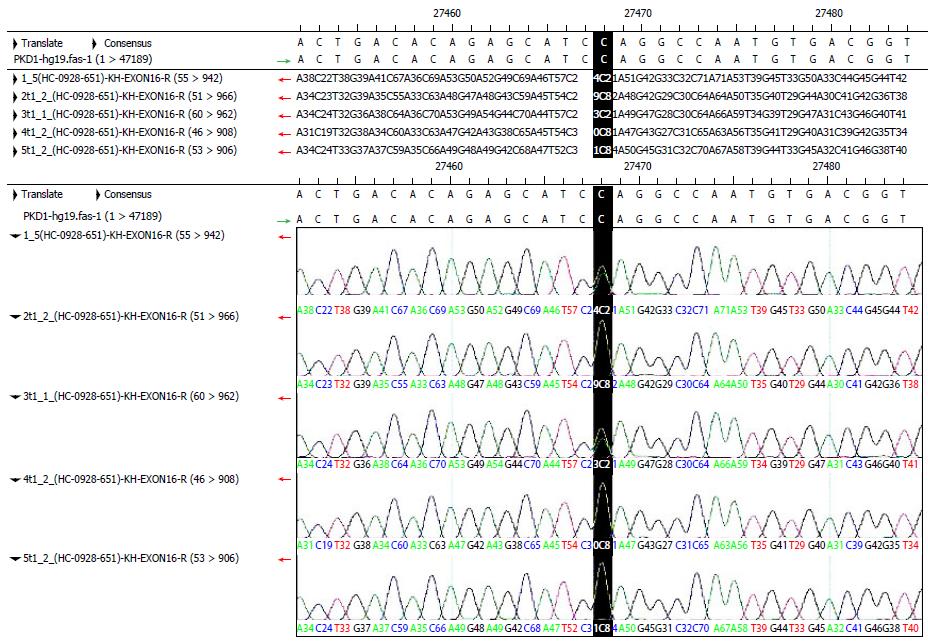
RESULTS: A total of 42 normal exons were identified in detecting mutations of the PKD1 gene. A synonymous mutation occurred in exon 5. The mutation was a homozygous T in the proband and was C in the reference sequence. This mutation was located in the third codon and did not change the amino acid encoded by the codon. Missense mutations occurred in exons 11 and 35. These mutations were located in the second codon; they changed the amino acid sequence and existed in the dbSNP library. A nonsense mutation occurred in exon 15. The mutation was a heterozygous CT in the proband and was C in the reference sequence. This mutation was located in the first codon and resulted in a termination codon. This mutation had an obvious influence on the encoded protein and changed the length of the protein from 4303 to 2246 amino acids. This was a new mutation that was not present in the dbSNP library.
CONCLUSION: The nonsense mutation of exon 15 existed in the proband and in the third individual. Additionally, the proband was heterozygous for this mutation, so the mutable site was a pathogenic mutation.
Core tip: This study explored the mutable sites in the PKD1 gene and found the potential mutable sites by detecting mutations of the candidate gene in a kindred with polycystic liver disease (PCLD). We analysed five blood samples obtained from the kindred by using long-range polymerase chain reaction and direct sequencing with different primers. We found synonymous mutations in exons 5, 11, 15, and 35 of the PKD1 gene. Only the mutation in exon 15 was not in the dbSNP library, and it was a heterozygous CT in the proband and was C in the reference sequence. This mutation was located in the first codon and resulted in a termination codon. This nonsense mutation existed also in the third individual, so the mutable site was a pathogenic mutation.
- Citation: Jin S, Cui K, Sun ZQ, Shen YY, Li P, Wang ZD, Li FF, Gong KN, Li S. Screening analysis of candidate gene mutations in a kindred with polycystic liver disease. World J Gastroenterol 2015; 21(8): 2343-2351
- URL: https://www.wjgnet.com/1007-9327/full/v21/i8/2343.htm
- DOI: https://dx.doi.org/10.3748/wjg.v21.i8.2343
Polycystic liver disease is a type of autosomal dominant polycystic liver disease. It can be divided into polycystic liver disease associated with autosomal dominant polycystic kidney disease (ADPKD) and the independent type of polycystic liver disease. Currently, four genes have been identified to be associated with the onset of this disease: the polycystic kidney disease genes PKD1 and PKD2 and the independent-type polycystic liver genes PRKCSH and SEC63. Polycystic liver disease often has a familial history. Simple polycystic liver disease is rare, and patients with polycystic liver disease account for more than half of the cases of ADPKD. The disease is common in women over 40 years old, and cysts can grow during pregnancy or from taking estrogen drugs. The patients with polycystic liver disease associated with ADPKD often have kidney damage. Renal complications are important factors that cause the death of patients. In the present study, we used long-range polymerase chain reaction (PCR) and direct sequencing with different sequencing primers for screening mutations in a kindred with polycystic liver disease, with aims to provide a theoretical basis to conduct genetic research of polycystic liver disease, assist in prenatal diagnosis in the future and clarify the patterns and characteristics of polycystic liver disease gene mutations.
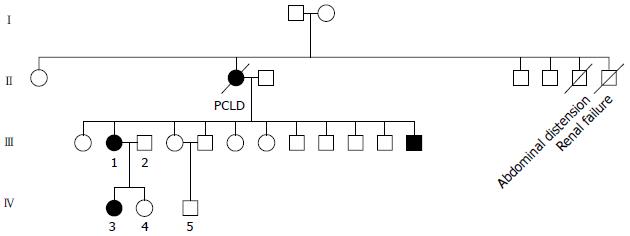
This study was approved by the medical Ethics Committee of the Shandong Cancer Hospital. We collected clinical data from the members of the polycystic liver disease pedigrees after they signed the informed consent form. Among 5 peripheral blood samples obtained, 2 were collected from patients (III1, IV3) diagnosed with polycystic liver disease and 3 from healthy subjects (III2, IV4, IV5). We stored the blood samples at -85 °C in an ultra-low temperature freezer.
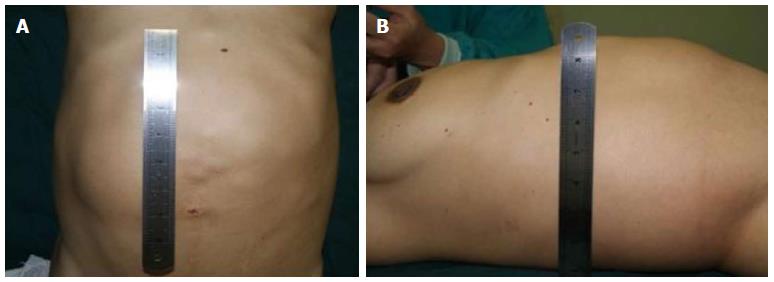
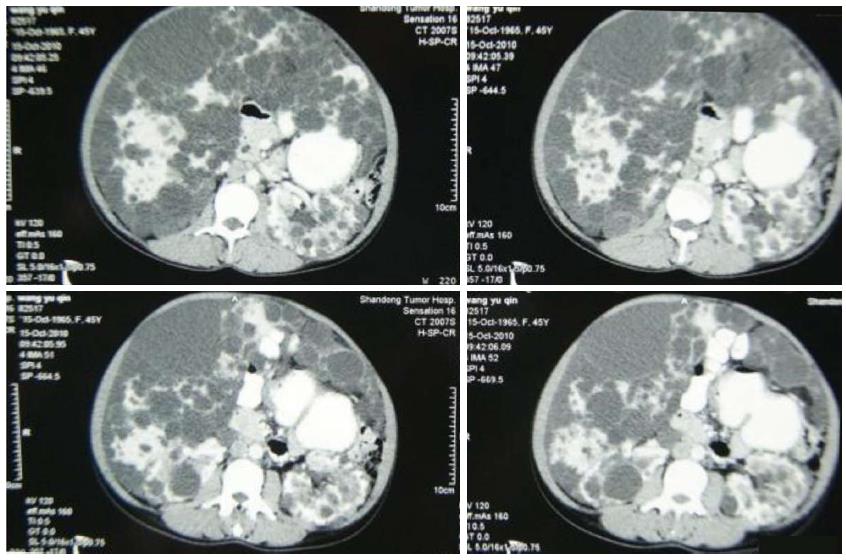
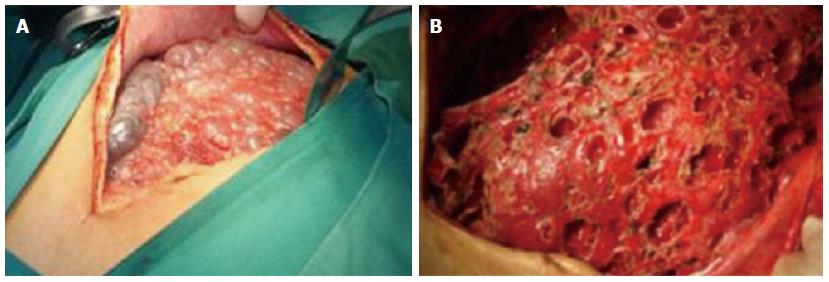
The clinical data of three generations of a kindred with polycystic liver disease are shown in Figure 1. The proband (III1) was a 44-year-old woman who was diagnosed with polycystic liver disease associated with ADPKD and had undergone an operation (Figures 2-4). Case III2 was the husband of the proband who was 46 years old and did not have the disease. Case IV3 was the older daughter of the proband who was 24 years old and also had polycystic liver disease. Case IV4 was the youngest daughter of the proband who did not have the disease. Case IV5 was the proband’s nephew who did not have the disease. The proband’s mother died from polycystic liver disease, and her two uncles both died from polycystic kidney disease.
We extracted the genomic DNA using the QIAamp DNA Extraction Kit (QIAGEN) from the peripheral venous blood samples of the members of the AKPLD kindred. The QIAamp DNA technique allows to extract genomic, mitochondrial, bacterial, parasitic and viral DNA from human tissue samples and can be used directly for PCR and marker detection. The method ensures obtaining highly purified DNA.
The PKD1 gene contains 46 exons, but the first 30 exons have repeat regions in the genome. The traditional method in which a single exon is amplified and sequenced cannot separate true genetic regions and repeat sequences. Therefore, the repeat regions were amplified twice by using nested PCR, but this was too troublesome. We used the method proposed by Ying-Cai Tan to synthesize all the PKD1 amplification and sequencing primers (Tables 1 and 2)[1]. The bidirectional sequencing of the PKD1 exons ensured the accuracy of the results.
| Fragment | Size (bp) | Exon | Forward primer | Reverse primer | Temperature(°C) |
| Exon 1 | 2278 | 1 | 5’-CGCAGCCTTACCATCCACCT-3’ | 5’-TCATCGCCCCTTCCTAAGCA-3’ | 64 |
| Exons 2-7 | 4041 | 2-7 | 5’-CCCCGAGTAGCTGGAACTACAGTTACACACT-3’ | 5’-CGTCCTGCTGTGCCAGAGGCG-3’ | 70 |
| Exons 8-12 | 3893 | 8-12 | 5’-ACGTCTGCGAGCTGCAGCCC-3’ | 5’-CTGCAGGGACAGGCGTCAGTGA-3’ | 70 |
| Exons 13-15 | 4391 | 13-15 | 5’-TGGAGGGAGGGACGCCAATC-3’ | 5’-GTCAACGTGGGCCTCCAAGT-3’ | 65 |
| Exons 15-21 | 5253 | 15-21 | 5’-ATCCCTGGGGGTCCTACCATCTCTTA-3’ | 5’-ACACAGGACAGAACGGCTGAGGCTA-3’ | 68 |
| Exons 22-26 | 3276 | 22-26 | 5’-ATGCTTAGTGAGGAGGCTGTGGGGGTCCA-3’ | 5’-GCTTAAAGGGGAATGGCTTAAACCCG-3’ | 70 |
| Exons 27-34 | 3916 | 27-34 | 5’-CGGGTCACCGGTTGTGGCA-3’ | 5’-ATGAGGCTCTTTCCACAGACAACAGAGGTT-3’ | 70 |
| Exons 35-41 | 2632 | 35-41 | 5’-CAAGAGGCTCAAGAAACTGCCCG-3’ | 5’-GGGCTGTGGAAGCCGCCTA-3’ | 68 |
| Exons 42-46 | 2370 | 42-46 | 5’-GAGTAGTTCTCCAGGAGTGCCG-3’ | 5’-ATTCTGCCTGGCCCTCGGCCTT-3’ | 63 |
| Primer name | Sequence | Genomic position (starting from the 5’ end) | Distance to exon-intron junction | Exons covered |
| PKD1-exon1-F | 5’-GCGTCGCTCAGCAGCAGGT-3’ | 2185830 | -140 Ex1 | 1 |
| PKD1-exon1-R | 5’-GCCCGCGTCCTGCTTCCC-3’ | 2185383 | 93 Ex1 | |
| PKD1-exon2-3-F | 5’-GGGATGCTGGCAATGTGTGGGAT-3’ | 2169468 | -89 Ex2 | 2-3 |
| PKD1-exon2-3-R | 5’-GGACCAACTGGGAGGGCAGAA-3’ | 2169038 | 77 Ex3 | |
| PKD1-exon4-F | 5’-GGCGGTGCTGTCAGGGTG-3’ | 2168948 | -102 Ex4 | 4 |
| PKD1-exon4-R | 5’-CCAGAGAGGCCTTCCTGAGC-3’ | 2168582 | 95 Ex4 | |
| PKD1-exon5A-F | 5’-GAACAGCATGGGAGCCTGTGAGT-3’ | 2168561 | -98 Ex5 | 5 |
| PKD1-exon5A-R | 5’-AGCCGGCCCAGCGGCATC-3’ | 2168033 | Within an exon | |
| PKD1-exon5B-F | 5’-GCCTGTCCCTCTGCTCCG-3’ | 2168262 | Within an exon | 5 |
| PKD1-exon5B-R | 5’-GTGTCAACGGTCAGTGTGGGC-3’ | 2167696 | 96 Ex5 | |
| PKD1-exon6-F | 5’-GTGTCTGCTGCCCACTCCC-3’ | 2167787 | -124 Ex6 | 6 |
| PKD1-exon6-R | 5’-CTCCTTCCTCCTGAGACTCCC-3’ | 2167397 | 93 Ex6 | |
| PKD1-exon7-F | 5’-GCTGCTGTGAGGGTGGGAGGA-3’ | 2167120 | -66 Ex7 | 7 |
| PKD1-exon7-R | 5’-TCCACCGCGGGCGCTCGGCA-3’ | 2166763 | 71 Ex7 | |
| PKD1-exon8-F | 5’-CTGGGCTGAGGAGGAGGG-3’ | 2166760 | -115 Ex8 | 8 |
| PKD1-exon8-R | 5’-GGGCACAAGCAACATTAAGGCCC-3’ | 2166413 | 117 Ex8 | |
| PKD1-exon9-F | 5’-CCTCTTCCTGGGAAGTTCGGGT-3’ | 2166206 | -87 Ex9 | 9 |
| PKD1-exon9-R | 5’-ACTCTGGTGGCCACAGGACCA-3’ | 2165895 | 98 Ex9 | |
| PKD1-exon10-F | 5’-GCAGGCAGTTGGGCATCTCTG-3’ | 2165697 | -71 Ex10 | 10 |
| PKD1-exon10-R | 5’-GACCCTGGGCAGCAGACAG-3’ | 2165309 | 70 Ex10 | |
| PKD1-exon11A-F | 5’-GTGTGGCTGACGAAGCGGG-3’ | 2165035 | -109 Ex11 | 11 |
| PKD1-exon11A-R | 5’-CCGTGGCGTTGGCACCAG-3’ | 2164455 | Within an exon | |
| PKD1-exon11B-F | 5’-CGCTATGAGGTCCGGGCAG-3’ | 2164641 | Within an exon | 11 |
| PKD1-exon11B-R | 5’-CCCTCACTGGGAAGCCAGG-3’ | 2164077 | 93 Ex11 | |
| PKD1-exon12-F | 5’-GGACTCTCCAGCCCGACG-3’ | 2163387 | -94 Ex12 | 12 |
| PKD1-exon12-R | 5’-GCAGAGGTGAAGGTGGAGC-3’ | 2163088 | 74 Ex12 | |
| PKD1-exon13-14-F | 5’-GTGGAGGGAGGGACGCCAA-3’ | 2163037 | -73 Ex13 | 13-14 |
| PKD1-exon13-14-R | 5’-GTCACAGTGAGGGCTGTTGGG-3’ | 2162230 | 111 Ex14 | |
| PKD1-exon15A-F | 5’-TTCTGCCGAGCGGGTGGG-3’ | 2161938 | -64 Ex15 | 15 |
| PKD1-exon15A-R | 5’-CATGTCGAAGGTCCACGTGATGT-3’ | 2161427 | Within an exon | |
| PKD1-exon15B-F | 5’-GACATGAGCCTGGCCGTGG-3’ | 2161516 | Within an exon | 15 |
| PKD1-exon15B-R | 5’-CCACCTCTGGCTCCACGCA-3’ | 2161027 | Within an exon | |
| PKD1-exon15C-F | 5’-CACGCGGAGCGGCACGTT-3’ | 2161121 | Within an exon | 15 |
| PKD1-exon15C-R | 5’-GGTGACCTCCGGACCCTC-3’ | 2160626 | Within an exon | |
| PKD1-exon15D-F | 5’-TCTGCTGTGGGCCGTGGG-3’ | 2160706 | Within an exon | 15 |
| PKD1-exon15D-R | 5’-CTGTACCGTGTGGTTGGTGGG-3’ | 2160215 | Within an exon | |
| PKD1-exon15E-F | 5’-ACAGCATCTTCGTCTATGTCCTG-3’ | 2160303 | Within an exon | 15 |
| PKD1-exon15E-R | 5’-GGTTCCCTGCCGTCATGGTG-3’ | 2159812 | Within an exon | |
| PKD1-exon15F-F | 5’-GGGCTGAGCTGGGAGACCT-3’ | 2159899 | Within an exon | 15 |
| PKD1-exon15F-R | 5’-GACAGCTGAGCCGGCAGC-3’ | 2159417 | Within an exon | |
| PKD1-exon15G-F | 5’-CTGTGGGCCAGCAGCAAGGT-3’ | 2159494 | Within an exon | 15 |
| PKD1-exon15G-R | 5’-CGTGCGGTTCTCACTGCCCA-3’ | 2159012 | Within an exon | |
| PKD1-exon15H-F | 5’-GACGTCACCTACACGCCCG-3’ | 2159095 | Within an exon | 15 |
| PKD1-exon15H-R | 5’-CCTCCCAGCGGTACTCAGTCT-3’ | 2158603 | Within an exon | |
| PKD1-exon15I-F | 5’-GATGCGGCGATCACAGCGCA-3’ | 2158688 | Within an exon | 15 |
| PKD1-exon15I-R | 5’-GGCCAGCCCTGGTGGCAA-3’ | 2158164 | 89 Ex15 | |
| PKD1-exon16-F | 5’-GGCCCGTCCTCAGTGCCT-3’ | 2158167 | -134 Ex16 | 16 |
| PKD1-exon16-R | 5’-GCGGCCTCCACCAGCACTA-3’ | 2157790 | 94 Ex16 | |
| PKD1-exon17-18-F | 5’-GAAACCTGGAGTTTGGGAGCAGC-3’ | 2157047 | -98 Ex17 | 17-18 |
| PKD1-exon17-18-R | 5’-TGACGTCACAGAGTCGGG-3’ | 2156344 | 55 Ex18 | |
| PKD1-exon19-20-F | 5’-GCACGGGTGAGTGCAGGC-3’ | 2156404 | -99 Ex19 | 19-20 |
| PKD1-exon19-20-R | 5’-CCGGGATGAGCCCTCTGCAA-3’ | 2155768 | 98 Ex20 | |
| PKD1-exon21-F | 5’-AGTCGTGGGCATCTGCTGGC-3’ | 2155550 | -75 Ex21 | 21 |
| PKD1-exon21-R | 5’-CAAGCTGCCCGTCTGCCCT-3’ | 2155240 | 83 Ex21 | |
| PKD1-exon22-F | 5’-CAGGTGAGGACCCGTGTAGAGA-3’ | 2154725 | -82 Ex22 | 22 |
| PKD1-exon22-R | 5’-GGGAGGAGGGAGGCAGAG-3’ | 2154431 | 68 Ex22 | |
| PKD1-exon23A-F | 5’-GCACCTCGCTCTCTGCCC-3’ | 2154024 | -128 Ex23 | 23 |
| PKD1-exon23A-R | 5’-GCCACCTTGGTGGAGACGG-3’ | 2153512 | Within an exon | |
| PKD1-exon23B-F | 5’-GGCTGCCACTTCTCCATCCC-3’ | 2153651 | Within an exon | 23 |
| PKD1-exon23B-R | 5’-GACACCCATGGAAGCCCTACG-3’ | 2153183 | 84 Ex23 | |
| PKD1-exon24-F | 5’-CGTGGCAGAGGGTGGGCT-3’ | 2153064 | -93 Ex24 | 24 |
| PKD1-exon24-R | 5’-CTCGCTGCCTGCCGTCCC-3’ | 2152721 | 94 Ex24 | |
| PKD1-exon25-26-F | 5’-GGCTCTGAGACTGCGACATCCAA-3’ | 2152702 | -68 Ex25 | 25-26 |
| PKD1-exon25-26-R | 5’-CTTGTTCTGACGCCTGCGACG-3’ | 2151964 | 98 Ex26 | |
| PKD1-exon27-28-F | 5’-GCTGAGATGACTTGCCTGGGATG-3’ | 2150644 | -77 Ex27 | 27-28 |
| PKD1-exon27-28-R | 5’-ACTGCAGGAGGCCACGGG-3’ | 2150094 | 73 Ex28 | |
| PKD1-exon29-30-F | 5’-CTCCGTGGGAGGTTGGGCA-3’ | 2150142 | -70 Ex29 | 29-30 |
| PKD1-exon29-30-R | 5’-CGCCTTTCCCTCTGGCTGC-3’ | 2149542 | 103 Ex30 | |
| PKD1-exon31-32F | 5’-CGGGCTCTGTCCTGTCTGC-3’ | 2148079 | -94 Ex31 | 31-32 |
| PKD1-exon31-32R | 5’-CCCAGCAAGGACACGCAGC-3’ | 2147642 | 87 Ex32 | |
| PKD1-exon33-34-F | 5’-GGAAGCCCAGGGTGTCCGT-3’ | 2147595 | -91 Ex33 | 33-34 |
| PKD1-exon33-34-R | 5’-CAGCCCTGCCCTGGCACC-3’ | 2147034 | 115 Ex34 | |
| PKD1-exon35-F | 5’-CAAGAGGCTCAAGAAACTGCCCG-3’ | 2144309 | -98 Ex35 | 35-36 |
| PKD1-exon36-R | 5’-GAGAAGTACAGGGCTTCCAGCAA-3’ | 2143714 | 98 Ex36 | |
| PKD1-exon37-F | 5’-CTCGGCTGGGAGCCACTG-3’ | 2143832 | -93 Ex37 | 37 |
| PKD1-exon37-R | 5’-GCCTTCTGAGGTGAGGAAAGGG-3’ | 2143439 | 106 Ex37 | |
| PKD1-exon38-F | 5’-CCACACCTGCCGCAGCCAT-3’ | 2143190 | -96 Ex38 | 38 |
| PKD1-exon38-R | 5’-CAAAGGTATCTACACATGTCCAC-3’ | 2142856 | 99 Ex38 | |
| PKD1-exon39-F | 5’-GCCAGCAGGGCAGTGGGA-3’ | 2142698 | -105 Ex39 | 39 |
| PKD1-exon39-R | 5’-CAGCTAGGGAGCAGGGCTGA-3’ | 2142385 | 96 Ex39 | |
| PKD1-exon40-F | 5’-GTGGCGCCGAACCAGAGC-3’ | 2142289 | -100 Ex40 | 40-41 |
| PKD1-exon41-R | 5’-GGGCTGTGGAAGCCGCCTA-3’ | 2141678 | 104 Ex41 | |
| PKD1-exon42-F | 5’-CCTCAGCCACGCCTGCACT-3’ | 2141678 | -80 Ex42 | 42 |
| PKD1-exon42-R | 5’-GGGTGAGACGCTGCCGGG-3’ | 2141338 | 86 Ex42 | |
| PKD1-exon43-F | 5’-CAGCGTCCCTCCCGCCCT-3’ | 2141213 | -38 Ex43 | 43-44 |
| PKD1-exon44-R | 5’-CAGGAAGACACGAGCTGCGG-3’ | 2140578 | 97 Ex44 | |
| PKD1-exon45-F | 5’-GCTGGCCATCCTGGTAGGTGA-3’ | 2140687 | -96 Ex45 | 45 |
| PKD1-exon45-R | 5’-GGACTTTGTGGCGGAACTGGG-3’ | 2140180 | 106 Ex45 | |
| PKD1-exon46-F | 5’-GGAGAGGGACACGCCCTG-3’ | 2140264 | -69 Ex46 | 46 |
| PKD1-exon46-R | 5’-ATTCTGCCTGGCCCTCGGCCTT-3’ | 2139635 | Within 3’UTR |
The PCR reaction setup is shown in Table 3. Because the amplified products were too long, we added dimethyl sulfoxide (DMSO) in the system to improve the efficiency. The PCR conditions were as follows: (1) fragment one: 94 °C for 4 min; followed by 35 cycles of 94 °C for 40 s, 62 °C for 1 min, and 65 °C for 150 s, with a final extension step at 65 °C for 10 min; (2) fragments two and three: Touchdown protocol composed of an initial step of 94 °C for 2 min; followed by 14 cycles of 94 °C for 30 s, 74 °C for 60 s, with a decrease of 0.5 °C per cycle, and 65 °C for 4 min; followed by 30 cycles of 94 °C for 30 s, 65 °C for 60 s, and 65 °C for 4 min, with a final extension step of 65 °C for 10 min; (3) fragment four: 94 °C for 2 min, followed by 35 cycles of 94 °C for 20 s and 65 °C for 5.5 min, with a final extension step at 65 °C for 10 min; (4) fragment five: 94 °C for 2 min, followed by 35 cycles of 94 °C for 30 s, 68 °C for 30 s, and 65 °C for 5 min, with a final extension step at 65 °C for 10 min; and (5) fragments six to nine: 94 °C for 2 min; followed by 14 cycles of 94 °C for 30 s, 66 °C for 30 s with a decrease of 0.5 °C per cycle, and 65 °C for 3.5 min; followed by 30 cycles of 94 °C for 30 s, 59 °C for 30 s, and 65 °C for 3.5 min, with a final extension step of 65 °C for 10 min.
| Polymerase chain reaction reaction system | Volume (μL) |
| LongAmp® Taq DNA polymerase | 1 |
| LongAmp Taq reaction buffer | 5 |
| Deoxynucleotide solution mix | 3 |
| Dimethyl sulfoxide | 1.25 |
| Forward primer (10 μmol/L) | 1 |
| Reverse primer (10 μmol/L) | 1 |
| DNA template | 1 |
| H2O | 11.75 |
| Total volume | 25 |
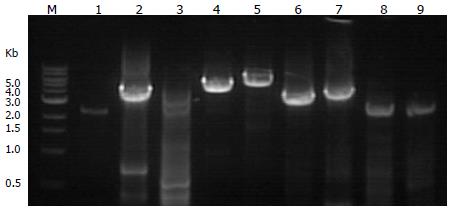
The long-range PCR amplification products were analyzed by 2% agarose gel electrophoresis. In Figure 5, PCR amplification products for fragments 1 to 9 are shown from the left to the right. The number and size of the fragments are shown in Table 1.
The DNA was sequenced by the DNA dideoxy sequencing method using a Biomek FX Beckman fluid moving workstation. We purified the PCR amplification products using the Zymoclean Gel DNA Recovery Kit (D4001), quantified the DNA with a NanoDrop 8000 spectrophotometer and sequenced the DNA with an ABI 3730XL automated DNA sequencer.
A total of 42 normal exons were identified in detecting mutations of the PKD1 gene. A synonymous mutation occurred in exon 5. The mutation was a homozygous T in the proband, which was C in the reference sequence. It was located in the third codon and did not change the amino acid encoded by the codon. Missense mutations occurred in exons 11 and 35. These mutations were located in the second codon; they changed the amino acid sequence and existed in the dbSNP library. A nonsense mutation occurred in exon 15. This mutation was a heterozygous CT in the proband, which was C in the reference sequence. It was located in the first codon and resulted in a termination codon. This was a new mutation that was not deposited in the dbSNP library.
Polycystic liver disease can be divided into two types according to its clinical manifestations: polycystic liver disease related to polycystic kidney disease and the independent type of polycystic liver disease[2]. At present, etiological studies have delved deep into the levels of molecules and genes, and four genes have been found to be clearly associated with the onset of this disease: PKD1 and PKD2 in polycystic kidney disease and PRKCSH and SEC63[3-6] in the independent type of polycystic liver disease. PKD1 is located on the short arm of chromosome 16 (16p13.3)[7], and PKD2 is located on the long arm of chromosome 4[8]; the incidence rate of mutations in these genes is approximately 1/500. The PKD1 gene contains 46 exons, is transcribed into an mRNA of 14.1 kb in length and encodes a protein consisting of 4303 amino acids; this protein is a membrane protein of the receptor type. The PKD2 gene contains 15 exons, is transcribed into an mRNA of 5.3 kb in length, and encodes a protein containing 968 amino acids. In total, 85% of the patients with polycystic kidney disease have the PKD1 mutation(s), and 15% have the PKD2 mutation(s). The mutations are evenly distributed in the two genes; there is no hotspot region of mutation. Some studies have shown that the location of the mutations may affect the severity of the clinical phenotype. Because the PKD1 gene is larger and there are homologous regions in the genome, genetic testing can be troublesome. The main tool for detection is based on PCR amplification, followed by sequencing of the corresponding exon region. The non-repetitive regions of the PKD2 and PKD1 genes can be directly amplified and sequenced, but amplification of the repetitive regions of PKD1 require nested PCR: primers for long-range PCR should be designed in the location where the homology of repetitive regions is not high so that a single exon could be amplified and sequenced. However, this method is not only cumbersome but also expensive. In this study, we used a method according to an article published this year that omitted the nested PCR step; thus, PKD1 can be effectively detected. Independent polycystic liver disease is caused by a mutation of the PRKCSH gene on chromosome 19 (19p13.2-13.1)[9] or the SEC63 gene on chromosome 6 (6q21-q23)[10]. PRKCSH[11] contains 18 exons, is transcribed into a 1.6 kb mRNA molecule and encodes a protein containing 527 amino acids; this protein is an endoplasmic reticulum protein. The SEC63 gene has 21 exons, is transcribed into an mRNA of 3.4 kb in length and encodes a protein containing 760 amino acids. Mutations of these two genes may affect the integrity of the membrane protein.
The possible pathogenic mechanisms of the mutations may be as follows: (1) Primary cilia are a core part of the signal transduction system. If there are mutations in the genes encoding the cilial proteins, this will change the cilial structure and disorder the reaction and transmission. These mutations will reduce the internal flow of Ca2+ and increase the concentration of cAMP[12], resulting in the hyperplasia of bile duct epithelial cells and the formation of a cyst[13]; (2) Liver cyst Sec proteins are involved in the synthesis of new proteins in the endoplasmic reticulum. The transport of translated and synthesized new proteins and the modification of the N-terminal glycosylation are mainly related to glucosidase II. Mutated liver cyst proteins cannot be bound to glucosidase, which results in mature defects of newly synthesized proteins[14]; (3) The further expansion after the formation of small cysts may be related to the following factors[15]: the pressure of the cystic cavity, the reconstruction of the surrounding matrix, the formation of new blood vessels, regulating factors, and so on. In the epithelial cyst model, when the secretion of cystic fluid and intracapsular pressure increase, the proliferation of epithelial cells is significantly sped up[16]. After the surrounding tissue is invaded by hyperplastic epithelial cells, metalloproteinases can remodel the matrix. At present, the enzyme has been separated from the epithelial cells of patients with the independent type of polycystic liver disease. The formation of new blood vessels can maintain the development of the epithelium and endothelium of the cyst[17]. The factor produced by the liver cyst epithelium can activate the vascular endothelial growth factor signaling pathway and lead to endothelial cell proliferation and differentiation. After estrogen stimulation, insulin-like growth factor I acts on the intrahepatic bile duct epithelium, and the bile duct epithelial cells proliferate rapidly[18]. A clinical investigation showed that multiparas were more likely to have a liver cyst; the patients who had used oral contraceptives or postmenopausal hormone replacement therapy may have had more and larger liver cysts[19-22]; and (4) The “two hit” theory[23]: in animal models, obvious cystic degeneration was present only when the animals had damage to both PKD alleles. This showed that cyst phenotypic development required the “two hit” condition. The first hit was the bud mutation of the initiative PKD1 or PKD2. The second hit was a single somatic mutation which caused the hyperplasia of luminal epithelial cells. At present, this theory has been confirmed in polycystic liver disease related to polycystic kidney disease but not in the independent type of polycystic liver disease.
The main surgical treatment methods for polycystic liver disease are: (1) percutaneous liver cyst puncture drainage and/or anhydrous alcohol injection treatment; (2) liver cyst opening the window to the top decompression; (3) liver resection or liver resection combined with cyst fenestration; (4) hepatic artery interventional embolization; and (5) liver transplantation. In recent years, the study of gene therapy has made remarkable achievements, especially in the treatment of genetic diseases. However, polycystic liver disease gene therapy research currently focuses on the in vitro study of cysts of liver cells; studies have not reported on these genes in vivo. At present, gene therapy is not yet fully mature[24-27].
According to the detection results, we found that the mutation[3] in exon 15 of the proband was a heterozygous CT mutation in the polycystic liver disease kindred. Through PCR amplification and DNA sequencing of this site in other individuals with this kindred, we found that the third individual with AKPKD also carried this nonsense mutation, which was not present in the remaining healthy individuals (Figure 6). The mutation was not included in the dbSNP library, but we discovered a record of it in the PKD mutation database. This mutation was located at the first position of the codon and changed the encoded amino acid into a termination codon; this obviously influenced the encoded protein. Therefore, the mutation was genetic with the disease in the kindred and was considered a pathogenic mutation. In the future, we can carry out an experiment to confirm the above results by developing an animal model. In addition, the characteristics of the clinical phenotype in this kindred showed that renal involvement mainly occurred in male patients, and female patients usually had the disease manifestation in the liver and kidney but primarily the liver. The results showed that hormones had a certain influence on the clinical phenotype, but the mechanism was not clear and requires further study. This study provided a new idea for investigating molecular genetic principles and mechanisms in polycystic liver disease and for operating polycystic liver disease screening, diagnosis and treatment in the future.
The authors thank the patient for her permission to report this case.
Polycystic liver disease (PCLD) is an autosomal dominant polycystic liver disease. It can be divided into polycystic liver disease associated with polycystic kidney and the independent type polycystic liver disease. Now four genes have been found to be associated with the onset of this disease and there are polycystic kidney disease genes PKD1 and PKD2 and the independent type polycystic liver genes PRKCSH and SEC63. There have been many studies on the biological and gene therapy of PCLD recently.
Recently the authors have made great achievements in the research of the genetic treatment, especially in the treatment of hereditary diseases. It has been a hotspot in the medical research. The gene therapy is the best treatment for hereditary diseases in theory. So the research of pathogenic genes provides a theoretical basis for the gene therapy.
The authors extracted the genomic DNA by using QIAamp DNA Extraction Kit from the peripheral venous blood samples of the members of the AKPLD family. The technique of QIAamp DNA can extract the genomic, mitochondrial, bacterial, parasitic and viral DNA from the human tissue samples and can be directly used for PCR and marker detection. The method can ensure obtaining high purity DNA. The five blood samples obtained from the family were analysed by using long-range polymerase chain reaction and direct sequencing with different primers.
A total of 42 normal exons were identified in detecting mutations of the PKD1 gene. The synonymous mutation occurred in the exon 5. The mutation was homozygous T in the proband and was C in the reference sequence. It was located at the third codon and did not change the amino acid coded by this codon. The missense mutation occurred in the exons 11 and 35. They were located at the second codon, changed amino acid sequence and exited in the dbSNP library. The nonsense mutation occurred in the exon 15. The mutation was CT heterozygous in the proband and was C in the reference sequence. It was located at the first codon and resulted in a termination codon. It was a new mutation and was not deposited in the dbSNP library.
Autosomal dominant inheritable disease is that the pathogenic genes are located in autosomes and a single gene mutation can cause the disease. The nonsense mutation is that the code which the base stands for becomes a termination codon due to the change of the base, and makes the peptide chain stop synthesis ahead of time.
This is a good retrospective study in which the authors analyzed the mutable sites in the polycystic liver disease gene and found the potential mutable sites by detecting mutations of the candidate gene in a family with PCLD. In the detection of the PCLD gene mutations, only 2158432(hg19) of the exon 15 was not in the dbSNP library and the proband was the heterozygosis. All were accord with the dominant mode of inheritance. By detecting other individuals’ the same mutable sites. We found the third individual also have it. So the mutable site was a pathogenic mutations and was genetic.
P- Reviewer: Celikbilek M, Sazci A S- Editor: Qi Y L- Editor: Wang TQ E- Editor: Zhang DN
| 1. | Tan YC, Michaeel A, Blumenfeld J, Donahue S, Parker T, Levine D, Rennert H. A novel long-range PCR sequencing method for genetic analysis of the entire PKD1 gene. J Mol Diagn. 2012;14:305-313. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 34] [Cited by in F6Publishing: 36] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 2. | Everson GT, Taylor MR. Management of polycystic liver disease. Curr Gastroenterol Rep. 2005;7:19-25. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 26] [Cited by in F6Publishing: 17] [Article Influence: 1.1] [Reference Citation Analysis (0)] |
| 3. | Reynolds DM, Falk CT, Li A, King BF, Kamath PS, Huston J, Shub C, Iglesias DM, Martin RS, Pirson Y. Identification of a locus for autosomal dominant polycystic liver disease, on chromosome 19p13.2-13.1. Am J Hum Genet. 2000;67:1598-1604. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 78] [Cited by in F6Publishing: 77] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 4. | Tahvanainen P, Tahvanainen E, Reijonen H, Halme L, Kääriäinen H, Höckerstedt K. Polycystic liver disease is genetically heterogeneous: clinical and linkage studies in eight Finnish families. J Hepatol. 2003;38:39-43. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 38] [Cited by in F6Publishing: 40] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 5. | Praetorius HA, Spring KR. The renal cell primary cilium functions as a flow sensor. Curr Opin Nephrol Hypertens. 2003;12:517-520. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 213] [Cited by in F6Publishing: 189] [Article Influence: 9.0] [Reference Citation Analysis (0)] |
| 6. | Everson GT, Taylor MR, Doctor RB. Polycystic disease of the liver. Hepatology. 2004;40:774-782. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 479] [Cited by in F6Publishing: 521] [Article Influence: 13.4] [Reference Citation Analysis (0)] |
| 7. | Reeders ST, Breuning MH, Davies KE, Nicholls RD, Jarman AP, Higgs DR, Pearson PL, Weatherall DJ. A highly polymorphic DNA marker linked to adult polycystic kidney disease on chromosome 16. Nature. 1985;317:542-544. [PubMed] [Cited in This Article: ] |
| 8. | Gabow PA. Autosomal dominant polycystic kidney disease. N Engl J Med. 1993;329:332-342. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 721] [Cited by in F6Publishing: 665] [Article Influence: 21.5] [Reference Citation Analysis (0)] |
| 9. | Tanner GA, McQuillan PF, Maxwell MR, Keck JK, McAteer JA. An in vitro test of the cell stretch-proliferation hypothesis of renal cyst enlargement. J Am Soc Nephrol. 1995;6:1230-1241. [PubMed] [Cited in This Article: ] |
| 10. | Saggar-Malik AK, Jeffery S, Patton MA. Autosomal dominant polycystic kidney disease. BMJ. 1994;308:1183-1184. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 3] [Cited by in F6Publishing: 3] [Article Influence: 0.1] [Reference Citation Analysis (0)] |
| 11. | Perrone RD, Grubman SA, Rogers LC, Lee DW, Moy E, Murray SL, Torres VE, Jefferson DM. Continuous epithelial cell lines from ADPKD liver cysts exhibit characteristics of intrahepatic biliary epithelium. Am J Physiol. 1995;269:G335-G345. [PubMed] [Cited in This Article: ] |
| 12. | Pei Y. A “two-hit” model of cystogenesis in autosomal dominant polycystic kidney disease? Trends Mol Med. 2001;7:151-156. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 87] [Cited by in F6Publishing: 79] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 13. | Drenth JP, Martina JA, Te Morsche RH, Jansen JB, Bonifacino JS. Molecular characterization of hepatocystin, the protein that is defective in autosomal dominant polycystic liver disease. Gastroenterology. 2004;126:1819-1827. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 51] [Cited by in F6Publishing: 49] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 14. | Drenth JP, Tahvanainen E, te Morsche RH, Tahvanainen P, Kääriäinen H, Höckerstedt K, van de Kamp JM, Breuning MH, Jansen JB. Abnormal hepatocystin caused by truncating PRKCSH mutations leads to autosomal dominant polycystic liver disease. Hepatology. 2004;39:924-931. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 40] [Cited by in F6Publishing: 39] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 15. | Davila S, Furu L, Gharavi AG, Tian X, Onoe T, Qian Q, Li A, Cai Y, Kamath PS, King BF. Mutations in SEC63 cause autosomal dominant polycystic liver disease. Nat Genet. 2004;36:575-577. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 213] [Cited by in F6Publishing: 180] [Article Influence: 9.0] [Reference Citation Analysis (0)] |
| 16. | Li A, Davila S, Furu L, Qian Q, Tian X, Kamath PS, King BF, Torres VE, Somlo S. Mutations in PRKCSH cause isolated autosomal dominant polycystic liver disease. Am J Hum Genet. 2003;72:691-703. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 152] [Cited by in F6Publishing: 132] [Article Influence: 6.3] [Reference Citation Analysis (0)] |
| 17. | Li TJ, Zhang HB, Lu JH, Zhao J, Yang N, Yang GS. Treatment of polycystic liver disease with resection-fenestration and a new classification. World J Gastroenterol. 2008;14:5066-5072. [PubMed] [DOI] [Cited in This Article: ] [Cited by in CrossRef: 18] [Cited by in F6Publishing: 13] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 18. | Dan AA, Younossi ZM. Quality of life and liver transplantation in patients with polycystic liver disease. Liver Transpl. 2006;12:1184-1185. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 8] [Cited by in F6Publishing: 8] [Article Influence: 0.4] [Reference Citation Analysis (0)] |
| 19. | Martin IJ, McKinley AJ, Currie EJ, Holmes P, Garden OJ. Tailoring the management of nonparasitic liver cysts. Ann Surg. 1998;228:167-172. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 88] [Cited by in F6Publishing: 92] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 20. | Everson GT, Helmke SM, Doctor B. Advances in management of polycystic liver disease. Expert Rev Gastroenterol Hepatol. 2008;2:563-576. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 20] [Cited by in F6Publishing: 19] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 21. | Torres VE, Harris PC. Autosomal dominant polycystic kidney disease: the last 3 years. Kidney Int. 2009;76:149-168. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 410] [Cited by in F6Publishing: 392] [Article Influence: 26.1] [Reference Citation Analysis (0)] |
| 22. | Masyuk T, Masyuk A, LaRusso N. Cholangiociliopathies: genetics, molecular mechanisms and potential therapies. Curr Opin Gastroenterol. 2009;25:265-271. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 76] [Cited by in F6Publishing: 75] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 23. | Spirli C, Okolicsanyi S, Fiorotto R, Fabris L, Cadamuro M, Lecchi S, Tian X, Somlo S, Strazzabosco M. ERK1/2-dependent vascular endothelial growth factor signaling sustains cyst growth in polycystin-2 defective mice. Gastroenterology. 2010;138:360-371.e7. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 74] [Cited by in F6Publishing: 83] [Article Influence: 5.9] [Reference Citation Analysis (0)] |
| 24. | Torrice A, Cardinale V, Gatto M, Semeraro R, Napoli C, Onori P, Alpini G, Gaudio E, Alvaro D. Polycystins play a key role in the modulation of cholangiocyte proliferation. Dig Liver Dis. 2010;42:377-385. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 13] [Cited by in F6Publishing: 13] [Article Influence: 0.9] [Reference Citation Analysis (0)] |
| 25. | Drenth JP, Martina JA, van de Kerkhof R, Bonifacino JS, Jansen JB. Polycystic liver disease is a disorder of cotranslational protein processing. Trends Mol Med. 2005;11:37-42. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 68] [Cited by in F6Publishing: 57] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 26. | Nauli SM, Alenghat FJ, Luo Y, Williams E, Vassilev P, Li X, Elia AE, Lu W, Brown EM, Quinn SJ. Polycystins 1 and 2 mediate mechanosensation in the primary cilium of kidney cells. Nat Genet. 2003;33:129-137. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1565] [Cited by in F6Publishing: 1466] [Article Influence: 69.8] [Reference Citation Analysis (0)] |
| 27. | Drenth JP, te Morsche RH, Smink R, Bonifacino JS, Jansen JB. Germline mutations in PRKCSH are associated with autosomal dominant polycystic liver disease. Nat Genet. 2003;33:345-347. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 174] [Cited by in F6Publishing: 156] [Article Influence: 7.4] [Reference Citation Analysis (0)] |