Published online Jan 21, 2015. doi: 10.3748/wjg.v21.i3.897
Peer-review started: June 3, 2014
First decision: June 27, 2014
Revised: August 28, 2014
Accepted: September 29, 2014
Article in press: September 30, 2014
Published online: January 21, 2015
AIM: To study the association of apolipoprotein E (APOE) polymorphisms with the susceptibility of inflammatory bowel disease (IBD) in Saudi patients.
METHODS: APOE genotyping was performed to evaluate the allele and genotype frequencies in 378 Saudi subjects including IBD patients with ulcerative colitis (n = 84) or Crohn’s disease (n = 94) and matched controls (n = 200) using polymerase chain reaction and reverse-hybridization techniques.
RESULTS: The frequencies of the APOE ε2 allele and ε2/ε3 and ε2/ε4 genotypes were significantly higher in IBD patients than in controls (P < 0.05), suggesting that the ε2 allele and its heterozygous genotypes may increase the susceptibility to IBD. On the contrary, the frequencies of the ε3 allele and ε3/ε3 genotype were lower in IBD patients as compared to controls, suggesting a protective effect of APOE ε3 for IBD. The prevalence of the ε4 allele was also higher in the patient group compared to controls, suggesting that the ε4 allele may also increase the risk of IBD. Our results also indicated that the APOE ε4 allele was associated with an early age of IBD onset. No effect of gender or type of IBD (familial or sporadic) on the frequency distribution of APOE alleles and genotypes was noticed in this study.
CONCLUSION: APOE polymorphism is associated with risk of developing IBD and early age of onset in Saudi patients, though further studies with a large-size population are warranted.
Core tip: This study shows apolipoprotein E (APOE) polymorphism is associated with risk of developing inflammatory bowel disease (IBD) in Saudi patients. Allele ε2 and its heterozygous genotypes increase the susceptibility to IBD, whereas the ε3 allele and ε3/ε3 genotype are protective for IBD. The APOE ε4 allele also increases the risk for IBD and is associated with early age at onset. The frequency distribution of APOE alleles and genotypes is not affected by gender or type of IBD (familial or sporadic).
- Citation: Al-Meghaiseeb ES, Al-Otaibi MM, Al-Robayan A, Al-Amro R, Al-Malki AS, Arfin M, Al-Asmari AK. Genetic association of apolipoprotein E polymorphisms with inflammatory bowel disease. World J Gastroenterol 2015; 21(3): 897-904
- URL: https://www.wjgnet.com/1007-9327/full/v21/i3/897.htm
- DOI: https://dx.doi.org/10.3748/wjg.v21.i3.897
The inflammatory bowel diseases (IBDs), encompassing Crohn’s disease (CD; OMIM 266600) and ulcerative colitis (UC; OMIM 191390), are chronic inflammatory disorders of the gastrointestinal tract. IBD has emerged as a global disease with increasing incidence and prevalence in different parts of the world[1-5]. The precise etiology of IBD is still unknown, but available evidence suggests that it is a complex multifactorial disease in which immune dysregulation caused by genetic and/or environmental factors plays an important role[6-8]. IBD appears to be caused by immunogenic responses against environmental factors and/or microbes inhabiting the distal ileum and colon of genetically susceptible hosts.
The incidence of IBD is higher in North America and Europe than in Asia and Africa, possibly due to the variation in environmental factors and genetic makeup. The hygiene hypothesis was suggested to be responsible for the rising prevalence of various autoimmune and inflammatory disorders in developed populations, which are thought to result from the lack of early exposure to bacterial infections due to good sanitary conditions[9]. The changes in dietary and intestinal microbial milieu are thought to play a key pathogenic role in the etiology of IBD, however the precise environmental factors influencing IBD prevalence have not been determined yet[10]. Intriguingly, the characteristics of Western and Asian IBD patients differ in epidemiology, phenotype and genetic susceptibility[11-14], highlighting ethnic variations.
Various epidemiologic and population-based studies have indicated that genetic factors contribute to the pathogenesis of IBD[15-17]. Apolipoprotein E (APOE) has an important role in cholesterol and lipid metabolism, and has also been shown to alter both innate and adaptive immune responses[18]. Several studies have indicated that APOE inhibits the production of T lymphocytes and regulates immune reactions by interacting with several cytokines[19-21]. Further, it has been suggested that APOE plays a key role in regulating immune response in various autoimmune diseases[22-24].
The gene encoding APOE is located on chromosome 19. It has 3 polymorphic alleles (ε2, ε3 and ε4) differing from one another by the presence of either a C or T nucleotide at codons 112 and 158. These alleles encode three different isoproteins differing significantly in structure and function, including receptor binding capacity and lipid metabolism[25]. By different combinations of these three alleles, six genotypes (ε2/ε2, ε3/ε3, ε2/ε3, ε3/ε4, ε2/ε4, and ε4/ε4) are formed[26,27]. Although the frequency of these alleles/genotypes varies significantly among different ethnic populations, APOE ε3/ε3 is the most common genotype and ε3 the most predominant allele in majority of the population[28,29]. Several studies have indicated an association between APOE alleles and genotypes with onset and severity of various autoimmune diseases[24,30-33]. Recently, association of APOE allele/genotype with UC has been reported in Chinese patients[34,35]. In this study, we examined the APOE allele/genotype frequencies in Saudi CD and UC patients and matched controls.
A total of 378 Saudi subjects including 178 IBD patients visiting the Gastroenterology Clinic and 200 age- and sex-matched healthy donors visiting the community health clinic of Prince Sultan Military Medical City, Riyadh were recruited in this study. Venous blood was collected from all the patients and controls. IBD patients were divided into familial (n = 20) and sporadic (n = 158) forms. They were grouped into patients with CD (n = 94, including 56 men and 38 women) with a mean age of 32 years (range: 17-65 years), and patients with UC (n = 84, including 34 men and 50 women) with mean age of 34 years (range: 22-68 years). Two hundred healthy Saudis (120 men and 80 women) were included in the study as controls. None of the controls had any history of IBD, diabetes, rheumatoid arthritis, systemic lupus erythematosus or other autoimmune diseases. The diagnoses of IBD (CD and UC) was based on the conventional endoscopic, radiologic, and histologic criteria as describe by Lennard-Jones[36]. Patient information such as age at diagnosis, disease location, disease characteristics, and extraintestinal location were used to divide the patients into groups. Patients with any other autoimmune disease or having clinical features of both UC and CD (intermediate colitis) were excluded from the study. Patients with CD were also assessed on the basis of the Montreal classification[37]. This study was approved by the ethical committee of PSMMC and written informed consent was obtained from all the subjects.
Genomic DNA was extracted from the blood of IBD patients and controls using QIAamp DNA mini kit (Qiagen, Venlo, Limburg, the Netherlands). APOE genotyping was performed using an APOE StripAssay kit based on a PCR and reverse-hybridization technique (ViennaLab Diagnostics GmbH, Vienna, Austria). To cross-check the results, the APOE genotyping was also performed by PCR and restriction fragment length polymorphism technique as previously described[38].
Briefly, genomic DNA (200-300 ng) was amplified in 25 μL reaction tubes for 40 cycles of 94 °C for 30 s, 68 °C for 10 s, 72 °C for 1 min; PCR products obtained were separated by electrophoresis on 1.5% agarose gel in TAE buffer, and visualized by ethidium bromide fluorescence. Fragments with the expected size were cut from the gel, purified using a GFX PCR DNA Gel band purification kit (GE Healthcare, Little Chalfont, Buckinghamshire, UK). Purified DNA was digested with HhaI enzyme and separated by agarose gel electrophoresis to identify the genotype. The frequencies of various genotypes in patients and controls were determined and compared. Both the above-mentioned procedures yielded completely matching results.
Frequencies of various alleles and genotypes for APOE polymorphism were analyzed by Fisher’s exact test and a P < 0.05 was considered as significant. The strength of the association of disease with respect to a particular allele/genotype is expressed by odd ratio interpreted as relative risk (RR) according to the method of Woolf as outlined by Schallreuter et al[39]. The RR was calculated only for those alleles and genotypes that were increased or decreased in IBD patients as compared to normal Saudis. RR was calculated using the following formula:
RR = (a×d)/(b×c)
Where a is number of patients expressing the allele or genotype; b is the number of patients without allele or genotype expression; c is number of controls expressing the allele or genotype; and d is the number of controls without allele or genotype expression.
The etiologic fraction (EF) indicates the hypothetical genetic component of the disease. EF values of > 0.00-0.99 are significant. It is calculated for positive associations (RR > 1) using the following formula proposed by Svejgaard et al[40]:
EF = (RR-1)f/RR, where f = a/(a +b)
Preventive fraction (PF) indicates the hypothetical protective effect of one allele/genotype for a disease. It is calculated for negative associations (RR < 1) using the following formula[40]:
PF = (1-RR)f/[RR (1-f)] + f , where f = a/(a +b)
Values of < 1.0 indicate the protective effect of an allele/genotype against the manifestation of disease.
The results of APOE genotyping in the IBD patients and the healthy controls are summarized in Tables 1, 2, 3 4 and 5. In both the IBD patient and control groups the genotype distributions were in Hardy-Weinberg equilibrium. The ε2 allele was present in 7.59% of IBD patients, while altogether absent in controls (P < 0.01) (Table 1). The frequency of allele ε4 was also significantly higher in patients compared with controls (P < 0.01), whereas the frequency of the ε3 allele was significantly lower (P < 0.01).
| Allele | IBD (n = 356) | Control (n = 400) | P value | RR | EF/PF |
| ε3 | 293 (82.30) | 383 (95.75) | < 0.01 | 0.206 | 0.549 |
| ε4 | 36 (10.11) | 17 (4.25) | < 0.01 | 2.531 | 0.411 |
| ε2 | 27 (7.59) | 0 (0) | < 0.01 | - | - |
The frequency of various genotypes of APOE also showed variations in patient and control groups. The prevalences of genotypes ε2/ε3, and ε2/ε4 were 13.48, and 6.18% in patients, while totally absent in the control group (P < 0.01) (Table 2). The difference in the frequencies of the ε3/ε4 genotype was not statistically significant between the patient and control groups, albeit that there is a trend towards a higher frequency in IBD patients. The frequency of the ε3/ε3 genotype was significantly higher in controls than that in IBD patients (P < 0.01). The genotypes ε2/ε2 and ε4/ε4 were absent in both patients and controls.
| Genotype | IBD (n = 178) | Control (n = 200) | P value | RR | EF/PF |
| ε3/ε3 | 118 (66.29) | 183 (91.5) | < 0.01 | 0.183 | 0.637 |
| ε3/ε4 | 25 (14.05) | 17 (8.5) | 0.10 | 1.759 | 0.256 |
| ε2/ε3 | 24 (13.48) | 0 (0) | < 0.01 | - | - |
| ε2/ε4 | 11 (6.18) | 0 (0) | < 0.01 | - | - |
| ε2/ε2 | 0 (0) | 0 (0) | - | - | - |
| ε4/ε4 | 0 (0) | 0 (0) | - | - | - |
The frequencies of alleles and genotypes of APOE polymorphism were not significantly different in male and female patients, except for the ε3 allele and homozygous ε3/ε3 genotype, which were present in significantly higher frequencies in female than male patients (P < 0.05) (Table 3).
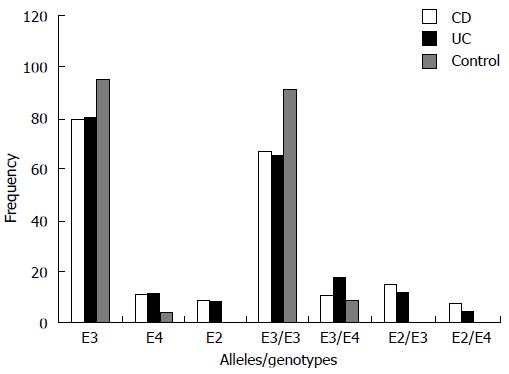
The difference in the frequencies of APOE alleles and genotypes in the CD and UC patients was not significant (Table 4, Figure 1). Moreover, when compared with controls separately, an almost similar pattern was noticed for both UC and CD, except that the frequency of genotype ε3/ε4 was significantly higher in UC patients (P = 0.03), but not in CD patients (P = 0.66), as compared to controls. However, the RR values calculated for the ε3/ε4 genotype in UC and CD (RR = 2.34 and 1.28, respectively) indicated a similar positive association for both. Similarly, the stratification of IBD patients into familial and sporadic forms showed no significant difference in the frequency distribution of either alleles or genotypes of APOE (Table 5). The APOE ε4 allele was significantly associated with an early age of onset in IBD (P≤ 0.05). The groups of patients with genotype ε3/ε4 (n = 25), and ε2/ε4 (n = 11) had lower age of onset than the patients with genotype ε3/ε3 and ε2/ε3.
| Genotype/allele | Familial (n = 20) | Sporadic (n = 158) | P value |
| ε3/ε3 | 14 (70.00) | 104 (65.82) | 0.80 |
| ε3/ε4 | 4 (20.00) | 21 (13.30) | 0.49 |
| ε2/ε3 | 2 (10.00) | 22 (13.92) | 1.00 |
| ε2/ε4 | 0 (0) | 11 (6.96) | 0.61 |
| ε3 | 34 (85.00) | 251 (79.43) | 0.52 |
| ε4 | 4 (10.00) | 32 (10.13) | 1.00 |
| ε2 | 2 (5.00) | 33 (10.44) | 0.40 |
Our results showed a higher frequency of the APOE ε2 allele and predominance of ε2/ε3 and ε2/ε4 genotypes in IBD patients in comparison with matched controls, suggesting that allele ε2 carriers are at a higher risk of developing IBD. The APOE ε2 isoprotein differs from the APOE ε3 isoprotein by one amino acid, at position 158, with ε2 containing cysteine and ε3 containing arginine. This single amino acid difference causes a marked reduction in binding capacity of APOE ε2 to the low density lipoprotein family of receptors[25], which in turn results in severe metabolic disturbances, particularly type III hyperlipidemia. Additionally, the two cysteines in APOE ε2 (positions 112 and 158) allow it to form disulfide-linked multimeric protein complexes[41]. These unique properties of APOE ε2 may contribute to its role in the etiology of IBD and other lipid-associated diseases.
Disturbances in the lipid, apolipoprotein, and lipoprotein profiles and cholesterol efflux in IBD patients have been reported[42-44]. Thus, genetic variations of apoproteins, essential in lipoprotein metabolism, may affect susceptibility to IBD. APOE is involved in transport and metabolism of cholesterol, triglyceride and other lipids. The lipid transporting and catabolic activity in APOE ε2 carriers is significantly slower compared to ε3 and ε4 carriers, due to low receptor binding affinity of ε2. Individuals with APOE ε2 are unable to efficiently clear lipids from plasma/tissues, which facilitates the accumulation of chylomicron, very low density lipoprotein and lipids[45]. It has been suggested that APOE protein might be involved in the pathogenesis of diseases via the sequestration of lipids contributing to the epidermal barrier function[46].
We also observed a significantly higher frequency of the ε2/ε3 genotype in Saudi IBD patients as compared to matched controls. This genotype has been associated with significant imbalance in lipids and lipoprotein metabolism, as well as with ischemic cerebrovascular diseases[47,48]. Parameters associated with atherosclerosis, such as inflammation, carotid intima media thickness, homocysteine and insulin resistance, are increased in IBD as reported by several researchers[49-53]. In addition, several studies have suggested that IBD is a risk factor for ischemic heart diseases, including atherosclerosis[49,53,54]. Furthermore, it has been reported that IBD is an independent predictor of hypertriglyceridemia[55] and hypocholesterolemia[56].
Results of this study showed a higher frequency of the ε4 allele in patients group compared to controls, suggesting that it also may increase the risk of IBD. Similarly, a higher frequency of the ε4 allele has been reported in Chinese UC patients[34]. These authors therefore suggested that APOE ε4 confers greater risk for the development of UC in Chinese. Our results indicate that allele ε4 increases the risk for both UC and CD in Saudi patients. The ε4 allele of the APOE gene is an established risk factor for low bone mineral density[57,58], and the high frequency of APOE ε4 in UC and CD patients may be responsible for low bone mineral density in patients with UC[34,59]. To the best of our knowledge, no published report has indicated any association of APOE polymorphism with CD, and this is the first report showing a significant association with both CD and UC.
APOE is multifunctional in nature, and the presence of APOE ε4 has been associated with an enhanced inflammatory immune response[60-62]. Though the exact mechanisms by which APOE ε4 regulates the innate immune response is far from clear. Significantly higher levels of the pro-inflammatory cytokines tumor necrosis factor-α and interleukin-6 have been reported in animals expressing the ε4 allele compared to those with the ε3 allele[60]. Increased oxidative stress in the APOE ε4 cells has been suggested to contribute to higher cytokine production by enhancing the activation of nuclear factor-κB[63]. Moreover, increased expression of interleukin-1β, macrophage inflammatory protein (MIP)-1α, and tumor necrosis factor-α, as well as the transactivation of nuclear factor-κB, have been observed in APOE ε4 macrophages[64]. Recently Li et al[34] postulated that the epistatic interaction of MIP-1α and APOE polymorphism may contribute to individual variation in MIP-1α levels in mucosa of UC patients.
Our results also indicate that the APOE ε4 allele is associated with early age at onset of IBD. Polymorphism in the APOE gene has been defined as a modifying factor for age at onset in neurodegenerative and autoimmune diseases[30,65,66]. Our results are also in accordance with various reports showing an association of the ε4 allele with early onset of some autoimmune and neurodegenerative diseases[30,64,67,68]. The APOE ε4 allele is believed to be responsible for reducing high-density lipoprotein and increasing low-density lipoprotein in high-fat intake individuals[69], which are critical risk factors for occlusive lipid disorders. The implication of APOE ε4 in lipid metabolism and development of immunologic responses to lipid antigens may contribute to IBD in Saudis with high-fat intake as reported earlier for psoriasis[70-72]. APOE ε4 has also been linked to lower C-reactive protein, and it has been suggested that this effect is a consequence of intrinsic functional differences among the ε2, ε3, and ε4 APOE proteins in plasma[73]. Our results also show that association of APOE polymorphism was not affected by the sex of the host and the association was similar in both CD and UC.
In conclusion, this study shows a significant relation between APOE polymorphisms and IBD. The ε2 allele is associated with increased susceptibility for IBD, whereas the ε3 allele may be protective for IBD in Saudis. In addition, the ε4 allele may be a risk factor of severity or early onset of IBD. However, this association of APOE polymorphisms with the risk of IBD warrants further studies with a larger population. Similar studies on different ethnic populations will be helpful in defining the role of APOE as a putative pharmacologic target for IBD.
The authors thank S. Sadaf Rizvi and Mohammad Al-Asmari for their help with laboratory work.
Inflammatory bowel diseases (IBD), including ulcerative colitis, and Crohn’s disease, are chronic inflammatory disorders of the gastrointestinal tract. The precise etiology of IBD is still unknown, but available data suggests a definite role of immune dysregulation caused by genetic and/or environmental factors. Apolipoprotein E (APOE) plays a pivotal role in immunogenic response by interacting with several cytokines and regulating macrophage functions. Therefore, the role of APOE polymorphism was studied in Saudi patients with IBD.
The gene encoding APOE is located on chromosome 19 and has three polymorphic alleles (ε2, ε3 and ε4) differing from one another by the presence of either a C or T nucleotide at codons 112 and 158. Alleles ε2, ε3 and ε4 encode different APOE isoproteins, which not only differ in structure, but also in function, including receptor binding capacity and lipid metabolism. The frequency of APOE alleles varies significantly among different ethnic populations. Several studies have indicated an association between APOE alleles and genotypes with onset and severity of various autoimmune diseases. Such association studies will help in the better prognosis and treatment of various autoimmune diseases.
There are increasing prevalences of obesity and lipid disorders in the Saudi population due to sedentary lifestyle, lack of exercise, and unique dietary habits of rich fat, sugar and red meat. Being a closed society with high rate of consanguinity, it is ideal for genetic association studies. However, the genetic studies on IBD/other autoimmune disorders in KSA and other Arab countries are scarce and inconclusive. This is the first report from a Saudi population showing the role of APOE polymorphism in the etiology of ulcerative colitis and Crohn’s disease.
The study results suggest that APOE polymorphism is associated with risk of developing IBD in Saudi patients. The ε2 allele and its heterozygous genotypes increase the susceptibility to IBD, whereas the ε3 allele and ε3/ε3 genotype are protective. The APOE ε4 allele also increases the risk for IBD and is associated with early age at onset. Similar studies on different ethnic populations will be helpful in defining the role of APOE as a putative pharmacologic target for IBD. Understanding this relationship may be potentially useful for predicting the vulnerability of individuals/populations to various autoimmune diseases.
In this study, the authors studied the association between APOE polymorphism and IBD in a Saudi Arabian population.
P- Reviewer: Il Kim T, Kochhar R, Sperti C S- Editor: Qi Y L- Editor: AmEditor E- Editor: Wang CH
| 1. | Cosnes J, Gower-Rousseau C, Seksik P, Cortot A. Epidemiology and natural history of inflammatory bowel diseases. Gastroenterology. 2011;140:1785-1794. [PubMed] [Cited in This Article: ] |
| 2. | Fadda MA, Peedikayil MC, Kagevi I, Kahtani KA, Ben AA, Al HI, Sohaibani FA, Quaiz MA, Abdulla M, Khan MQ. Inflammatory bowel disease in Saudi Arabia: a hospital-based clinical study of 312 patients. Ann Saudi Med. 2012;32:276-282. [PubMed] [DOI] [Cited in This Article: ] |
| 3. | Gunisetty S, Tiwari S, Bardia A, Phanibhushan M, Satti V, Habeeb M, Khan A. The epidemiology and prevalence of Ulcerative colitis in the South of India. O J Immunol. 2012;2:144-148. [DOI] [Cited in This Article: ] [Cited by in Crossref: 3] [Cited by in F6Publishing: 3] [Article Influence: 0.3] [Reference Citation Analysis (0)] |
| 4. | Molodecky NA, Soon IS, Rabi DM, Ghali WA, Ferris M, Chernoff G, Benchimol EI, Panaccione R, Ghosh S, Barkema HW. Increasing incidence and prevalence of the inflammatory bowel diseases with time, based on systematic review. Gastroenterology. 2012;142:46-54.e42; quiz e30. [PubMed] [Cited in This Article: ] |
| 5. | Zeng Z, Zhu Z, Yang Y, Ruan W, Peng X, Su Y, Peng L, Chen J, Yin Q, Zhao C. Incidence and clinical characteristics of inflammatory bowel disease in a developed region of Guangdong Province, China: a prospective population-based study. J Gastroenterol Hepatol. 2013;28:1148-1153. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 108] [Cited by in F6Publishing: 125] [Article Influence: 11.4] [Reference Citation Analysis (0)] |
| 6. | Podolsky DK. Inflammatory bowel disease. N Engl J Med. 2002;347:417-429. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 2693] [Cited by in F6Publishing: 2685] [Article Influence: 122.0] [Reference Citation Analysis (2)] |
| 7. | Strober W, Fuss I, Mannon P. The fundamental basis of inflammatory bowel disease. J Clin Invest. 2007;117:514-521. [PubMed] [Cited in This Article: ] |
| 8. | Xavier RJ, Podolsky DK. Unravelling the pathogenesis of inflammatory bowel disease. Nature. 2007;448:427-434. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 2894] [Cited by in F6Publishing: 3117] [Article Influence: 183.4] [Reference Citation Analysis (8)] |
| 9. | Gent AE, Hellier MD, Grace RH, Swarbrick ET, Coggon D. Inflammatory bowel disease and domestic hygiene in infancy. Lancet. 1994;343:766-767. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 236] [Cited by in F6Publishing: 240] [Article Influence: 8.0] [Reference Citation Analysis (1)] |
| 10. | Matricon J, Barnich N, Ardid D. Immunopathogenesis of inflammatory bowel disease. Self Nonself. 2010;1:299-309. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 137] [Cited by in F6Publishing: 150] [Article Influence: 15.0] [Reference Citation Analysis (0)] |
| 11. | Yang SK, Hong WS, Min YI, Kim HY, Yoo JY, Rhee PL, Rhee JC, Chang DK, Song IS, Jung SA. Incidence and prevalence of ulcerative colitis in the Songpa-Kangdong District, Seoul, Korea, 1986-1997. J Gastroenterol Hepatol. 2000;15:1037-1042. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 140] [Cited by in F6Publishing: 141] [Article Influence: 5.9] [Reference Citation Analysis (0)] |
| 12. | Ling KL, Ooi CJ, Luman W, Cheong WK, Choen FS, Ng HS. Clinical characteristics of ulcerative colitis in Singapore, a multiracial city-state. J Clin Gastroenterol. 2002;35:144-148. [PubMed] [Cited in This Article: ] |
| 13. | Inoue N, Tamura K, Kinouchi Y, Fukuda Y, Takahashi S, Ogura Y, Inohara N, Núñez G, Kishi Y, Koike Y. Lack of common NOD2 variants in Japanese patients with Crohn’s disease. Gastroenterology. 2002;123:86-91. [PubMed] [Cited in This Article: ] |
| 14. | Leong RW, Lau JY, Sung JJ. The epidemiology and phenotype of Crohn’s disease in the Chinese population. Inflamm Bowel Dis. 2004;10:646-651. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 162] [Cited by in F6Publishing: 166] [Article Influence: 8.3] [Reference Citation Analysis (0)] |
| 15. | Kim ES, Kim WH. Inflammatory bowel disease in Korea: epidemiological, genomic, clinical, and therapeutic characteristics. Gut Liver. 2010;4:1-14. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 68] [Cited by in F6Publishing: 69] [Article Influence: 4.9] [Reference Citation Analysis (0)] |
| 16. | Yun J, Xu CT, Pan BR. Epidemiology and gene markers of ulcerative colitis in the Chinese. World J Gastroenterol. 2009;15:788-803. [PubMed] [Cited in This Article: ] |
| 17. | Waterman M, Xu W, Stempak JM, Milgrom R, Bernstein CN, Griffiths AM, Greenberg GR, Steinhart AH, Silverberg MS. Distinct and overlapping genetic loci in Crohn‘s disease and ulcerative colitis: correlations with pathogenesis. Inflamm Bowel Dis. 2011;17:1936-1942. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 82] [Cited by in F6Publishing: 94] [Article Influence: 7.2] [Reference Citation Analysis (0)] |
| 18. | Laskowitz DT, Lee DM, Schmechel D, Staats HF. Altered immune responses in apolipoprotein E-deficient mice. J Lipid Res. 2000;41:613-620. [PubMed] [Cited in This Article: ] |
| 19. | Yin M, Zhang L, Sun XM, Mao LF, Pan J. Lack of apoE causes alteration of cytokines expression in young mice liver. Mol Biol Rep. 2010;37:2049-2054. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 17] [Cited by in F6Publishing: 15] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 20. | Baitsch D, Bock HH, Engel T, Telgmann R, Müller-Tidow C, Varga G, Bot M, Herz J, Robenek H, von Eckardstein A. Apolipoprotein E induces antiinflammatory phenotype in macrophages. Arterioscler Thromb Vasc Biol. 2011;31:1160-1168. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 209] [Cited by in F6Publishing: 229] [Article Influence: 17.6] [Reference Citation Analysis (0)] |
| 21. | Zhang H, Wu LM, Wu J. Cross-talk between apolipoprotein E and cytokines. Mediators Inflamm. 2011;2011:949072. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 96] [Cited by in F6Publishing: 118] [Article Influence: 9.1] [Reference Citation Analysis (0)] |
| 22. | Zhang HL, Wu J. Apolipoprotein E4 and psoriasis. Arch Dermatol Res. 2010;302:151. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 3] [Cited by in F6Publishing: 3] [Article Influence: 0.2] [Reference Citation Analysis (0)] |
| 23. | Postigo J, Genre F, Iglesias M, Fernández-Rey M, Buelta L, Carlos Rodríguez-Rey J, Merino J, Merino R. Exacerbation of type II collagen-induced arthritis in apolipoprotein E-deficient mice in association with the expansion of Th1 and Th17 cells. Arthritis Rheum. 2011;63:971-980. [PubMed] [Cited in This Article: ] |
| 24. | Song LJ, Liu WW, Fan YC, Qiu F, Chen QL, Li XF, Ding F. The positive correlations of apolipoprotein E with disease activity and related cytokines in systemic lupus erythematosus. Diagn Pathol. 2013;8:175. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 10] [Cited by in F6Publishing: 11] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 25. | Artiga MJ, Bullido MJ, Sastre I, Recuero M, García MA, Aldudo J, Vázquez J, Valdivieso F. Allelic polymorphisms in the transcriptional regulatory region of apolipoprotein E gene. FEBS Lett. 1998;421:105-108. [PubMed] [Cited in This Article: ] |
| 26. | Utermann G, Hees M, Steinmetz A. Polymorphism of apolipoprotein E and occurrence of dysbetalipoproteinaemia in man. Nature. 1977;269:604-607. [PubMed] [Cited in This Article: ] |
| 27. | Hatters DM, Peters-Libeu CA, Weisgraber KH. Apolipoprotein E structure: insights into function. Trends Biochem Sci. 2006;31:445-454. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 371] [Cited by in F6Publishing: 385] [Article Influence: 21.4] [Reference Citation Analysis (0)] |
| 28. | Yin R, Pan S, Wu J, Lin W, Yang D. Apolipoprotein E gene polymorphism and serum lipid levels in the Guangxi Hei Yi Zhuang and Han populations. Exp Biol Med (Maywood). 2008;233:409-418. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 10] [Cited by in F6Publishing: 13] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 29. | Al-Dabbagh NM, Al-Dohayan N, Arfin M, Tariq M. Apolipoprotein E polymorphisms and primary glaucoma in Saudis. Mol Vis. 2009;15:912-919. [PubMed] [Cited in This Article: ] |
| 30. | Pertovaara M, Lehtimäki T, Rontu R, Antonen J, Pasternack A, Hurme M. Presence of apolipoprotein E epsilon4 allele predisposes to early onset of primary Sjogren’s syndrome. Rheumatology (Oxford). 2004;43:1484-1487. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 22] [Cited by in F6Publishing: 24] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 31. | Mooyaart AL, Valk EJ, van Es LA, Bruijn JA, de Heer E, Freedman BI, Dekkers OM, Baelde HJ. Genetic associations in diabetic nephropathy: a meta-analysis. Diabetologia. 2011;54:544-553. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 168] [Cited by in F6Publishing: 167] [Article Influence: 12.8] [Reference Citation Analysis (0)] |
| 32. | Maehlen MT, Provan SA, de Rooy DP, van der Helm-van Mil AH, Krabben A, Saxne T, Lindqvist E, Semb AG, Uhlig T, van der Heijde D. Associations between APOE genotypes and disease susceptibility, joint damage and lipid levels in patients with rheumatoid arthritis. PLoS One. 2013;8:e60970. [PubMed] [Cited in This Article: ] |
| 33. | Al Harthi F, Huraib GB, Zouman A, Arfin M, Tariq M, Al-Asmari A. Apolipoprotein E gene polymorphism and serum lipid profile in Saudi patients with psoriasis. Dis Markers. 2014;2014:239645. [PubMed] [Cited in This Article: ] |
| 34. | Li K, Wang B, Sui H, Liu S, Yao S, Guo L, Mao D. Polymorphisms of the macrophage inflammatory protein 1 alpha and ApoE genes are associated with ulcerative colitis. Int J Colorectal Dis. 2009;24:13-17. [PubMed] [Cited in This Article: ] |
| 35. | Liang WD, Yang JF, Yan J, Jin J, Li KS, Li JS, Bi YT. [Association of combined polymorphisms in MIP-1α and ApoE genes with the susceptibility of inflammatory bowel disease]. Zhonghua Yixue Zazhi. 2011;91:1250-1253. [PubMed] [Cited in This Article: ] |
| 36. | Lennard-Jones JE. Classification of inflammatory bowel disease. Scand J Gastroenterol Suppl. 1989;170:2-6; discussion 16-19. [PubMed] [Cited in This Article: ] |
| 37. | Silverberg MS, Satsangi J, Ahmad T, Arnott ID, Bernstein CN, Brant SR, Caprilli R, Colombel JF, Gasche C, Geboes K. Toward an integrated clinical, molecular and serological classification of inflammatory bowel disease: report of a Working Party of the 2005 Montreal World Congress of Gastroenterology. Can J Gastroenterol. 2005;19 Suppl A:5A-36A. [PubMed] [Cited in This Article: ] |
| 38. | Al- Dabbagh NM, Al-Saleh S, Al-Dohayan N, Al-Asmari AK, Arfin M, Tariq M. The role of Apolipoprotein E gene polymorphisms in Primary Glaucoma and Pseudoexfoliation Syndrome. Glaucoma - Basic and Clinical Aspects. INTECH. 2013;130-156. [DOI] [Cited in This Article: ] |
| 39. | Schallreuter KU, Levenig C, Kühnl P, Löliger C, Hohl-Tehari M, Berger J. Histocompatibility antigens in vitiligo: Hamburg study on 102 patients from northern Germany. Dermatology. 1993;187:186-192. [PubMed] [Cited in This Article: ] |
| 40. | Svejgaard A, Platz P, Ryder LP. HLA and disease 1982--a survey. Immunol Rev. 1983;70:193-218. [PubMed] [Cited in This Article: ] |
| 41. | Halford J, Mazeika G, Slifer S, Speer M, Saunders AM, Strittmatter WJ, Morgenlander JC. APOE2 allele increased in tardive dyskinesia. Mov Disord. 2006;21:540-542. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 7] [Cited by in F6Publishing: 8] [Article Influence: 0.4] [Reference Citation Analysis (0)] |
| 42. | Agouridis AP, Elisaf M, Milionis HJ. An overview of lipid abnormalities in patients with inflammatory bowel disease. Ann Gastroenterol. 2011;24:181-187. [PubMed] [Cited in This Article: ] |
| 43. | Koutroubakis IE, Malliaraki N, Vardas E, Ganotakis E, Margioris AN, Manousos ON, Kouroumalis EA. Increased levels of lipoprotein (a) in Crohn’s disease: a relation to thrombosis? Eur J Gastroenterol Hepatol. 2001;13:1415-1419. [PubMed] [Cited in This Article: ] |
| 44. | Ripollés Piquer B, Nazih H, Bourreille A, Segain JP, Huvelin JM, Galmiche JP, Bard JM. Altered lipid, apolipoprotein, and lipoprotein profiles in inflammatory bowel disease: consequences on the cholesterol efflux capacity of serum using Fu5AH cell system. Metabolism. 2006;55:980-988. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 54] [Cited by in F6Publishing: 56] [Article Influence: 3.1] [Reference Citation Analysis (0)] |
| 45. | Miyauchi H. [Immunohistochemical study for the localization of apolipoprotein AI, B100, and E in normal and psoriatic skin]. Igaku Kenkyu. 1991;61:79-86. [PubMed] [Cited in This Article: ] |
| 46. | Furumoto H, Nakamura K, Imamura T, Hamamoto Y, Shimizu T, Muto M, Asagami C. Association of apolipoprotein allele epsilon 2 with psoriasis vulgaris in Japanese population. Arch Dermatol Res. 1997;289:497-500. [PubMed] [Cited in This Article: ] |
| 47. | Couderc R, Mahieux F, Bailleul S, Fenelon G, Mary R, Fermanian J. Prevalence of apolipoprotein E phenotypes in ischemic cerebrovascular disease. A case-control study. Stroke. 1993;24:661-664. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 95] [Cited by in F6Publishing: 98] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 48. | Hsia SH, Connelly PW, Hegele RA. Restriction isotyping of apolipoprotein E R145C in type III hyperlipoproteinemia. J Investig Med. 1995;43:187-194. [PubMed] [Cited in This Article: ] |
| 49. | Danesh J, Wheeler JG, Hirschfield GM, Eda S, Eiriksdottir G, Rumley A, Lowe GD, Pepys MB, Gudnason V. C-reactive protein and other circulating markers of inflammation in the prediction of coronary heart disease. N Engl J Med. 2004;350:1387-1397. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 2136] [Cited by in F6Publishing: 2072] [Article Influence: 103.6] [Reference Citation Analysis (0)] |
| 50. | Papa A, Santoliquido A, Danese S, Covino M, Di Campli C, Urgesi R, Grillo A, Guglielmo S, Tondi P, Guidi L. Increased carotid intima-media thickness in patients with inflammatory bowel disease. Aliment Pharmacol Ther. 2005;22:839-846. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 64] [Cited by in F6Publishing: 48] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 51. | Danese S, Sgambato A, Papa A, Scaldaferri F, Pola R, Sans M, Lovecchio M, Gasbarrini G, Cittadini A, Gasbarrini A. Homocysteine triggers mucosal microvascular activation in inflammatory bowel disease. Am J Gastroenterol. 2005;100:886-895. [PubMed] [Cited in This Article: ] |
| 52. | Bregenzer N, Hartmann A, Strauch U, Schölmerich J, Andus T, Bollheimer LC. Increased insulin resistance and beta cell activity in patients with Crohn‘s disease. Inflamm Bowel Dis. 2006;12:53-56. [PubMed] [Cited in This Article: ] |
| 53. | Dagli N, Poyrazoglu OK, Dagli AF, Sahbaz F, Karaca I, Kobat MA, Bahcecioglu IH. Is inflammatory bowel disease a risk factor for early atherosclerosis? Angiology. 2010;61:198-204. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 53] [Cited by in F6Publishing: 64] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 54. | Rungoe C, Basit S, Ranthe MF, Wohlfahrt J, Langholz E, Jess T. Risk of ischaemic heart disease in patients with inflammatory bowel disease: a nationwide Danish cohort study. Gut. 2013;62:689-694. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 117] [Cited by in F6Publishing: 123] [Article Influence: 11.2] [Reference Citation Analysis (0)] |
| 55. | Visschers RG, Olde Damink SW, Schreurs M, Winkens B, Soeters PB, van Gemert WG. Development of hypertriglyceridemia in patients with enterocutaneous fistulas. Clin Nutr. 2009;28:313-317. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 11] [Cited by in F6Publishing: 11] [Article Influence: 0.7] [Reference Citation Analysis (0)] |
| 56. | Crook MA, Velauthar U, Moran L, Griffiths W. Hypocholesterolaemia in a hospital population. Ann Clin Biochem. 1999;36:613-616. [PubMed] [Cited in This Article: ] |
| 57. | Shiraki M, Shiraki Y, Aoki C, Hosoi T, Inoue S, Kaneki M, Ouchi Y. Association of bone mineral density with apolipoprotein E phenotype. J Bone Miner Res. 1997;12:1438-1445. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 137] [Cited by in F6Publishing: 141] [Article Influence: 5.2] [Reference Citation Analysis (0)] |
| 58. | Wong SY, Lau EM, Li M, Chung T, Sham A, Woo J. The prevalence of Apo E4 genotype and its relationship to bone mineral density in Hong Kong Chinese. J Bone Miner Metab. 2005;23:261-265. [PubMed] [Cited in This Article: ] |
| 59. | Ulivieri FM, Lisciandrano D, Ranzi T, Taioli E, Cermesoni L, Piodi LP, Nava MC, Vezzoli M, Bianchi PA. Bone mineral density and body composition in patients with ulcerative colitis. Am J Gastroenterol. 2000;95:1491-1494. [PubMed] [Cited in This Article: ] |
| 60. | Lynch JR, Tang W, Wang H, Vitek MP, Bennett ER, Sullivan PM, Warner DS, Laskowitz DT. APOE genotype and an ApoE-mimetic peptide modify the systemic and central nervous system inflammatory response. J Biol Chem. 2003;278:48529-48533. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 253] [Cited by in F6Publishing: 272] [Article Influence: 13.0] [Reference Citation Analysis (0)] |
| 61. | Jofre-Monseny L, Minihane AM, Rimbach G. Impact of apoE genotype on oxidative stress, inflammation and disease risk. Mol Nutr Food Res. 2008;52:131-145. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 210] [Cited by in F6Publishing: 225] [Article Influence: 14.1] [Reference Citation Analysis (0)] |
| 62. | Vitek MP, Brown CM, Colton CA. APOE genotype-specific differences in the innate immune response. Neurobiol Aging. 2009;30:1350-1360. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 248] [Cited by in F6Publishing: 243] [Article Influence: 16.2] [Reference Citation Analysis (0)] |
| 63. | Ophir G, Amariglio N, Jacob-Hirsch J, Elkon R, Rechavi G, Michaelson DM. Apolipoprotein E4 enhances brain inflammation by modulation of the NF-kappaB signaling cascade. Neurobiol Dis. 2005;20:709-718. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 101] [Cited by in F6Publishing: 112] [Article Influence: 5.9] [Reference Citation Analysis (0)] |
| 64. | Jofre-Monseny L, Loboda A, Wagner AE, Huebbe P, Boesch-Saadatmandi C, Jozkowicz A, Minihane AM, Dulak J, Rimbach G. Effects of apoE genotype on macrophage inflammation and heme oxygenase-1 expression. Biochem Biophys Res Commun. 2007;357:319-324. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 80] [Cited by in F6Publishing: 77] [Article Influence: 4.5] [Reference Citation Analysis (0)] |
| 65. | Kwon OD, Khaleeq A, Chan W, Pavlik VN, Doody RS. Apolipoprotein E polymorphism and age at onset of Alzheimer’s disease in a quadriethnic sample. Dement Geriatr Cogn Disord. 2010;30:486-491. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 15] [Cited by in F6Publishing: 19] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 66. | Peng H, Wang C, Chen Z, Sun Z, Jiao B, Li K, Huang F, Hou X, Wang J, Shen L. The APOE ε2 allele may decrease the age at onset in patients with spinocerebellar ataxia type 3 or Machado-Joseph disease from the Chinese Han population. Neurobiol Aging. 2014;35:2179.e15-2179.e18. [PubMed] [DOI] [Cited in This Article: ] [Cited by in F6Publishing: 26] [Reference Citation Analysis (0)] |
| 67. | Harwood DG, Barker WW, Ownby RL, St George-Hyslop P, Mullan M, Duara R. Apolipoprotein E polymorphism and age of onset for Alzheimer’s disease in a bi-ethnic sample. Int Psychogeriatr. 2004;16:317-326. [PubMed] [Cited in This Article: ] |
| 68. | Kampman O, Anttila S, Illi A, Mattila KM, Rontu R, Leinonen E, Lehtimäki T. Apolipoprotein E polymorphism is associated with age of onset in schizophrenia. J Hum Genet. 2004;49:355-359. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 17] [Cited by in F6Publishing: 17] [Article Influence: 0.9] [Reference Citation Analysis (0)] |
| 69. | Grant WB. A multicountry ecological study of risk-modifying factors for prostate cancer: apolipoprotein E epsilon4 as a risk factor and cereals as a risk reduction factor. Anticancer Res. 2010;30:189-199. [PubMed] [Cited in This Article: ] |
| 70. | Kelly ME, Clay MA, Mistry MJ, Hsieh-Li HM, Harmony JA. Apolipoprotein E inhibition of proliferation of mitogen-activated T lymphocytes: production of interleukin 2 with reduced biological activity. Cell Immunol. 1994;159:124-139. [PubMed] [Cited in This Article: ] |
| 71. | Koga T, Duan H, Urabe K, Furue M. In situ localization of CD83-positive dendritic cells in psoriatic lesions. Dermatology. 2002;204:100-103. [PubMed] [Cited in This Article: ] |
| 72. | Oestreicher JL, Walters IB, Kikuchi T, Gilleaudeau P, Surette J, Schwertschlag U, Dorner AJ, Krueger JG, Trepicchio WL. Molecular classification of psoriasis disease-associated genes through pharmacogenomic expression profiling. Pharmacogenomics J. 2001;1:272-287. [PubMed] [Cited in This Article: ] |
| 73. | Chasman DI, Kozlowski P, Zee RY, Kwiatkowski DJ, Ridker PM. Qualitative and quantitative effects of APOE genetic variation on plasma C-reactive protein, LDL-cholesterol, and apoE protein. Genes Immun. 2006;7:211-219. [PubMed] [DOI] [Cited in This Article: ] [Cited by in F6Publishing: 1] [Reference Citation Analysis (0)] |