Published online Jun 28, 2015. doi: 10.3748/wjg.v21.i24.7400
Peer-review started: January 24, 2015
First decision: February 10, 2015
Revised: February 24, 2015
Accepted: May 2, 2015
Article in press: May 4, 2015
Published online: June 28, 2015
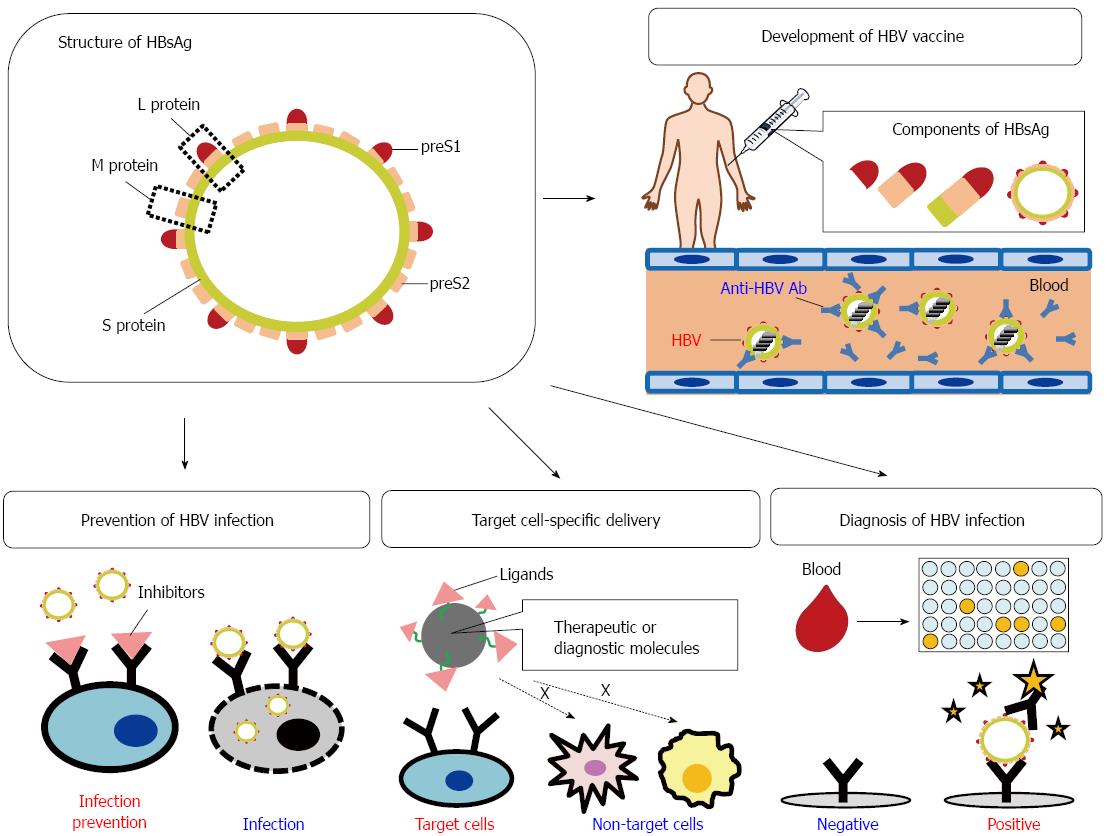
Human hepatitis B virus (HBV) is a member of the family Hepadnaviridae, and causes acute and chronic infections of the liver. The hepatitis B surface antigen (HBsAg) contains the large (L), middle (M), and small (S) surface proteins. The L protein consists of the S protein, preS1, and preS2. In HBsAg, the preS domain (preS1 + preS2) plays a key role in the infection of hepatocytic cells by HBV and has several immunogenic epitopes. Based on these characteristics of preS, several preS-based diagnostic and therapeutic materials and systems have been developed. PreS1-specific monoclonal antibodies (e.g., MA18/7 and KR127) can be used to inhibit HBV infection. A myristoylated preS1 peptide (amino acids 2-48) also inhibits the attachment of HBV to HepaRG cells, primary human hepatocytes, and primary tupaia hepatocytes. Antibodies and antigens related to the components of HBsAg, preS (preS1 + preS2), or preS1 can be available as diagnostic markers of acute and chronic HBV infections. Hepatocyte-targeting delivery systems for therapeutic molecules (drugs, genes, or proteins) are very important for increasing the clinical efficacy of these molecules and in reducing their adverse effects on other organs. The selective delivery of diagnostic molecules to target hepatocytic cells can also improve the efficiency of diagnosis. In addition to the full-length HBV vector, preS (preS1 + preS2), preS1, and preS1-derived fragments can be useful in hepatocyte-specific targeting. In this review, we discuss the literature concerning the applications of the HBV preS domain in bio- and nanotechnology.
Core tip: The hepatitis B surface antigen (HBsAg) of human hepatitis B virus (HBV) contains the large (L), middle (M), and small (S) surface proteins. The L protein consists of the S protein, preS1, and preS2. In HBsAg, the preS domain (preS1 + preS2) plays a key role in the infection of hepatocytic cells by HBV and has several immunogenic epitopes. Therefore, the preS domain can act as a diagnostic or therapeutic target or as material for developing inhibitors of HBV infection, HBV vaccines, diagnostic tools for HBV infection, and hepatocyte-targeting delivery systems for diagnostic or therapeutic molecules.
- Citation: Toita R, Kawano T, Kang JH, Murata M. Applications of human hepatitis B virus preS domain in bio- and nanotechnology. World J Gastroenterol 2015; 21(24): 7400-7411
- URL: https://www.wjgnet.com/1007-9327/full/v21/i24/7400.htm
- DOI: https://dx.doi.org/10.3748/wjg.v21.i24.7400
Human hepatitis B virus (HBV) is a double-stranded circular DNA virus that causes acute and chronic infections of the liver. Most adults with acute HBV infections make a full recovery, but about 5% suffer chronic hepatitis B. The occurrence of acute HBV infection in neonates and infants (< 4 years) dramatically increases the risk of chronic HBV infection by up to 90%. Chronic HBV infection causes severe liver diseases, including liver cirrhosis and hepatocellular carcinoma (HCC), and HCC is associated with significant morbidity and mortality[1-4].
The HBV virus is subdivided into eight genotypes (A-H), which show distinct geographic distributions. In brief, genotypes A, D, and H are prevalent in Western countries (e.g., Central and North America and northwestern Europe) and India, and genotypes B and C are prevalent in Asian countries (e.g., Japan, Taiwan, Korea, China, and East Asia). Genotype E has been detected in Africa, genotype F in Central and South America, and genotype G in France and North America[2,3,5].
The HBV particle contains the hepatitis B surface antigen (HBsAg), which specifically recognizes human hepatocytes and HCC cells. In HBsAg, the preS domain (preS1 + preS2) plays a key role in HBV infection and the host immune responses (see below). Because of these functions, the preS domain can act as a diagnostic or therapeutic target or as material for developing inhibitors of HBV infection, HBV vaccines, diagnostic tools for HBV infection, and hepatocyte-targeting delivery systems for diagnostic or therapeutic molecules (Figure 1).
In this review, we discuss the literature concerning the applications of the HBV preS domain in bio- and nanotechnology.
The HBV particle contains HBsAg and the hepatitis B core antigen (HBcAg), together with a double-stranded DNA molecule and the hepatitis B envelope antigen (HBeAg)[1,4,6-8]. HBeAg shares its residue 1-149 region with HBcAg[9,10]. The sizes of the HBV particle and HBsAg are about 44 and 22 nm, respectively[1,11].
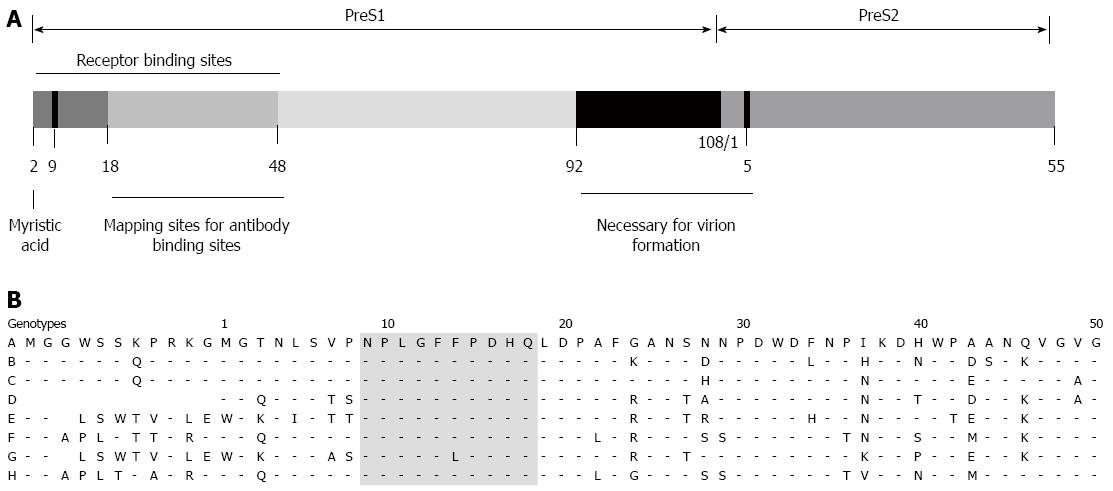
HBsAg consists of the large (L), middle (M), and small (S) surface proteins. The L and M proteins contain the S protein (226 amino acids [aa]) and 55 hydrophilic aa at the N-terminus of the S protein (called “preS2”). The L protein has an extension of hydrophilic residues (108 or 119, depending on the genotype), called “preS1”, at the N-terminus of preS2[6-8] (Figure 1 and 2A).
The preS1 domain has receptor binding sites (aa 1-48), containing essential residues at aa 9-18 in genotype D. The eight HBV genotypes show very similar sequences in the essential residues[7,12,13]. The preS1 domain is myristoylated at the glycine residue at position 2 and this N-terminal myristoylation dramatically increases HBV infectivity[6,7,12]. Previous studies have also reported that residues 21-47 of preS1 are important in the attachment of HBV to the receptors on hepatocytes[14,15] (Figure 2B). (Note that residues 21-47 correspond to aa 10-36 in genotype D).
The asialoglycoprotein receptor on the surfaces of human hepatocytes and HCC cells is considered to be a functional preS1-specific receptor[7,16,17]. Recently, the sodium taurocholate cotransporting polypeptide (NTCP) was identified as a novel preS1-specific receptor[18,19], and inhibitors that block its metabolic functions may be able to prevent HBV entry (e.g., cyclosporine A[20] and irbesartan[21]).
Human and tupaia hepatocytes can be reproducibly infected with HBV, but HBV infection in tupaia hepatocytes causes only mild and transient infection, with low viral titers. Because mice are not infected by HBV, transgenic or human-liver-chimeric mice are used as animal models of HBV infection[22-24]. However, a recent study reported that the myristoylated preS1 peptide (aa 2-48) can bind to primary hepatocytes derived from nonsusceptible species, such as the mouse, rat, dog, and rabbit[6]. However, the previous study of these researchers reported the conflicting result that the myristoylated preS1 peptide (aa 2-48) cannot bind to primary rodent hepatocytes or the mouse liver cell line AML-12[12]. Furthermore, after an intravenous injection of the preS1 lipopeptide (aa 2-48) into mice, rats, and dogs, its accumulation in the livers was detected[25].
NTCP is regarded as a receptor that binds the myristoylated preS1 peptide (aa 2-48) to the hepatocytes of nonsusceptible hosts (e.g., mouse)[26,27]. In fact, the preS1 lipopeptide (aa 2-48) can bind to mouse NTCP and human NTCP, but its affinity is higher for human hepatocyte NTCP than in mouse hepatocyte NTCP[26,27]. In contrast, the expression of NTCP is higher for mouse (five times), rat (four times), and monkey livers (twice) than in human liver. Rat hepatocytes show three times higher levels of NTCP than human hepatocytes. However, the expression of NTCP in HCC is half that in the normal human liver[28]. Therefore, the higher binding of the myristoylated preS1 peptide (aa 2-48) to mouse and rat hepatocytes may be associated with their higher levels of NTCP.
However, the higher binding of myristoylated preS1 peptide (aa 2-48) to mouse and rat hepatocytes does not mean high levels of HBV infection in the rat and mouse, because HBV entry is restricted by mouse NTCP, and efficient HBV infection does not even occur in mouse hepatocytes expressing human NTCP[26,27,29]. Although aa 84-87 and aa 157-165 of human NTCP play important roles in preS1 binding and HBV entry, the region at residues 84-87 is a major host infection-determining area. In fact, although HBV entry into mouse hepatocytes through mouse NTCP is restricted, HBV entry is possible when mouse NTCP aa 84-87 are substituted with their human counterparts[26,27].
The preS domain (preS1 + preS2) also contains several immunogenic epitopes[30-32]. These epitopes may be potential targets for HBV vaccines and antibodies that inhibit HBV infection (see below).
PreS1-specific antibodies can be used to inhibit HBV infection. The very N-terminal part of preS1 (aa 1-21) and the C-terminal part (aa 78-116) are nonneutralizing epitopes[32] and the region at residues 21-48 is considered a virus-neutralizing epitope[32-34] (Figure 2A). For example, the monoclonal antibody (mAb) MA18/7, which specifically recognizes a preS1 epitope (aa 20-23; DPAF)[35], can block the HBV infection of primary tupaia hepatocytes[36]. The mAb KR127, which binds to aa 37-45 of the preS1 domain[37], shows virus-neutralizing activity and confers efficient protection against HBV infection in chimpanzees[38]. In addition to MA18/7 and KR127, an mAb targeting preS1 aa 32-47 (F35.25)[39] or preS1 aa 30-35[40] has been constructed, but its efficiency in inhibiting HBV infection of hepatocytes or animals has not yet been demonstrated.
A preS1-derived lipopeptide (myristoylated preS1 peptide; also known as Myrcludex B) containing aa 2-48 inhibits the attachment of HBV to HepaRG cells[6,41,42], primary human hepatocytes[6,41], and primary tupaia hepatocytes[6,12], but the nonmyristoylated preS1 peptide failed to inhibit HBV infection of these cells[6,12,42]. Interestingly, the replacement of myristic acid (tetradecanoic acid) in the lipopeptide with palmitic acid (hexadecanoic acid) or a cholesteryl moiety enhanced its inhibition of HBV infection, whereas caprylic acid (octanoic acid) or valeric acid (pentanoic acid) reduced its inhibition of HBV infection[12].
In an in vivo experiment using urokinase-type plasminogen activator (uPA+/-) transgenic mice crossed with RAG-2-/-/perforin-/- mice lacking mature T, B, and NK cells, the injection of human-hepatocyte-transplanted mice with the myristoylated preS1 peptide (aa 2-48) efficiently prevented HBV infection[43]. The results of a recent phase IIa trial of the myristoylated preS1 lipopeptide (aa 2-48) are available from the company website (http://www.hepatera.ru//node/53?ln=en).
In the preS1 sequence at aa 2-48 of genotype D, the region defined by residues 2-18 is essential for the inhibition of HBV infection, whereas the aa 19-48 region has no or little effect on this inhibition[12,41,42]. A deletion or mutation between residues 2 and 18 dramatically reduces the efficiency of inhibition of HBV infection[6,42]. Recently, a modified myristoylated preS1 peptide aa 2-19 showed significantly higher affinity for HepG2 and HuH-7 cells than two unmodified myristoylated peptides derived from preS1 aa 2-19 of genotypes A-D[44]. However, the palmitoylated preS1 sequence aa 11-31 of genotype C, which corresponds to sequence aa 1-19 of genotype D, potentially inhibits HBV infection[45] (Figure 2B).
Several preS1-binding peptides obtained from a phage display library have been developed to inhibit HBV infection[46-49]. Theoretically, they block the binding of the HBV preS1 domain to target cells. However, there are very few in vitro or in vivo data regarding their efficiency in inhibiting HBV infection, other than a recent study in which two preS1-binding peptides (LKKKW and LRNIR) inhibited HBV attachment to HepG2 cells[49].
The levels of serum HBV-related antibodies and antigens, HBV DNA, and aminotransferases (e.g., alanine aminotransferase and aspartate aminotransferase) are commonly assayed for the diagnosis of acute and chronic HBV infections[1,3,4].
HBV-related antibodies and antigens, such as anti-hepatitis B core antibody (anti-HBc), HBeAg and anti-HBeAg antibody (anti-HBe), and HBsAg and anti-HBsAg antibody (anti-HBs), are potential markers for diagnosing HBV infection. HBsAg appears in the first stage after HBV infection and anti-HBc is detected within 2 wk of the appearance of HBsAg. For these reasons, HBsAg and anti-HBc can be considered useful markers for diagnosing the acute phase of HBV infection[1,4]. However, an HBsAg-targeting immunoassay often produces false-negative results in patients infected with mutant HBsAg[50,51] or in patients with low HBsAg levels, because HBsAg can disappear at any stage of the disease[1,3].
HBsAg can be detected as a diagnostic marker in serum and saliva samples from patients infected with HBV[52-54]. However, during recovery from illness, HBsAg disappears more rapidly from the saliva than from the sera of patients with acute HBV infections[52]. To detect HBsAg in serum and saliva samples, an enzyme immunoassay[52,55], ELISA[55], time-resolved fluoroimmunoassay (TRFIA)[56], or chemiluminescence immunoassay[50,53-55] is commonly used. Recently, modified versions of these assays for the more sensitive detection of HBsAg have been reported, such as assays based on magnetic beads and TRFIA[57] and a bioluminescent enzyme immunoassay using firefly luciferase[58].
Antibodies and antigens related to the components of HBsAg, preS (preS1 + preS2), or preS1 are also available as diagnostic markers of acute and chronic HBV infections[59-61]. Serum preS1-related antibodies are detected with a typical ELISA microplate coated with one or two copies of preS1 (e.g., aa 21-47 or 21-119) fusion proteins[62,63]. In another assay, an indirect ELISA was constructed by coating the plates with two copies of the preS1 peptide (aa 21-47) fused to glutathione S-transferase through a flexible linker[64]. However, these assays targeting serum preS1-related antibodies show relatively low positive rates in diagnoses using sera from HBV-infected patients[62,63].
ELISA and TRFIA are applicable to the detection of serum preS1 antigen[65-67]. However, an analysis of serum preS1 antigen showed that TRFIA is better than ELISA in its precision, specificity, and sensitivity[67].
An immunoassay in which a plate is coated with a polyclonal antibody directed against preS (preS1 + preS2) has been used to detect serum HBV preS antigen. Its sensitivity, specificity, and accuracy in serum samples from patients with chronic HBV infection were 81% (72%-89%), 65% (58%-70%), and 72%, respectively[66]. A double-sandwich immunoassay in a plate coated with two mAbs, anti-PreS1 mAb and anti-HBc mAb, has also been developed. In HBsAg-positive serum samples infected with the wild-type viruses of four genotypes (A-D), the assay using two mAbs displayed higher sensitivity than the assay using anti-preS1 mAb, anti-HBc mAb, or anti-HBe mAb alone, but lower sensitivity than the assay using anti-HBs mAb. However, the sensitivity of the assay based on the anti-preS1 and anti-HBc mAbs for the serum samples of 10 patients infected with an HBsAg-mutant virus was 80% (8/10, 95%CI: 44.4%-97.5%)[68].
The HBV surface and core proteins are regarded as potential targets for the development of HBV vaccines. Although the S protein is the major component of HBsAg, the preS1 and preS2 regions are more immunogenic targets within HBsAg[2,32,69].
To prevent HBV infection, two types of vaccines have been developed, HBV protein vaccines and HBV DNA vaccines. Protein-based HBV vaccines containing recombinant HBsAg usually require adjuvants. They also induce ineffective T-cell responses, but these problems can be improved by their formulation with novel adjuvants, e.g., 3-deacylated monophosphoryl lipid A or saponins purified from Quillaja saponaria Molina[70,71]. HBV DNA vaccines expressing HBsAg are produced by inserting the HBsAg gene into an expression vector. The efficiency of DNA immunization can be enhanced by combining it with an optimized delivery technology, such as electroporation[72] or the gene gun[73].
Conventional yeast-derived HBV vaccines (second generation) contain the S protein of HBV. These vaccines induce protective antibody responses in healthy adult recipients (about 90%), but fail to elicit adequate antibody production in up to 10% of individuals, who may become chronic HBV carriers and develop liver disease (e.g., liver cirrhosis or HCC)[2,74,75]. Mutations in the S gene may be associated with inadequate control of HBV infection after vaccination with conventional HBV vaccines[2,76]. However, when combined with the core antigen, conventional vaccines can exert more potent and efficient therapeutic effects[77,78].
In contrast, a third-generation HBV vaccine, Sci-B-Vac™ (also known as Hepimmune or Bio-Hep-B), containing the S, preS1, and preS2 protein components, is safe and more immunogenic than conventional HBV vaccines[79-81] because the preS1 and preS2 domains contain T- and B-cell-specific epitopes[30-32]. An HBV vaccine containing the preS1 and preS2 domains can elicit protective antibody levels in non- or low responders to conventional HBV vaccines[82,83]. HBV-specific T-cell proliferation and interferon γ (IFN-γ) production are induced in non- or low responders after the injection of Sci-B-Vac™, whereas non- or low responders display no proliferation of T cells, which are important for an efficient B-cell response[83].
HBsAg DNA vaccines expressing either the L, M, or S protein can induce anti-HBs antibodies, but interestingly, HBsAg DNA vaccines expressing the L protein was less immunogenic than the HBsAg DNA vaccine with the M or S protein. However, HBsAg-specific interleukin 2 (IL-2) and IFN-γ secretion in mouse splenocytes did not differ significantly after vaccination with either of the three HBsAg DNA vaccines[84]. In contrast, the modified HBsAg DNA vaccine that expresses the truncated N-terminal sequence (aa 1-18) within the preS1 region, enhances anti-HBs antibody levels in mice earlier and with higher titers than the HBsAg DNA vaccine expressing the full L protein. However, the two vaccines do not differ in the levels of IFN-γ and IL-4 secreted from mouse splenocytes after vaccination[81,84]. These data suggest that the HBsAg DNA vaccine expressing N-terminal-truncated preS1/preS2/S proteins can induce both anti-HBs antibodies and T- and B-cell immune responses more efficiently than full-length preS1/preS2/S proteins.
HBV vaccines constructed by fusing the preS1 peptide to the core protein or S protein induce efficient cellular and humoral immune responses[72,85,86], as mentioned above, because preS1 contains immunogenic T- and B-cell epitopes, e.g., preS1 aa 21-30 contains a T-cell epitope[31] and aa 12-32 and 32-53 contain B-cell epitopes[30].
Generally, the immune system is weakened in patients with chronic HBV infection. Therefore, therapeutic vaccines that can efficiently stimulate the immune responses will be useful in treating chronic HBV infection[87]. According to the results of several studies, HBV vaccines containing the preS1 domain can be used to treat chronic HBV infection because they induce highly potent immune responses[81,86,88,89]. Furthermore, a combined therapy with the preS1/preS2/S vaccine and antiviral drugs (e.g., lamivudine and clevudine) induces more efficient viral suppression and enhanced immune responses than the antiviral drug or vaccine alone[89-91].
Hepatocyte-targeting delivery systems for therapeutic molecules (drugs, genes, or proteins) are very important for increasing the clinical efficacy of these molecules and in reducing their adverse effects on other organs. The selective delivery of diagnostic molecules to target hepatocytic cells can also improve the efficiency of diagnosis[92].
Recombinant HBV is used as a vector for delivering genes to hepatocytes or HCC cells[93-96]. For example, the delivery of small interfering RNA (siRNA) by recombinant HBV successfully inhibited HBsAg expression in primary tree shrew hepatocytes infected with wild-type HBV[96].
In addition to the full-length HBV vector, preS (preS1 + preS2), preS1, and preS1-derived fragments can be useful in hepatocyte-specific targeting. As a simple gene delivery system, the preS1 peptide (aa 21-47) fused to an arginine 9-mer, which binds to anionic genes, efficiently delivered siRNA into HepG2 cells[97].
The conjugation of viral vectors (e.g., bacteriophages)[98] with a preS1 peptide makes it possible to deliver genes into hepatocytic cells. Bacteriophage T7 displaying a preS1 fragment (aa 60-108) was constructed as a vector for delivering genes into HepG2 cells. Recombinant T7 phage particles containing preS1 aa 60-108 displayed higher recovery in HepG2 cells than T7 phage particle containing preS1 aa 1-47[99]. The incorporation of preS1 peptides into a mutant herpes simplex virus type 1 (HSV-1) virus, from which the glycoprotein that binds to the host cell glycosaminoglycans was deleted, increased their binding activity to HepG2 cells compared with that of the mutant HSV-1 virus alone, but no data are available for nonhepatocytic cells[100].
A preS-conjugated liposome has been constructed, simply by mixing a His-tagged C-terminal recombinant preS protein with cationic liposomes. In a transfection experiment using different cells, the conjugate containing plasmid DNA (pDNA) expressed more β-galactosidase in human hepatocytes than in human pulmonary artery smooth muscle cells, human renal epithelial cells, human dermal fibroblasts (fetal), or human cardiac fibroblasts. The β-galactosidase activity was higher in the livers of immunocompromised mice treated with the preS-conjugated liposome/pDNA complex than in mice treated with the liposome/pDNA complex[101]. In a recent study, the incorporation of a preS1-derived lipopeptide (aa 2-48 with myristic acid) into a PEGylated liposome enhanced its uptake by primary mice hepatocytes and its accumulation in the liver. The livers of mice treated with a PEG-preS1-liposome containing silybin were more efficiently protected from carbon tetrachloride (CCl4)-induced acute hepatitis than were those of mice treated with the PEG-liposome containing silybin or with silybin alone[102].
An L-protein-based bionanocapsule that contains the preS1 region has been developed to deliver genes into human hepatocytes[103]. A complex of the bionanocapsule and a cationic liposome (EL-01-A) significantly increased its transfection efficiency in HepG2 cells compared with that of the bionanocapsule alone[104]. The injection of a bionanocapsule/liposome complex containing the GFP gene into mice carrying tumor cells induced GFP expression in HCCs (NuE tumors), but in neither mouse liver nor human epidermoid carcinoma (A431)[104]. In another study, mice bearing NuE tumors were injected with GFP fused with preS (preS1 + pesS2), and no GFP fluorescence was found in the mouse liver, but was observed in the NuE tumors[105]. These results contradict those of recent studies in which a myristoylated preS1 peptide (aa 4-48) accumulated in the livers of mice and rats after its intravenous injection, and bound to mouse hepatocytes[6,25-27]. Therefore, further studies are required to establish definitively whether myristoylated preS1 peptides (aa 4-48), full preS1, and preS (preS1 + preS2) differ in their affinity for human and mouse hepatocytes.
Mixing liposomes with preS1 or preS (preS1 + preS2) is a simple method of constructing hepatocyte-targeting gene delivery systems. However, according to a recent study, a mixture of myristoylated preS1 (aa 4-48) and liposomes caused myristic acid to be inserted into the lipid layer of the liposomes, markedly reducing the efficiency of liver targeting[102].
A protein-based nanocage composed of HSP16.5 can be fused to several peptides and proteins, and is used as a cell-targeting delivery system for genes and drugs[106-109]. A nanocage fused to preS1 increased its specificity for HCC cells (HepG2 and Huh-7) more significantly than for human breast cancer cells (MCF-7) or human epithelial carcinoma cells (HeLa)[110]. The myristoylated-preS1-fused nanocage also displays higher affinity for HepaRG cells than the nonmyristoylated preS1-fused nanocage[111].
A construct in which technetium-99m (99mTc) is conjugated to a stearoylated preS1 peptide (aa 2-48) through a mercaptoacetyltriglycerin linker has been synthesized as a single-photon emission computed tomography (SPECT) tracer. After the tracer was injected intravenously into rats, its accumulation was higher in their livers than in other tissues (heart, lung, spleen, kidney, muscle, brain, intestine, duodenum, and tail)[112]. In that study, stearic acid was used instead of myristic acid. In a previous study, peptides conjugated with a palmitoyl moiety (C16) with a longer carbon chain or a cholesteryl moiety (C27) with more carbon atoms than the myristoyl moiety (C14) increased its affinity for primary tupaia hepatocytes, whereas fatty acids with shorter carbon chains (e.g., caprylic acid [C8] and valeric acid [C5]) markedly reduced its affinity for hepatocytes[12]. Stearic acid is a fatty acid with 18 carbon atoms. Therefore, the affinities of stearoylated preS1 aa 2-48 and myristoylated preS1 aa 2-48 for hepatocytes may differ.
Although preS (preS1 + preS2)- and preS1-conjugated delivery systems can specifically target hepatocytes and HCC cells, they cannot distinguish between normal and abnormal hepatocytic cells (e.g., cirrhotic liver and HCC cells). A novel gene delivery system has been reported that responds to the hyperactivated intracellular signals of tumor cells (e.g., protein kinase A [PKA] and PKCα), but not to the normal intracellular signals of normal cells or tissues[113-115]. Combining this system with nanoparticles containing preS1 makes it possible to distinguish between normal human hepatocytes and HCC cells[116]. The combined system also increases the transfection efficiency and selectivity for HCC cells (e.g., HepG2 and Huh-7 cells) with hyperactivated PKA or PKCα, but shows no gene expression in human epidermoid carcinoma cells (A431), human colon carcinoma cells (WiDr), or human lung adenocarcinoma cells (A549), which also contain hyperactivated PKA or PKCα[116,117].
Recently, a research group reported an interesting relationship between endocytosis and the lengths of the preS1 peptide, using virus-like particles (VLPs) derived from the bacteriophage AP205 coat protein and fused with different lengths of preS1 peptide at the C-terminus of the coat protein sequence. The VLP containing preS1 aa 10-36 bound strongly to HepG2 cells, but was not found in the cytosol of HepG2 cells. However, a VLP containing preS1 aa 2-108 was endocytosed and was observed in the cytosol[118]. This study suggests that preS1 fragments, like full-length preS1, can specifically bind to hepatocytic cells, but has not shown satisfactory results for their cellular uptake by hepatocytic cells.
The preS domain (preS1 + preS2) of HBV HBsAg plays a key role in HBV infection by attaching to hepatocytic cells and interacting with receptors, and contains several immunogenic epitopes. Based on these characteristics of preS, several preS-based diagnostic and therapeutic materials and systems have been developed, including inhibitors of HBV infection, HBV vaccines, diagnostic tools for HBV infection, and hepatocyte-targeting delivery systems for diagnostic or therapeutic molecules. As a good example, the myristoylated preS1 peptide (aa 4-48) is highly potent in inhibiting HBV entry into human hepatocytes, and the clinical trial results for this peptide have been reported. However, there are still several opportunities for the development of more preS1-fragment-based inhibitors using the myristoylated preS1 peptide (aa 4-48), e.g., by replacing myristic acid with other fatty acids or by altering the amino acid sequence at receptor binding sites[12,44].
The HBV virus is classified into at least eight genotypes (A-H) which have distinct geographic distributions. The eight HBV genotypes show very similar sequences in essential residues (aa 9-18) within receptor binding sites located in the preS domain (Figure 2). These results, however, do not mean that bio- and nanotechnological approach using a HBV genotype-derived preS domain can be applicable to other HBV genotypes, due to high genetic diversity at the preS domain.
Recent studies suggest that preS1-derived fragments can recognize receptors on rodent hepatocytes. Mice and rats have been used to evaluate hepatocyte-targeting delivery systems based on preS1 or preS (preS1 + preS2). However, HBV entry into mouse hepatocytes is restricted and mouse hepatocytes are not efficiently infected by HBV. Several studies have also reported that preS1-derived peptides and full-length preS1 or preS (preS1 + preS2) differ in their binding affinities for the mouse liver. Therefore, as stated above, further studies are required to clarify whether preS1-derived fragments, full-length preS1, and preS (preS1 + preS2) differ in their affinities for human and mouse hepatocytes.
In a recent study[118], the intracellular transfer of VLPs conjugated with preS1 fragments clearly differed from that of VLPs conjugated with full-length preS1. The preS1-fragment-fused VLP bound strongly to HepG2 cells, but was not transferred into the cytosol. Although further studies are needed, this study suggests that preS1-fragment-conjugated delivery systems cannot be used to deliver therapeutic molecules into the cytosol of hepatocytic cells.
Delivery systems using preS1 or preS (preS1 + preS2) are also unable to distinguish between normal and abnormal hepatocytes (e.g., cirrhotic liver cells and HCC cells). However, the development of abnormal-hepatocyte-targeting delivery systems (for example, by combining abnormal-cell-targeting delivery systems with delivery systems based on preS1 or preS [preS1 + preS2]) should enhance the therapeutic efficacy against abnormal hepatocytic cells and reduce the risk of adverse effects on normal hepatocytes.
P- Reviewer: Bock CT, Osna NA, Panduro A S- Editor: Ma YJ L- Editor: A E- Editor: Zhang DN
| 1. | Gitlin N. Hepatitis B: diagnosis, prevention, and treatment. Clin Chem. 1997;43:1500-1506. [PubMed] [Cited in This Article: ] |
| 2. | Kao JH, Chen DS. Global control of hepatitis B virus infection. Lancet Infect Dis. 2002;2:395-403. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 596] [Cited by in F6Publishing: 595] [Article Influence: 27.0] [Reference Citation Analysis (0)] |
| 3. | Aspinall EJ, Hawkins G, Fraser A, Hutchinson SJ, Goldberg D. Hepatitis B prevention, diagnosis, treatment and care: a review. Occup Med (Lond). 2011;61:531-540. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 83] [Cited by in F6Publishing: 92] [Article Influence: 7.7] [Reference Citation Analysis (0)] |
| 4. | Hoofnagle JH, Doo E, Liang TJ, Fleischer R, Lok AS. Management of hepatitis B: summary of a clinical research workshop. Hepatology. 2007;45:1056-1075. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 438] [Cited by in F6Publishing: 420] [Article Influence: 24.7] [Reference Citation Analysis (0)] |
| 5. | Kramvis A, Kew M, François G. Hepatitis B virus genotypes. Vaccine. 2005;23:2409-2423. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 256] [Cited by in F6Publishing: 255] [Article Influence: 13.4] [Reference Citation Analysis (0)] |
| 6. | Meier A, Mehrle S, Weiss TS, Mier W, Urban S. Myristoylated PreS1-domain of the hepatitis B virus L-protein mediates specific binding to differentiated hepatocytes. Hepatology. 2013;58:31-42. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 96] [Cited by in F6Publishing: 103] [Article Influence: 9.4] [Reference Citation Analysis (0)] |
| 7. | Glebe D, Urban S. Viral and cellular determinants involved in hepadnaviral entry. World J Gastroenterol. 2007;13:22-38. [PubMed] [DOI] [Cited in This Article: ] [Cited by in CrossRef: 207] [Cited by in F6Publishing: 198] [Article Influence: 11.6] [Reference Citation Analysis (0)] |
| 8. | Urban S, Bartenschlager R, Kubitz R, Zoulim F. Strategies to inhibit entry of HBV and HDV into hepatocytes. Gastroenterology. 2014;147:48-64. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 242] [Cited by in F6Publishing: 233] [Article Influence: 23.3] [Reference Citation Analysis (0)] |
| 9. | Schödel F, Peterson D, Zheng J, Jones JE, Hughes JL, Milich DR. Structure of hepatitis B virus core and e-antigen. A single precore amino acid prevents nucleocapsid assembly. J Biol Chem. 1993;268:1332-1337. [PubMed] [Cited in This Article: ] |
| 10. | Kenney JM, von Bonsdorff CH, Nassal M, Fuller SD. Evolutionary conservation in the hepatitis B virus core structure: comparison of human and duck cores. Structure. 1995;3:1009-1019. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 107] [Cited by in F6Publishing: 112] [Article Influence: 3.9] [Reference Citation Analysis (0)] |
| 11. | Gilbert RJ, Beales L, Blond D, Simon MN, Lin BY, Chisari FV, Stuart DI, Rowlands DJ. Hepatitis B small surface antigen particles are octahedral. Proc Natl Acad Sci USA. 2005;102:14783-14788. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 81] [Cited by in F6Publishing: 75] [Article Influence: 3.9] [Reference Citation Analysis (0)] |
| 12. | Glebe D, Urban S, Knoop EV, Cag N, Krass P, Grün S, Bulavaite A, Sasnauskas K, Gerlich WH. Mapping of the hepatitis B virus attachment site by use of infection-inhibiting preS1 lipopeptides and tupaia hepatocytes. Gastroenterology. 2005;129:234-245. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 194] [Cited by in F6Publishing: 196] [Article Influence: 10.3] [Reference Citation Analysis (0)] |
| 13. | Chi SW, Kim DH, Lee SH, Chang I, Han KH. Pre-structured motifs in the natively unstructured preS1 surface antigen of hepatitis B virus. Protein Sci. 2007;16:2108-2117. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 43] [Cited by in F6Publishing: 46] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 14. | Neurath AR, Kent SB, Strick N, Parker K. Identification and chemical synthesis of a host cell receptor binding site on hepatitis B virus. Cell. 1986;46:429-436. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 412] [Cited by in F6Publishing: 402] [Article Influence: 10.6] [Reference Citation Analysis (0)] |
| 15. | Dash S, Rao KV, Panda SK. Receptor for pre-S1(21-47) component of hepatitis B virus on the liver cell: role in virus cell interaction. J Med Virol. 1992;37:116-121. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 36] [Cited by in F6Publishing: 38] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 16. | Engelke M, Mills K, Seitz S, Simon P, Gripon P, Schnölzer M, Urban S. Characterization of a hepatitis B and hepatitis delta virus receptor binding site. Hepatology. 2006;43:750-760. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 118] [Cited by in F6Publishing: 123] [Article Influence: 6.8] [Reference Citation Analysis (0)] |
| 17. | Zhang X, Lin SM, Chen TY, Liu M, Ye F, Chen YR, Shi L, He YL, Wu LX, Zheng SQ. Asialoglycoprotein receptor interacts with the preS1 domain of hepatitis B virus in vivo and in vitro. Arch Virol. 2011;156:637-645. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 21] [Cited by in F6Publishing: 22] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 18. | Yan H, Zhong G, Xu G, He W, Jing Z, Gao Z, Huang Y, Qi Y, Peng B, Wang H. Sodium taurocholate cotransporting polypeptide is a functional receptor for human hepatitis B and D virus. Elife. 2012;1:e00049. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1280] [Cited by in F6Publishing: 1428] [Article Influence: 119.0] [Reference Citation Analysis (0)] |
| 19. | König A, Döring B, Mohr C, Geipel A, Geyer J, Glebe D. Kinetics of the bile acid transporter and hepatitis B virus receptor Na+/taurocholate cotransporting polypeptide (NTCP) in hepatocytes. J Hepatol. 2014;61:867-875. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 109] [Cited by in F6Publishing: 113] [Article Influence: 11.3] [Reference Citation Analysis (0)] |
| 20. | Watashi K, Sluder A, Daito T, Matsunaga S, Ryo A, Nagamori S, Iwamoto M, Nakajima S, Tsukuda S, Borroto-Esoda K. Cyclosporin A and its analogs inhibit hepatitis B virus entry into cultured hepatocytes through targeting a membrane transporter, sodium taurocholate cotransporting polypeptide (NTCP). Hepatology. 2014;59:1726-1737. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 192] [Cited by in F6Publishing: 195] [Article Influence: 19.5] [Reference Citation Analysis (0)] |
| 21. | Blanchet M, Sureau C, Labonté P. Use of FDA approved therapeutics with hNTCP metabolic inhibitory properties to impair the HDV lifecycle. Antiviral Res. 2014;106:111-115. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 59] [Cited by in F6Publishing: 58] [Article Influence: 5.8] [Reference Citation Analysis (0)] |
| 22. | Dandri M, Burda MR, Török E, Pollok JM, Iwanska A, Sommer G, Rogiers X, Rogler CE, Gupta S, Will H. Repopulation of mouse liver with human hepatocytes and in vivo infection with hepatitis B virus. Hepatology. 2001;33:981-988. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 321] [Cited by in F6Publishing: 331] [Article Influence: 14.4] [Reference Citation Analysis (0)] |
| 23. | Bissig KD, Wieland SF, Tran P, Isogawa M, Le TT, Chisari FV, Verma IM. Human liver chimeric mice provide a model for hepatitis B and C virus infection and treatment. J Clin Invest. 2010;120:924-930. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 272] [Cited by in F6Publishing: 267] [Article Influence: 19.1] [Reference Citation Analysis (0)] |
| 24. | Inuzuka T, Takahashi K, Chiba T, Marusawa H. Mouse models of hepatitis B virus infection comprising host-virus immunologic interactions. Pathogens. 2014;3:377-389. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 18] [Cited by in F6Publishing: 20] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 25. | Schieck A, Schulze A, Gähler C, Müller T, Haberkorn U, Alexandrov A, Urban S, Mier W. Hepatitis B virus hepatotropism is mediated by specific receptor recognition in the liver and not restricted to susceptible hosts. Hepatology. 2013;58:43-53. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 90] [Cited by in F6Publishing: 94] [Article Influence: 8.5] [Reference Citation Analysis (0)] |
| 26. | Ni Y, Lempp FA, Mehrle S, Nkongolo S, Kaufman C, Fälth M, Stindt J, Königer C, Nassal M, Kubitz R. Hepatitis B and D viruses exploit sodium taurocholate co-transporting polypeptide for species-specific entry into hepatocytes. Gastroenterology. 2014;146:1070-1083. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 540] [Cited by in F6Publishing: 548] [Article Influence: 54.8] [Reference Citation Analysis (0)] |
| 27. | Yan H, Peng B, He W, Zhong G, Qi Y, Ren B, Gao Z, Jing Z, Song M, Xu G. Molecular determinants of hepatitis B and D virus entry restriction in mouse sodium taurocholate cotransporting polypeptide. J Virol. 2013;87:7977-7991. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 134] [Cited by in F6Publishing: 147] [Article Influence: 13.4] [Reference Citation Analysis (0)] |
| 28. | Qiu X, Bi YA, Balogh LM, Lai Y. Absolute measurement of species differences in sodium taurocholate cotransporting polypeptide (NTCP/Ntcp) and its modulation in cultured hepatocytes. J Pharm Sci. 2013;102:3252-3263. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 40] [Cited by in F6Publishing: 43] [Article Influence: 3.9] [Reference Citation Analysis (0)] |
| 29. | Li H, Zhuang Q, Wang Y, Zhang T, Zhao J, Zhang Y, Zhang J, Lin Y, Yuan Q, Xia N. HBV life cycle is restricted in mouse hepatocytes expressing human NTCP. Cell Mol Immunol. 2014;11:175-183. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 63] [Cited by in F6Publishing: 80] [Article Influence: 8.0] [Reference Citation Analysis (0)] |
| 30. | Milich DR. T- and B-cell recognition of hepatitis B viral antigens. Immunol Today. 1988;9:380-386. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 103] [Cited by in F6Publishing: 104] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 31. | Ferrari C, Cavalli A, Penna A, Valli A, Bertoletti A, Pedretti G, Pilli M, Vitali P, Neri TM, Giuberti T. Fine specificity of the human T-cell response to the hepatitis B virus preS1 antigen. Gastroenterology. 1992;103:255-263. [PubMed] [Cited in This Article: ] |
| 32. | Bremer CM, Sominskaya I, Skrastina D, Pumpens P, El Wahed AA, Beutling U, Frank R, Fritz HJ, Hunsmann G, Gerlich WH. N-terminal myristoylation-dependent masking of neutralizing epitopes in the preS1 attachment site of hepatitis B virus. J Hepatol. 2011;55:29-37. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 41] [Cited by in F6Publishing: 44] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 33. | Pontisso P, Ruvoletto MG, Gerlich WH, Heermann KH, Bardini R, Alberti A. Identification of an attachment site for human liver plasma membranes on hepatitis B virus particles. Virology. 1989;173:522-530. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 105] [Cited by in F6Publishing: 117] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 34. | De Falco S, Ruvoletto MG, Verdoliva A, Ruvo M, Raucci A, Marino M, Senatore S, Cassani G, Alberti A, Pontisso P. Cloning and expression of a novel hepatitis B virus-binding protein from HepG2 cells. J Biol Chem. 2001;276:36613-36623. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 60] [Cited by in F6Publishing: 62] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 35. | Germaschewski V, Murray K. Screening a monoclonal antibody with a fusion-phage display library shows a discontinuity in a linear epitope within PreS1 of hepatitis B virus. J Med Virol. 1995;45:300-305. [PubMed] [Cited in This Article: ] |
| 36. | Glebe D, Aliakbari M, Krass P, Knoop EV, Valerius KP, Gerlich WH. Pre-s1 antigen-dependent infection of Tupaia hepatocyte cultures with human hepatitis B virus. J Virol. 2003;77:9511-9521. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 148] [Cited by in F6Publishing: 146] [Article Influence: 7.0] [Reference Citation Analysis (0)] |
| 37. | Maeng CY, Ryu CJ, Gripon P, Guguen-Guillouzo C, Hong HJ. Fine mapping of virus-neutralizing epitopes on hepatitis B virus PreS1. Virology. 2000;270:9-16. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 61] [Cited by in F6Publishing: 65] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 38. | Hong HJ, Ryu CJ, Hur H, Kim S, Oh HK, Oh MS, Park SY. In vivo neutralization of hepatitis B virus infection by an anti-preS1 humanized antibody in chimpanzees. Virology. 2004;318:134-141. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 53] [Cited by in F6Publishing: 57] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 39. | Petit MA, Strick N, Dubanchet S, Capel F, Neurath AR. Inhibitory activity of monoclonal antibody F35.25 on the interaction between hepatocytes (HepG2 cells) and preS1-specific ligands. Mol Immunol. 1991;28:517-521. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 16] [Cited by in F6Publishing: 19] [Article Influence: 0.6] [Reference Citation Analysis (0)] |
| 40. | Küttner G, Kramer A, Schmidtke G, Giessmann E, Dong L, Roggenbuck D, Scholz C, Seifert M, Stigler RD, Schneider-Mergener J. Characterization of neutralizing anti-pre-S1 and anti-pre-S2 (HBV) monoclonal antibodies and their fragments. Mol Immunol. 1999;36:669-683. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 25] [Cited by in F6Publishing: 27] [Article Influence: 1.1] [Reference Citation Analysis (0)] |
| 41. | Gripon P, Cannie I, Urban S. Efficient inhibition of hepatitis B virus infection by acylated peptides derived from the large viral surface protein. J Virol. 2005;79:1613-1622. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 262] [Cited by in F6Publishing: 267] [Article Influence: 14.1] [Reference Citation Analysis (0)] |
| 42. | Schulze A, Schieck A, Ni Y, Mier W, Urban S. Fine mapping of pre-S sequence requirements for hepatitis B virus large envelope protein-mediated receptor interaction. J Virol. 2010;84:1989-2000. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 162] [Cited by in F6Publishing: 173] [Article Influence: 11.5] [Reference Citation Analysis (0)] |
| 43. | Petersen J, Dandri M, Mier W, Lütgehetmann M, Volz T, von Weizsäcker F, Haberkorn U, Fischer L, Pollok JM, Erbes B. Prevention of hepatitis B virus infection in vivo by entry inhibitors derived from the large envelope protein. Nat Biotechnol. 2008;26:335-341. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 309] [Cited by in F6Publishing: 325] [Article Influence: 20.3] [Reference Citation Analysis (0)] |
| 44. | Kang JH, Toita R, Asai D, Yamaoka T, Murata M. Liver cell-specific peptides derived from the preS1 domain of human hepatitis B virus. J Virol Methods. 2014;201:20-23. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 2] [Cited by in F6Publishing: 2] [Article Influence: 0.2] [Reference Citation Analysis (0)] |
| 45. | Kim DH, Ni Y, Lee SH, Urban S, Han KH. An anti-viral peptide derived from the preS1 surface protein of hepatitis B virus. BMB Rep. 2008;41:640-644. [PubMed] [Cited in This Article: ] |
| 46. | Deng Q, Zhuang M, Kong YY, Xie YH, Wang Y. Screening for PreS specific binding ligands with a phage displayed peptides library. World J Gastroenterol. 2005;11:4018-4023. [PubMed] [Cited in This Article: ] |
| 47. | Deng Q, Zhai JW, Michel ML, Zhang J, Qin J, Kong YY, Zhang XX, Budkowska A, Tiollais P, Wang Y. Identification and characterization of peptides that interact with hepatitis B virus via the putative receptor binding site. J Virol. 2007;81:4244-4254. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 33] [Cited by in F6Publishing: 36] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 48. | Wang W, Liu Y, Zu X, Jin R, Xiao G. Blocking peptides against HBV: preS1 protein selected from a phage display library. Biochem Biophys Res Commun. 2011;412:633-637. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 8] [Cited by in F6Publishing: 8] [Article Influence: 0.6] [Reference Citation Analysis (0)] |
| 49. | He Y, Ye X, Tiollais P, Zhang J, Zhang J, Liu J, Xie Y. Selection of HBV preS1-binding penta-peptides by phage display. Acta Biochim Biophys Sin (Shanghai). 2014;46:691-698. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 10] [Cited by in F6Publishing: 10] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 50. | Coleman PF. Detecting hepatitis B surface antigen mutants. Emerg Infect Dis. 2006;12:198-203. [PubMed] [DOI] [Cited in This Article: ] [Cited by in F6Publishing: 6] [Reference Citation Analysis (0)] |
| 51. | Foy MC, Thio CL, Hwang HS, Saulynas M, Hamilton JP, Fine DM, Atta MG. False-negative hepatitis B virus (HBV) surface antigen in a vaccinated dialysis patient with a high level of HBV DNA in the United States. Clin Vaccine Immunol. 2012;19:820-822. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 9] [Cited by in F6Publishing: 10] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 52. | Zhevachevsky NG, Nomokonova NY, Beklemishev AB, Belov GF. Dynamic study of HBsAg and HBeAg in saliva samples from patients with hepatitis B infection: diagnostic and epidemiological significance. J Med Virol. 2000;61:433-438. [PubMed] [DOI] [Cited in This Article: ] [Cited by in F6Publishing: 1] [Reference Citation Analysis (0)] |
| 53. | Deguchi M, Yamashita N, Kagita M, Asari S, Iwatani Y, Tsuchida T, Iinuma K, Mushahwar IK. Quantitation of hepatitis B surface antigen by an automated chemiluminescent microparticle immunoassay. J Virol Methods. 2004;115:217-222. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 154] [Cited by in F6Publishing: 162] [Article Influence: 8.1] [Reference Citation Analysis (0)] |
| 54. | Shinkai N, Matsuura K, Sugauchi F, Watanabe T, Murakami S, Iio E, Ogawa S, Nojiri S, Joh T, Tanaka Y. Application of a newly developed high-sensitivity HBsAg chemiluminescent enzyme immunoassay for hepatitis B patients with HBsAg seroclearance. J Clin Microbiol. 2013;51:3484-3491. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 50] [Cited by in F6Publishing: 51] [Article Influence: 4.6] [Reference Citation Analysis (0)] |
| 55. | Ly TD, Servant-Delmas A, Bagot S, Gonzalo S, Férey MP, Ebel A, Dussaix E, Laperche S, Roque-Afonso AM. Sensitivities of four new commercial hepatitis B virus surface antigen (HBsAg) assays in detection of HBsAg mutant forms. J Clin Microbiol. 2006;44:2321-2326. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 75] [Cited by in F6Publishing: 79] [Article Influence: 4.4] [Reference Citation Analysis (0)] |
| 56. | Myyryläinen T, Talha SM, Swaminathan S, Vainionpää R, Soukka T, Khanna N, Pettersson K. Simultaneous detection of Human Immunodeficiency Virus 1 and Hepatitis B virus infections using a dual-label time-resolved fluorometric assay. J Nanobiotechnology. 2010;8:27. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 15] [Cited by in F6Publishing: 15] [Article Influence: 1.1] [Reference Citation Analysis (0)] |
| 57. | Ren ZQ, Liu TC, Hou JY, Chen MJ, Chen ZH, Lin GF, Wu YS. A rapid and sensitive method based on magnetic beads for the detection of hepatitis B virus surface antigen in human serum. Luminescence. 2014;29:591-597. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 9] [Cited by in F6Publishing: 9] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 58. | Minekawa T, Takehara S, Takahashi M, Okamoto H. Development of a highly sensitive bioluminescent enzyme immunoassay for hepatitis B virus surface antigen capable of detecting divergent mutants. Clin Vaccine Immunol. 2013;20:1255-1265. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 8] [Cited by in F6Publishing: 8] [Article Influence: 0.7] [Reference Citation Analysis (0)] |
| 59. | Klinkert MQ, Theilmann L, Pfaff E, Schaller H. Pre-S1 antigens and antibodies early in the course of acute hepatitis B virus infection. J Virol. 1986;58:522-525. [PubMed] [Cited in This Article: ] |
| 60. | Taliani G, Rapicetta M, Francisci D, Xiang J, Sarrecchia B, De Bac C, Stagni G. Correlation of preS antigens and clinical status during chronic hepatitis B virus infection. Med Microbiol Immunol. 1991;180:239-248. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 3] [Cited by in F6Publishing: 3] [Article Influence: 0.1] [Reference Citation Analysis (0)] |
| 61. | Trépo C, Zoulim F, Alonso C, Petit MA, Pichoud C, Vitvitski L. Diagnostic markers of viral hepatitis B and C. Gut. 1993;34:S20-S25. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 15] [Cited by in F6Publishing: 18] [Article Influence: 0.6] [Reference Citation Analysis (0)] |
| 62. | Wei J, Wang YQ, Lu ZM, Li GD, Wang Y, Zhang ZC. Detection of anti-preS1 antibodies for recovery of hepatitis B patients by immunoassay. World J Gastroenterol. 2002;8:276-281. [PubMed] [Cited in This Article: ] |
| 63. | Wei J, Liu XJ, Wang YQ, Lu ZM, Li GD, Wang Y, Zhang ZC. Development of the diagnostic immunoassay to detect anti-PreS1(21-47aa) antibody--a marker suggesting the health improvement of hepatitis B patients. Clin Chim Acta. 2002;317:159-169. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 7] [Cited by in F6Publishing: 8] [Article Influence: 0.4] [Reference Citation Analysis (0)] |
| 64. | Hu W, Li F, Yang X, Li Z, Xia H, Li G, Wang Y, Zhang Z. A flexible peptide linker enhances the immunoreactivity of two copies HBsAg preS1 (21-47) fusion protein. J Biotechnol. 2004;107:83-90. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 28] [Cited by in F6Publishing: 30] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 65. | Le Guillou DB, Duclos-Vallée JC, Eberle F, Capel F, Petit MA. Evaluation of an enzyme-linked immunosorbent assay for detection and quantification of hepatitis B virus PreS1 envelope antigen in serum samples: comparison with two commercial assays for monitoring hepatitis B virus DNA. J Viral Hepat. 2000;7:387-392. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 22] [Cited by in F6Publishing: 23] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 66. | Lian M, Zhou X, Wei L, Qiu S, Zhou T, Li L, Gu X, Luo M, Zheng X. Serum levels of preS antigen (HBpreSAg) in chronic hepatitis B virus infected patients. Virol J. 2007;4:93. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 4] [Cited by in F6Publishing: 4] [Article Influence: 0.2] [Reference Citation Analysis (0)] |
| 67. | Hu Z, Li M, Huang B, Liu J, Yu L, Chen G. Detection of hepatitis B virus PreS1 antigen using a time-resolved fluoroimmunoassay. J Immunoassay Immunochem. 2012;33:156-165. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 12] [Cited by in F6Publishing: 13] [Article Influence: 1.1] [Reference Citation Analysis (0)] |
| 68. | Yuan Q, Ge S, Xiong J, Yan Q, Li Z, Hao X, Tian D, Niu J, Su Z, Chen C. A novel immunoassay for PreS1 and/or core-related antigens for detection of HBsAg variants. J Virol Methods. 2010;168:108-113. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 15] [Cited by in F6Publishing: 16] [Article Influence: 1.1] [Reference Citation Analysis (0)] |
| 69. | Jeulin H, Velay A, Murray J, Schvoerer E. Clinical impact of hepatitis B and C virus envelope glycoproteins. World J Gastroenterol. 2013;19:654-664. [PubMed] [DOI] [Cited in This Article: ] [Cited by in CrossRef: 13] [Cited by in F6Publishing: 11] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 70. | Bienzle U, Günther M, Neuhaus R, Vandepapeliere P, Vollmar J, Lun A, Neuhaus P. Immunization with an adjuvant hepatitis B vaccine after liver transplantation for hepatitis B-related disease. Hepatology. 2003;38:811-819. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 5] [Cited by in F6Publishing: 4] [Article Influence: 0.2] [Reference Citation Analysis (0)] |
| 71. | Bauer T, Günther M, Bienzle U, Neuhaus R, Jilg W. Vaccination against hepatitis B in liver transplant recipients: pilot analysis of cellular immune response shows evidence of HBsAg-specific regulatory T cells. Liver Transpl. 2007;13:434-442. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 34] [Cited by in F6Publishing: 32] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 72. | Chen H, Wen B, Deng Y, Wang W, Yin X, Guan J, Ruan L, Tan W. Enhanced effect of DNA immunization plus in vivo electroporation with a combination of hepatitis B virus core-PreS1 and S-PreS1 plasmids. Clin Vaccine Immunol. 2011;18:1789-1795. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 13] [Cited by in F6Publishing: 15] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 73. | Schirmbeck R, Reimann J. Modulation of gene-gun-mediated Th2 immunity to hepatitis B surface antigen by bacterial CpG motifs or IL-12. Intervirology. 2001;44:115-123. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 38] [Cited by in F6Publishing: 39] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 74. | Sjogren MH. Prevention of hepatitis B in nonresponders to initial hepatitis B virus vaccination. Am J Med. 2005;118 Suppl 10A:34S-39S. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 57] [Cited by in F6Publishing: 57] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 75. | Filippelli M, Lionetti E, Gennaro A, Lanzafame A, Arrigo T, Salpietro C, La Rosa M, Leonardi S. Hepatitis B vaccine by intradermal route in non responder patients: an update. World J Gastroenterol. 2014;20:10383-10394. [PubMed] [DOI] [Cited in This Article: ] [Cited by in CrossRef: 57] [Cited by in F6Publishing: 53] [Article Influence: 5.3] [Reference Citation Analysis (0)] |
| 76. | Zuckerman AJ. Effect of hepatitis B virus mutants on efficacy of vaccination. Lancet. 2000;355:1382-1384. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 119] [Cited by in F6Publishing: 124] [Article Influence: 5.2] [Reference Citation Analysis (0)] |
| 77. | Aguilar JC, Lobaina Y, Muzio V, García D, Pentón E, Iglesias E, Pichardo D, Urquiza D, Rodríguez D, Silva D. Development of a nasal vaccine for chronic hepatitis B infection that uses the ability of hepatitis B core antigen to stimulate a strong Th1 response against hepatitis B surface antigen. Immunol Cell Biol. 2004;82:539-546. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 56] [Cited by in F6Publishing: 57] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 78. | Betancourt AA, Delgado CA, Estévez ZC, Martínez JC, Ríos GV, Aureoles-Roselló SR, Zaldívar RA, Guzmán MA, Baile NF, Reyes PA. Phase I clinical trial in healthy adults of a nasal vaccine candidate containing recombinant hepatitis B surface and core antigens. Int J Infect Dis. 2007;11:394-401. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 69] [Cited by in F6Publishing: 76] [Article Influence: 4.5] [Reference Citation Analysis (0)] |
| 79. | Shouval D, Ilan Y, Adler R, Deepen R, Panet A, Even-Chen Z, Gorecki M, Gerlich WH. Improved immunogenicity in mice of a mammalian cell-derived recombinant hepatitis B vaccine containing pre-S1 and pre-S2 antigens as compared with conventional yeast-derived vaccines. Vaccine. 1994;12:1453-1459. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 90] [Cited by in F6Publishing: 93] [Article Influence: 3.1] [Reference Citation Analysis (0)] |
| 80. | Raz R, Koren R, Bass D. Safety and immunogenicity of a new mammalian cell-derived recombinant hepatitis B vaccine containing Pre-S1 and Pre-S2 antigens in adults. Isr Med Assoc J. 2001;3:328-332. [PubMed] [Cited in This Article: ] |
| 81. | Ge G, Wang S, Han Y, Zhang C, Lu S, Huang Z. Removing N-terminal sequences in pre-S1 domain enhanced antibody and B-cell responses by an HBV large surface antigen DNA vaccine. PLoS One. 2012;7:e41573. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 9] [Cited by in F6Publishing: 9] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 82. | Rendi-Wagner P, Shouval D, Genton B, Lurie Y, Rümke H, Boland G, Cerny A, Heim M, Bach D, Schroeder M. Comparative immunogenicity of a PreS/S hepatitis B vaccine in non- and low responders to conventional vaccine. Vaccine. 2006;24:2781-2789. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 106] [Cited by in F6Publishing: 102] [Article Influence: 5.7] [Reference Citation Analysis (0)] |
| 83. | Krawczyk A, Ludwig C, Jochum C, Fiedler M, Heinemann FM, Shouval D, Roggendorf M, Roggendorf H, Lindemann M. Induction of a robust T- and B-cell immune response in non- and low-responders to conventional vaccination against hepatitis B by using a third generation PreS/S vaccine. Vaccine. 2014;32:5077-5082. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 42] [Cited by in F6Publishing: 41] [Article Influence: 4.1] [Reference Citation Analysis (0)] |
| 84. | Shen M, Wang S, Ge G, Xing Y, Ma X, Huang Z, Lu S. Profiles of B and T cell immune responses elicited by different forms of the hepatitis B virus surface antigen. Vaccine. 2010;28:7288-7296. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 12] [Cited by in F6Publishing: 10] [Article Influence: 0.7] [Reference Citation Analysis (0)] |
| 85. | Yue Q, Hu X, Yin W, Xu X, Wei S, Lei Y, Lü X, Yang J, Su M, Xu Z. Immune responses to recombinant Mycobacterium smegmatis expressing fused core protein and preS1 peptide of hepatitis B virus in mice. J Virol Methods. 2007;141:41-48. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 8] [Cited by in F6Publishing: 8] [Article Influence: 0.5] [Reference Citation Analysis (0)] |
| 86. | Chen X, Li M, Le X, Ma W, Zhou B. Recombinant hepatitis B core antigen carrying preS1 epitopes induce immune response against chronic HBV infection. Vaccine. 2004;22:439-446. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 32] [Cited by in F6Publishing: 31] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 87. | Michel ML, Deng Q, Mancini-Bourgine M. Therapeutic vaccines and immune-based therapies for the treatment of chronic hepatitis B: perspectives and challenges. J Hepatol. 2011;54:1286-1296. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 99] [Cited by in F6Publishing: 105] [Article Influence: 8.1] [Reference Citation Analysis (0)] |
| 88. | Safadi R, Israeli E, Papo O, Shibolet O, Melhem A, Bloch A, Rowe M, Alper R, Klein A, Hemed N. Treatment of chronic hepatitis B virus infection via oral immune regulation toward hepatitis B virus proteins. Am J Gastroenterol. 2003;98:2505-2515. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 34] [Cited by in F6Publishing: 38] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 89. | Hoa PT, Huy NT, Thu le T, Nga CN, Nakao K, Eguchi K, Chi NH, Hoang BH, Hirayama K. Randomized controlled study investigating viral suppression and serological response following pre-S1/pre-S2/S vaccine therapy combined with lamivudine treatment in HBeAg-positive patients with chronic hepatitis B. Antimicrob Agents Chemother. 2009;53:5134-5140. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 42] [Cited by in F6Publishing: 45] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 90. | Lo CM, Lau GK, Chan SC, Fan ST, Wong J. Efficacy of a pre-S containing vaccine in patients receiving lamivudine prophylaxis after liver transplantation for chronic hepatitis B. Am J Transplant. 2007;7:434-439. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 49] [Cited by in F6Publishing: 44] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 91. | Menne S, Tennant BC, Gerin JL, Cote PJ. Chemoimmunotherapy of chronic hepatitis B virus infection in the woodchuck model overcomes immunologic tolerance and restores T-cell responses to pre-S and S regions of the viral envelope protein. J Virol. 2007;81:10614-10624. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 21] [Cited by in F6Publishing: 23] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 92. | Kang JH, Toita R, Murata M. Liver cell-targeted delivery of therapeutic molecules. Crit Rev Biotechnol. 2014;1-12. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 27] [Cited by in F6Publishing: 30] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 93. | Protzer U, Nassal M, Chiang PW, Kirschfink M, Schaller H. Interferon gene transfer by a hepatitis B virus vector efficiently suppresses wild-type virus infection. Proc Natl Acad Sci USA. 1999;96:10818-10823. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 86] [Cited by in F6Publishing: 97] [Article Influence: 3.9] [Reference Citation Analysis (0)] |
| 94. | Wang L, Kaneko S, Honda M, Kobayashi K. Approach to establishing a liver targeting gene therapeutic vector using naturally occurring defective hepatitis B viruses devoid of immunogenic T cell epitope. Virus Res. 2002;85:187-197. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 6] [Cited by in F6Publishing: 6] [Article Influence: 0.3] [Reference Citation Analysis (0)] |
| 95. | Shlomai A, Lubelsky Y, Har-Noy O, Shaul Y. The “Trojan horse” model-delivery of anti-HBV small interfering RNAs by a recombinant HBV vector. Biochem Biophys Res Commun. 2009;390:619-623. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 11] [Cited by in F6Publishing: 12] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 96. | Hong R, Bai W, Zhai J, Liu W, Li X, Zhang J, Cui X, Zhao X, Ye X, Deng Q. Novel recombinant hepatitis B virus vectors efficiently deliver protein and RNA encoding genes into primary hepatocytes. J Virol. 2013;87:6615-6624. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 35] [Cited by in F6Publishing: 38] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 97. | Huang W, Li X, Yi M, Zhu S, Chen W. Targeted delivery of siRNA against hepatitis B virus by preS1 peptide molecular ligand. Hepatol Res. 2014;44:897-906. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 11] [Cited by in F6Publishing: 11] [Article Influence: 1.1] [Reference Citation Analysis (0)] |
| 98. | Bakhshinejad B, Sadeghizadeh M. Bacteriophages and their applications in the diagnosis and treatment of hepatitis B virus infection. World J Gastroenterol. 2014;20:11671-11683. [PubMed] [DOI] [Cited in This Article: ] [Cited by in CrossRef: 20] [Cited by in F6Publishing: 19] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 99. | Tang KH, Yusoff K, Tan WS. Display of hepatitis B virus PreS1 peptide on bacteriophage T7 and its potential in gene delivery into HepG2 cells. J Virol Methods. 2009;159:194-199. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 21] [Cited by in F6Publishing: 21] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 100. | Argnani R, Boccafogli L, Marconi PC, Manservigi R. Specific targeted binding of herpes simplex virus type 1 to hepatocytes via the human hepatitis B virus preS1 peptide. Gene Ther. 2004;11:1087-1098. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 21] [Cited by in F6Publishing: 24] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 101. | Wang Z, Yuan Z, Jin L. Gene delivery into hepatocytes with the preS/liposome/DNA system. Biotechnol J. 2008;3:1286-1295. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 5] [Cited by in F6Publishing: 6] [Article Influence: 0.4] [Reference Citation Analysis (0)] |
| 102. | Zhang X, Zhang Q, Peng Q, Zhou J, Liao L, Sun X, Zhang L, Gong T. Hepatitis B virus preS1-derived lipopeptide functionalized liposomes for targeting of hepatic cells. Biomaterials. 2014;35:6130-6141. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 38] [Cited by in F6Publishing: 40] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 103. | Yamada T, Iwasaki Y, Tada H, Iwabuki H, Chuah MK, VandenDriessche T, Fukuda H, Kondo A, Ueda M, Seno M. Nanoparticles for the delivery of genes and drugs to human hepatocytes. Nat Biotechnol. 2003;21:885-890. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 204] [Cited by in F6Publishing: 209] [Article Influence: 10.0] [Reference Citation Analysis (0)] |
| 104. | Jung J, Matsuzaki T, Tatematsu K, Okajima T, Tanizawa K, Kuroda S. Bio-nanocapsule conjugated with liposomes for in vivo pinpoint delivery of various materials. J Control Release. 2008;126:255-264. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 58] [Cited by in F6Publishing: 62] [Article Influence: 3.6] [Reference Citation Analysis (0)] |
| 105. | Kasuya T, Yamada T, Uyeda A, Matsuzaki T, Okajima T, Tatematsu K, Tanizawa K, Kuroda S. In vivo protein delivery to human liver-derived cells using hepatitis B virus envelope pre-S region. J Biosci Bioeng. 2008;106:99-102. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 11] [Cited by in F6Publishing: 11] [Article Influence: 0.7] [Reference Citation Analysis (0)] |
| 106. | Sao K, Murata M, Fujisaki Y, Umezaki K, Mori T, Niidome T, Katayama Y, Hashizume M. A novel protease activity assay using a protease-responsive chaperone protein. Biochem Biophys Res Commun. 2009;383:293-297. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 7] [Cited by in F6Publishing: 9] [Article Influence: 0.6] [Reference Citation Analysis (0)] |
| 107. | Toita R, Murata M, Tabata S, Abe K, Narahara S, Piao JS, Kang JH, Hashizume M. Development of human hepatocellular carcinoma cell-targeted protein cages. Bioconjug Chem. 2012;23:1494-1501. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 33] [Cited by in F6Publishing: 37] [Article Influence: 3.1] [Reference Citation Analysis (0)] |
| 108. | Toita R, Murata M, Abe K, Narahara S, Piao JS, Kang JH, Hashizume M. A nanocarrier based on a genetically engineered protein cage to deliver doxorubicin to human hepatocellular carcinoma cells. Chem Commun (Camb). 2013;49:7442-7444. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 35] [Cited by in F6Publishing: 40] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 109. | Toita R, Murata M, Abe K, Narahara S, Piao JS, Kang JH, Ohuchida K, Hashizume M. Biological evaluation of protein nanocapsules containing doxorubicin. Int J Nanomedicine. 2013;8:1989-1999. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 5] [Cited by in F6Publishing: 10] [Article Influence: 0.9] [Reference Citation Analysis (0)] |
| 110. | Murata M, Narahara S, Umezaki K, Toita R, Tabata S, Piao JS, Abe K, Kang JH, Ohuchida K, Cui L. Liver cell specific targeting by the preS1 domain of hepatitis B virus surface antigen displayed on protein nanocages. Int J Nanomedicine. 2012;7:4353-4362. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 21] [Cited by in F6Publishing: 23] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 111. | Murata M, Piao JS, Narahara S, Kawano T, Hamano N, Kang JH, Asai D, Ugawa R, Hashizume M. Expression and characterization of myristoylated preS1-conjugated nanocages for targeted cell delivery. Protein Expr Purif. 2015;110:52-56. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 3] [Cited by in F6Publishing: 4] [Article Influence: 0.4] [Reference Citation Analysis (0)] |
| 112. | Müller T, Mehrle S, Schieck A, Haberkorn U, Urban S, Mier W. Liver imaging with a novel hepatitis B surface protein derived SPECT-tracer. Mol Pharm. 2013;10:2230-2236. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 6] [Cited by in F6Publishing: 6] [Article Influence: 0.5] [Reference Citation Analysis (0)] |
| 113. | Kang JH, Asai D, Kim JH, Mori T, Toita R, Tomiyama T, Asami Y, Oishi J, Sato YT, Niidome T. Design of polymeric carriers for cancer-specific gene targeting: utilization of abnormal protein kinase Calpha activation in cancer cells. J Am Chem Soc. 2008;130:14906-14907. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 70] [Cited by in F6Publishing: 64] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 114. | Kang JH, Toita R, Katayama Y. Bio and nanotechnological strategies for tumor-targeted gene therapy. Biotechnol Adv. 2010;28:757-763. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 17] [Cited by in F6Publishing: 18] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 115. | Toita R, Kang JH, Tomiyama T, Kim CW, Shiosaki S, Niidome T, Mori T, Katayama Y. Gene carrier showing all-or-none response to cancer cell signaling. J Am Chem Soc. 2012;134:15410-15417. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 25] [Cited by in F6Publishing: 26] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 116. | Kang JH, Oishi J, Kim JH, Ijuin M, Toita R, Jun B, Asai D, Mori T, Niidome T, Tanizawa K. Hepatoma-targeted gene delivery using a tumor cell-specific gene regulation system combined with a human liver cell-specific bionanocapsule. Nanomedicine. 2010;6:583-589. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 12] [Cited by in F6Publishing: 14] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 117. | Oishi J, Jung J, Tsuchiya A, Toita R, Kang JH, Mori T, Niidome T, Tanizawa K, Kuroda S, Katayama Y. A gene-delivery system specific for hepatoma cells and an intracellular kinase signal based on human liver-specific bionanocapsules and signal-responsive artificial polymer. Int J Pharm. 2010;396:174-178. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 6] [Cited by in F6Publishing: 6] [Article Influence: 0.4] [Reference Citation Analysis (0)] |
| 118. | Kalniņš G, Cielēns I, Renhofa R. Virus-like particles addressed by HBV preS1 sequences. Env Exp Biol. 2013;11:1-8. [Cited in This Article: ] |