INTRODUCTION
Hepatitis B virus (HBV) infection is a major global health problem, despite the availability of effective vaccines. Approximately 2 billion people are infected with HBV, and over 350 million patients are chronic HBV carriers[1]. Chronic hepatitis B leads to a risk of liver cirrhosis and hepatocellular carcinoma, and more than 700000 people die from HBV-associated diseases each year[2].
Nucleos(t)ide analogs and interferon (IFN) are the currently available treatments for chronic hepatitis B. Although nucleos(t)ide analogs efficiently inhibit HBV replication, they do not completely eliminate HBV, and stopping treatment can result in viral relapse, which is why patients must be administered nucleos(t)ide analog for life once treatment has begun. Moreover, HBV can develop resistance to drugs over the long term. High-dose INF-α may help degrade nuclear viral DNA[3], but the effect is limited, and the adverse systemic events of high doses are not well tolerated by most patients. Therefore, novel therapeutic options to completely eradicate HBV infection are needed.
HBV LIFE CYCLE
HBV is a DNA virus with a 3.2 kb-long partially double-stranded relaxed circular DNA (rcDNA) as its genome (Figure 1). Covalently closed circular DNA (cccDNA) is formed from the conversion of rcDNA after HBV infection. HBV cccDNA exists persistently in hepatocyte nuclei in an episomal state, where it acts as a viral transcription template. Transcribed RNA, known as pregenomic RNA (pgRNA), serves as mRNA for the viral proteins or serves as a template for the viral genome DNA through reverse transcription. Cytoplasmic pgRNA and the P protein (viral reverse transcriptase) are co-packaged into viral envelope proteins, and rcDNA is produced from the reverse transcription of pgRNA. Nucleocapsids containing rcDNA are released from the host cell as virions or are converted to cccDNA in the nucleus (Figure 2).
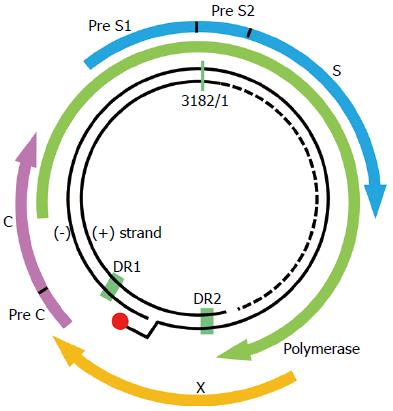
Figure 1 Hepatitis B virus genome.
Hepatitis B virus is of the Hepadnaviridae family and has a 3.2 kb partially double-stranded relaxed circular DNA (rcDNA) as its genome. The genome consists of four genes: C, P, S and X, which partially overlap. The genes encode core protein, DNA polymerase (reverse transcriptase), surface envelope protein, and X protein.
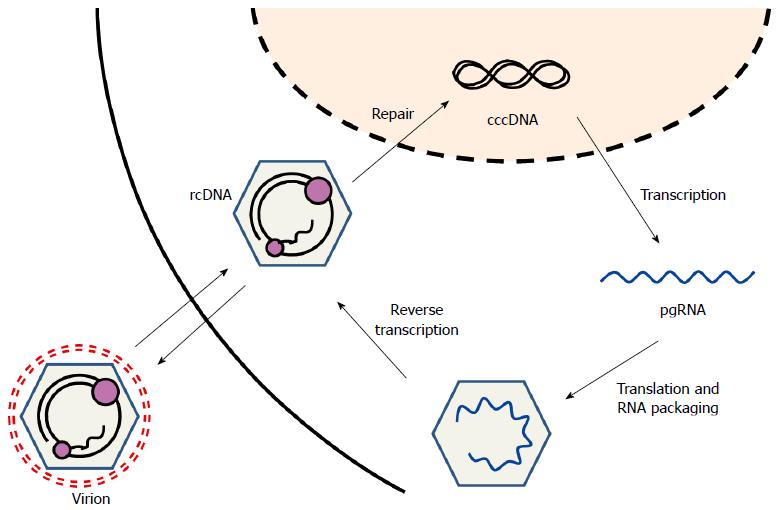
Figure 2 Life cycle of hepatitis B virus.
After hepatitis B virus (HBV) virions infect hepatocytes, relaxed circular DNA (rcDNA) enters the nucleus and is converted to covalently closed circular HBV DNA (cccDNA). HBV cccDNA persists in the nucleus as a mini-chromosome. HBV pregenomic RNA (pgRNA) is transcribed from cccDNA and acts as the viral protein mRNA. HBV pgRNA is co-packaged with reverse transcriptase in capsids and is reverse-transcribed into rcDNA. Nucleocapsids containing rcDNA are released from the host cell as virions.
Nucleos(t)ide analogs, the primary drugs that are currently used to treat patients with chronic hepatitis B, restrain HBV replication by inhibiting pgRNA reverse transcription into rcDNA. However, they do not eliminate nuclear cccDNA, which is responsible for the persistence of an HBV infection. Therefore, novel therapeutic approaches to target HBV cccDNA are urgently needed.
MEASURING CCCDNA LEVELS
The cccDNA levels in HBV-infected human liver cells are low, ranging from 1 to 50 copies per hepatocyte[4]. Therefore, stable and simple methods to measure cccDNA are necessary to evaluate HBV infection status and to assess the effects of antiviral treatment. Two methods are mainly used to measure cccDNA levels. Real-time polymerase chain reaction (PCR) amplification with specific primers detects cccDNA. However, PCR amplification can also include noise derived from other co-extracted viral DNA, leading to unreliable results. Southern blotting detects cccDNA directly and can distinguish cccDNA from other viral DNAs via the migration distance in electrophoresis[5,6]. However, Southern blotting is not convenient and is time-consuming. Furthermore, supercoiled cccDNA may be nicked during the DNA extraction procedure, which can change the cccDNA structure and its electrophoretic pattern; thus, the extraction step must be performed carefully. These procedures are frequently utilized in research, but they need to be refined to establish more reliable and convenient protocols.
IMMUNE-BASED THERAPIES
Immune-based therapies have been applied to eradicate persistent HBV infections. IFN-α exerts immunomodulatory and antiviral effects leading to HBeAg seroconversion or normalization of ALT. However, its effect is limited and associated with many adverse events, such as flu-like symptoms, bone marrow suppression, fatigue, impaired glucose tolerance, and depression. A recent study reported that IFN-α and lymphotoxin(LT)-β receptor activation pathways can degrade cccDNA through deamination by APOBEC3A and 3B, and the LT-β receptor agonist is effective at low doses without toxicity[3]. These results indicate that cytokines or cytokine receptor agonists that can induce cccDNA deamination and degradation may be potential therapeutic candidates against HBV with minimal side effects.
GENOME EDITING TECHNOLOGIES
For the complete eradication of HBV, it is necessary to target persistent cccDNA. Genome editing technologies, such as zinc finger nucleases (ZFNs), transcription activator-like effector nucleases (TALENs), and the clustered regularly interspaced short palindromic repeats/CRISPR associated (CRISPR/Cas) system, have attracted attention as potential therapeutic tools[7-9]. ZFNs and TALENs are engineered chimeric nucleases with sequence-specific binding domains and Fok1 nuclease domains that function as dimers and induce DNA double-strand breaks at specific target sites. These technologies have been used in genetic engineering in various fields, including HBV therapeutics, although certain limitations remain. ZFN specificity can be altered by context-dependent effects caused by interactions among neighboring zinc fingers of the DNA binding domain[10]. TALENs are better than ZFNs in regard to DNA specificity, but the large size of TALENs is a disadvantage, that makes delivery by viral vectors difficult[11]. ZFNs and TALENs have been used to target HBV cccDNA and have yielded some favorable effects[12,13]. However, both of ZFNs and TALENs require a pair of site-specific nucleases for each target, and thus, substantial efforts are needed to produce customized proteins.
The CRISPR/Cas system has rapidly developed as a novel genome editing tool, which is more easily customizable than ZFNs and TALENs[8]. It is derived from the acquired immune system of bacteria and archaea against invading foreign DNA. The type II CRISPR/Cas system of Streptococcus pyogenes has been utilized for research. It consists of a Cas9 nuclease and a single guide RNA (sgRNA), which leads Cas9 to the target site. A 20-nt complementary sequence to the target DNA is positioned at the 5′ end of the sgRNA, which is adjacent to the 5′ part of a proto spacer adjacent motif (PAM) matching the 5′-NGG sequence. The Cas9/sgRNA complex is guided to the target site via RNA-DNA complementary base pairing, and induces site-specific double-strand breaks (Figure 3). This system can be used flexibly by designing sgRNA to any sequences with the PAM form.
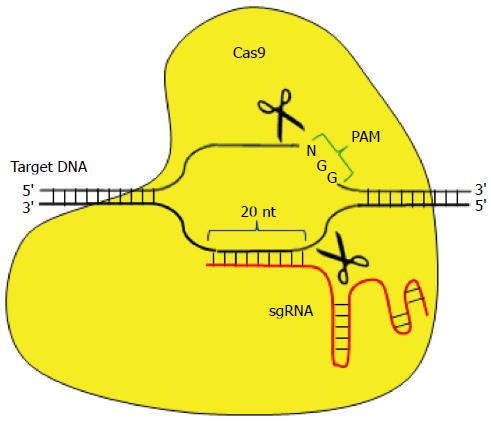
Figure 3 Clustered regularly interspaced short palindromic repeats/Cas9 system.
The clustered regularly interspaced short palindromic repeats (CRISPR)/Cas system consists of a Cas9 nuclease and a single guide RNA (sgRNA), which leads Cas9 to the target site. A 20-nt complementary sequence to the target DNA is positioned at the 5′ end of the sgRNA, which is adjacent to the 5′ part of a proto spacer adjacent motif (PAM) matching the 5′-NGG sequence. The Cas9/sgRNA complex is guided to the target site via RNA-DNA complementary base pairing and induces site-specific double-strand breaks.
Although the CRISPR/Cas9 system has been simulated using mathematical models as a therapeutic option against HBV cccDNA[14] and is being used to target various sites of cccDNA in some recent reports[15-17], the protocols need to be improved. The efficiency is still limited, as the appropriate methods to deliver the nuclease into target cells have not been established. Delivery methods via viral vectors, plasmid DNA expressing Cas9 and sgRNAs, in vitro transcribed Cas9 mRNA and sgRNAs are currently used[18]. However, the conventional transfection methods of plasmid DNA or mRNA, such as electroporation or cationic lipids, are inefficient, transient, and confined to certain cell lines. Viral vectors, including adenoviral and lentiviral vectors, can more efficiently deliver the nucleases compared to the transfection methods. Up to now, some studies targeting cccDNA via the CRISPR/Cas9 system have been performed using lentiviral vectors[16,17]. Lentiviral vectors enable the constitutive expression of Cas9 and/or sgRNAs. However, these can be integrated to the host genome, which may cause insertional mutagenesis. It is reported that integrase-deficient lentiviral vectors can deliver ZFNs[19], which may also be applicable to the CRISPR/Cas9 system. In the future, the development of improved delivery methods would lead to further progress in this field.
It is also important to select effective and specific target sequences in the viral DNA. It may be more effective to target several sequences in the HBV genome in parallel for better results. In addition, the target sequences are desirable to be well-conserved among virus isolates, and contain non-homologous sequences in the human genomes to minimize toxicity. In the CRISPR/Cas9 system, off-target effects can be observed if the sequence has up to five mismatches to the sgRNA[20]. Because HBV genomic sequences are not highly homologous with the human genome, there are many possible candidate sequences for HBV-specific sgRNAs in the HBV genome. However, these settings of sgRNAs may result in differences in the editing efficiency, which should be precisely verified in the future. After the cleavage of cccDNA, DNA repair can occur with accompanying mutations, which may be harmful to the host. Previous reports have claimed that some specific mutations of the HBV genome are associated with exacerbation of malignancy[21-23]. Thus, it may be better to carefully avoid such sites whose mutations can accelerate carcinogenesis when selecting the targets.
For practical use, there is still plenty room for the improvements to achieve safer and more effective therapeutic options using genome editing technology.