Published online Dec 21, 2014. doi: 10.3748/wjg.v20.i47.17830
Revised: July 14, 2014
Accepted: July 24, 2014
Published online: December 21, 2014
Recently, single nucleotide polymorphisms, in the vicinity of the interferon lambda 3 (IFNL3) gene have been identified as the strongest predictor of spontaneous and treatment induced clearance of hepatitis C virus (HCV) infection. Since then, increasing evidence has implicated the innate immune response in mediating the IFNL3 genotype effect. Dendritic cells (DCs) are key to the host immune response in HCV infection and their vital role in the IFNL3 genotype effect is emerging. Reports have identified subclasses of DCs, particularly myeloid DC2s and potentially plasmacytoid DCs as the major producers of IFNL3 in the setting of HCV infection. Given the complexities of dendritic cell biology and the conflicting current available data, this review aims to summarize what is currently known regarding the role of dendritic cells in HCV infection and to place it into context of what is know about lambda interferons and dendritic cells in general.
Core tip: Increasing evidence implicates the innate immune response in mediating the interferon lambda 3 (IFNL3) genotype effect in hepatitis C virus (HCV) infection. Dendritic cells (DCs) are essential players in the host immune response to HCV infection, especially with respect to the IFNL3 genotype effect. Subsets of DCs, myeloid DC2s and potentially plasmacytoid DCs, appear to particularly important in orchestrating the IFNL3 genotype effect.
-
Citation: O’Connor KS, George J, Booth D, Ahlenstiel G. Dendritic cells in hepatitis C virus infection: Key players in the
IFNL3 -genotype response. World J Gastroenterol 2014; 20(47): 17830-17838 - URL: https://www.wjgnet.com/1007-9327/full/v20/i47/17830.htm
- DOI: https://dx.doi.org/10.3748/wjg.v20.i47.17830
An estimated 3% of the world’s population is infected with hepatitis C virus (HCV)[1]. With low spontaneous clearance rates, 80% of individuals go on to develop chronic infection, which is associated with long term complications including cirrhosis, hepatocellular carcinoma and death from chronic liver failure[2]. Recently, single nucleotide polymorphisms (SNPs) in the region of the interferon lambda 3 (IFNL3; formerly known as IL28B) gene were identified to strongly predict spontaneous and treatment-induced clearance of HCV of HCV infection[3-7]. IFNL3 encodes IFNL3 a member of the type III interferon (IFN) family and thus belongs to the group of innate immune cytokines. Dendritic cells (DC) are recognized as the major producers of IFNs and central players in the host immune response against HCV[8]. In this review we explore the role of DC in chronic hepatitis C (CHC) in the context of IFNL3 and its polymorphism.
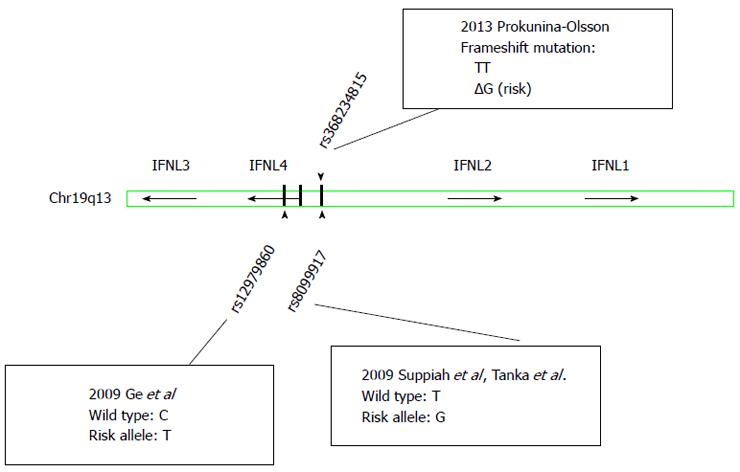
The poor treatment response rates, high economic burden and significant adverse effects associated with traditional antiviral therapy for CHC consisting of pegylated IFN-alpha and ribavirin (Peg-IFNα/RBV) motivated research into host genetic factors associated with successful HCV clearance. In 2009, four landmark genome wide association studies (GWAS) independently described SNPs in the vicinity of IFNL3 that were dramatically predictive of response to Peg-IFNα/RBV therapy in patients with genotype 1 HCV[3-5,9]. The favourable variants of the two most widely studied SNPs, rs12979860 and rs8099917, are the strongest pre-treatment predictors of SVR in genotype 1 HCV infection, but clearly also affect treatment response to Peg-IFNα/RBV in HCV genotype 2 and 3 infections[10]. Subsequently, this genetic variation has also been associated with spontaneous clearance of HCV[6,7] and liver inflammation in chronic HCV infection[11-13], strongly implicating the innate immune response in the IFNL3 genetic response.
Three classes of IFNs are now recognized (type I, II and III) and these cytokines are crucial to the establishment of an antiviral immune response. They are classified based on differences in structure, receptor and biological function: Type I IFNs include IFNA and IFN-beta (IFN-β), whereas the only type II IFN is IFN-gamma (IFN-γ)[14]. The type III or lambda IFNs were more recently identified in 2003 by two independent research groups[15,16]. Initially three members in this family were described: IFNL1 or IL29, IFNL2 or IL28A and IFNL3 or IL28B. Interestingly, lambda IFNL share similarities with both the IL10 family of cytokines and type I IFNs[17]. They signal through the same janus tyrosine kinase (JAK)/signal transducers and activators of transcription (STAT) pathway leading to induction of interferon-stimulated genes (ISGs) and antiviral activity[15,18]. Signaling through common pathways facilitates type I and type III IFNs to induce similar biological activities, mediated by the induction of nearly identical sets of more than 300 ISGs[19]. Lambda IFN induced antiviral activity has been reported against many different viruses including inhibition of HCV replication in vitro[19,20]; IFNL3 has been shown to inhibit HCV replication in three independent HCV models by the JAK/STAT pathway[21]. There is also in vitro evidence that IFNA induces expression of IFNL genes and that both cytokines appear to enhance the activity of the other[22].
In addition to their antiviral properties, lambda IFNLs exert complex and varied effects on immune cell function that are likely context dependent: Briefly, lambda IFNs have been shown to reduce the production of T helper 2 (Th2) cytokines (IL-4, IL-13, IL-14 and IL-15) thus potentially favouring Th1 driven immune response[23,24]. Further, enhanced adaptive immunity has been suggested by IFNL3 induced reduction in regulatory T cells, increased CD8+ T cell numbers[25] and augmentation of CD8+ T cell cytotoxicity[26].
In 2013, a new polymorphism (rs368234815) located between the genes for IFNL2 and IFNL3 (Figure 1) was identified and found to induce a frame shift mutation resulting in transient expression of an IFN analogue, IFN-lamda 4 (IFNL4), in stimulated human hepatocytes[27]. The authors postulated that the genotype dependent production of the protein IFNL4 resulted in altered ISG expression and thus may explain the effects on viral clearance. rs368234815 is in high linkage disequilibrium with rs12979860, but more strongly associated with spontaneous and treatment induced HCV clearance, especially in individuals of African ancestry. The exact mechanisms molecular functions of IFNL4 remain to be clarified[28,29].
All IFNLs signal through the same heterodimeric receptor complex. This is composed of a unique interferon lambda receptor 1 (IFNLR1) [also known as IL28R alpha or cytokine receptor family 2 member 12 (CRF2-12)] component and IL10R2[15,16]. Both subunits are required for signalling. IL10R2 is ubiquitously expressed while IFNLR1 displays restricted tissue expression, predominately on cells of epithelial origin (including: keratinocytes and cells of kidney, lung and gastrointestinal tract origin) as well as specific subsets of immune cells[30-32]. The IFNLR1 gene generates several splice variants including: a full length, membrane bound IFNLR1 and secreted, soluble IFNLR1 protein[33].
Expression of IFNLR1 mRNA in human immune cells, especially B, T and NK-cells, has been previously demonstrated[30]. However, these immune cells were shown to express relatively more soluble receptor, which is postulated to act as an inhibitor to IFNL activity[30]. In contrast to these reports high levels of IFNLR1 have been detected on plasmacytoid dendritic cells (pDCs) relative to other cell populations in peripheral blood mononuclear cells (PBMCs) by us and others[30,34,35]. Furthermore, we have demonstrated significant up-regulation of IFNLR1 expression after IFNA stimulation in pDCs suggesting that IFNA may enhance IFNL receptor expression and sensitivity to IFNL. Analysis of the ratio of membrane-bound receptor (IFNLR1-mem) to soluble isoforms (IFNLR1-sol) for pDCs, demonstrated that the majority was the isoform encoding the membrane-associated or functional form of IFNLR1[36]. We have also demonstrated that IFNLR1 expression was not significantly higher in HCV-infected liver biopsies compared with unstimulated pDCs[36].
Previous work has produced conflicting evidence on whether or not immune cells are a target for Type III IFNs. Several studies have failed to show a response to IFN-lambdas (IFNL1 and/or IFNL2) by a variety of immune cells including B, T and natural killer cells (NK cells) as well as monocytes[30,37]. In contrast, several other human studies have revealed a direct effect of IFNLs on monocytes[38,39], dendritic cells[37] and T cells[23,38,40]. Work in pDCs has shown that IFNL1 results in altered expression of costimulatory molecules such as CD80[41]. We and others have demonstrated that pDCs are responsive to IFNL3 as detected by up-regulation of the ISG MxA[36] and increased production of IFNα[42]. IFNL may indeed act as an autocrine signal for pDCs with the ability to improve survival and enhance antiviral response[35]. This suggests a positive feedback loop for the production of IFNL3, particularly by DCs within HCV infected livers and the potential for an augmented response with IFNA therapy.
DCs are professional antigen presenting cells and play a major role in orchestrating the innate immune response against hepatitis C virus[43]. They are a rare cell population representing 0.3%-0.5% of normal human peripheral blood mononuclear cells[44,45]. They can be broadly categorized into two major subsets: pDCs and conventional myeloid DCs (mDCs). pDC and mDc differ significantly in terms of their morphology, phenotype and function; their individual features are summarized in Table 1.
| Plasmacytoid dendritic cells | Myeloid dendritic cells | ||
| Morphology | Round, resemble plasma cells | Prominent cytoplasmic protrusions | |
| Phenotype | CD11C- CD1a+ | CD11c+ CD1a+ | |
| CD123high | CD123low | ||
| mDC1 | mDC2 | ||
| BDCA-4+ (CD304+) | |||
| BDCA-2+ (CLEC4C) | BDCA-1+ (CD1c+) | BDCA-3+ (CD141+) | |
| CLEC9A | |||
| TLR receptor expression | TLR1, TLR6, TLR7, TLR9, TLR10 | TLR1, TLR2, TLR3, TLR4, TLR5, TLR6, TLR7, TLR8,TLR10 | TLR1, TLR2, TLR3,TLR6, TLR8, TLR10 |
| MHC I | + | ++ | ++ |
| MHC II | + | +++ | ++ |
| CD86 | + | +++ | ++ |
| CD40 | + | ++ | +++ |
| CXCR3 | +++ | + | ++ |
| ICOS L | ++ | + | +++ |
mDCs originate from myeloid precursors in the bone marrow and display classic DC morphology with branched protrusions or dendrites[46]. mDCs are classical antigen presenting cells and have the ability to activate naive and effector T cells[43]. mDC can be further subdivided into mDC1 CD1c+ (blood antigen 1+; BDCA1+) or mDC2 CD141+ (blood antigen 3+; BDCA3+)[47]. Human mDC2s are reported to be a counterpart of murine CD8a+ DC[48]. mDCs express a variety of toll like receptors (TLRs) such as TLR2 recognizing viral ligands (including HCV core and NS3) and TLR3 recognizing double stranded RNA viruses[49]. mDC2s express higher levels of TLR3 than mDC1s and lack TLR4 expression. mDC2s are the rarest DC population in bone marrow and peripheral blood[50]. mDC1s are the most potent producers of IL-12 thus rendering them more efficient than mDC2s at promoting cytotoxic CD8+ T-cell responses[51].
In contrast, pDC display a plasma cell morphology and under steady state conditions express lower levels of MHC class I, MHC class II and co-stimulatory molecules such as CD86[8]. pDCs strongly express the pattern recognition receptors, TLR7 and TLR9, but not TLR3 and are thus capable of recognizing single stranded RNA and unmethlyated CpG-containing DNA ligands respectively[52]. Upon exposure to viral stimuli they are well recognized to produce massive amounts of type I interferons and acquire the capacity to present antigen[53]. In addition, they provide help to natural killer cells[54], regulate cell trafficking through the production of chemokines[55] and alter Th1/Th2 responses[56].
In CHC numbers of circulating pDCs and mDCs are reduced in peripheral blood compared with healthy controls[57-61] but both populations of DCs are significantly increased in the livers of CHC patients[61,62]. Furthermore in CHC, circulating numbers of DCs are inversely correlated with the serum alanine aminotransferase levels and severity of liver disease[63]. This suggests that immune cell trafficking to the liver may be the reason for reduced peripheral DCs numbers. Enriched mDC2 numbers in CHC infected livers have also recently been demonstrated[42,64]. Apart from enrichment in the liver, hepatic mDC2s display higher expression of CD40, CD80, CD83 and CD86 than those seen in the circulating peripheral blood compartment suggesting a more mature phenotype[65].
Both the innate and adaptive arms of the immune system contribute to the host's ability to resolve HCV infection. The first line of defense against viral infections is the innate immune response with the IFNs playing a key role in induction of the antiviral state and control of HCV replication[66]. Specific viral motifs known as pathogen-associated molecular patterns are recognized by pattern recognition receptors (PRRs). Two groups of PRRs sense viral infection, RIG-I like-receptors and TLRs (TLR3, 7, 8 or 9)[67]. Downstream signalling leads to translocation of IFN regulatory factor 3 and synthesis of IFNs and pro-inflammatory cytokines[68].
Human pDCs recognize HCV predominantly through a TLR7 medicated pathway[59]. mDCs recognize HCV infection and mediate IFNL induction by the dsRNA sensing, TLR3-mediatd pathway[42]. Subsequently secreted IFNs bind to the IFN receptors and activate the JAK/STAT pathway leading to the induction of ISGs[69]. The expression of ISGs establishes an antiviral state including in neighboring uninfected hepatocytes. However, the induction of the endogenous IFN system in the liver has limited antiviral efficiency with, persistence of HCV observed for decades despite the expression of hundreds of ISGs[70,71] In fact, it is now well established that patients with an activated endogenous IFN system are poor responders to IFNA based therapies[70-72]. Interestingly, there is evidence that hepatic IFNL rather than type I IFN induction is more closely correlated with the strength of the ISG response[73].
HCV has the ability to impede the IFN response at several levels including: NS3/4A protein cleaving adapter molecules and blocking PRR signalling; HCV core protein interfering with JAK/STAT signalling and ISG expression; NS5A inhibiting the function of several ISGs and HCV may interact directly with pDC to impair IFN production and promote apoptosis[74]. In this context, HCV has the ability to evade the host antiviral response in hepatocytes through cleavage of key molecules involved in RIG-I and TLR3 signalling hence, interfering with the induction of endogenous IFNs and ISGs. pDC by contrast have the ability to overcome this evasion through direct cell contact and transfer of viral RNA from hepatocytes recognized by TLR7. This leads to the synthesis of interferon stimulated genes and secretion of IFNs[59]. There is some evidence that in CHC infection DC function is impaired as a result of reduced antigen presentation to CD4+ T cells mediated through interference by HCV proteins[43].
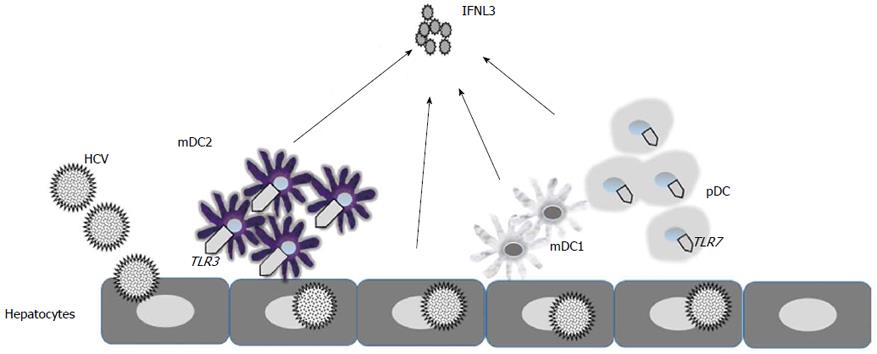
There is evidence that human hepatocytes, DCs and macrophages all produce IFNL in response to HCV infection[41,75-79]. Importantly, mDC2 peripheral blood DCs have been identified by several groups as major producers of IFNL in HCV infection[42,65]. Data suggests that IFNL induction is dependent on direct cell to cell interaction with HCV infected hepatoma cells mediated through TLR3 signalling[42]. In comparison to other DC subsets mDC2s produced large amounts of IFNL when stimulated with cell-cultured HCV and HCV-transfected Huh7.5.1 cells[65]. This is further supported by evidence that the mouse homologue for human mDC2, CD8 alpha+ cells, are potent producers of IFNL2 and IFNL3 following TLR3 activation[80].
In contrast to these studies Murata et al[81] found that mDC2s stimulated by TLR3 agonists and pDCs stimulated by TLR7 agonists both produce large amounts of IFNL3. Importantly, detectable levels of IFNL3 were only demonstrated by TLR7 agonists and not TLR3 agonists in PBMCs. There was evidence of a more robust production of IFNL3 mRNA in PBMC of patients with hepatitis C with the favorable IFNL3 genotype (rs8099917 TT) after stimulation with TLR7 agonists[81]. Importantly, this study detected that induction of IFNL3 protein was strongly correlated with Peg-IFNα/RBV treatment response in CHC[81] and that this measurement was a more accurate predictor of treatment response (95.7%) than IFNL3 genotyping (65.2%)[81] (Figure 2).
To date, the literature has been conflicting as to whether IFNL3 genotype alters IFNL3 expression. Early work suggested higher expression in whole blood in the responder genotype[4,5]. However, similar studies in PBMCs did not confirm this association[3,42]. In several independent reports examining liver biopsies from subjects with CHC, no association between IFNL3 expression and IFNL3 genotype was noted[82-84]. Yoshio et al[65] identified greater IFNL3 production by mDC2s of patients with IFNL3 responder genotypes in vitro with HCV co-culture but not TLR3 agonists. Other reports have found no IFNL3 genotype association with IFNL production in mDC2s[42] or pDCs[36]. Thus, it is possible that IFNL3 production is temporally regulated, cell type and context dependent and that genotype differences may only be observed in particular cell populations at crucial stages of infection. Further complicating DC analysis is evidence that peripheral blood DC differ from tissue resident DC[85,86].
Since the identification IFNL3 polymorphisms as important predictors of HCV clearance in 2009, signficant progress has been made relating to the underlying mechanisms: Particularly, dendritic cell subsets such as mDC2s and potentially pDCs seem to be important to IFNL3 phenomena. Given their pivotal roles in innate and adaptive immunity, genetically determined, differential regulation of IFNL3 expression is thus likely to control disease outcomes. It has remained difficult, however, to pinpoint exactly, how the polymorphism translates into different regulation. Further research is required to clarify the genetic association with clinical outcomes and how IFNL3 medicated DC responses change the ultimate outcome of HCV infection.
P- Reviewer: Conti B, Lukacs-Kornek V, Pawlowska M S- Editor: Gou SX L- Editor: A E- Editor: Liu XM
| 1. | WHO. Hepatitis C. Fact Sheet No. 164, Revised October. 2000; Available from: http://www.who.int/mediacentre/factsheets/fs164/en/. [Cited in This Article: ] |
| 2. | Hoofnagle JH. Course and outcome of hepatitis C. Hepatology. 2002;36:S21-S29. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 104] [Cited by in F6Publishing: 361] [Article Influence: 16.4] [Reference Citation Analysis (0)] |
| 3. | Ge D, Fellay J, Thompson AJ, Simon JS, Shianna KV, Urban TJ, Heinzen EL, Qiu P, Bertelsen AH, Muir AJ. Genetic variation in IL28B predicts hepatitis C treatment-induced viral clearance. Nature. 2009;461:399-401. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 2776] [Cited by in F6Publishing: 2666] [Article Influence: 177.7] [Reference Citation Analysis (0)] |
| 4. | Suppiah V, Moldovan M, Ahlenstiel G, Berg T, Weltman M, Abate ML, Bassendine M, Spengler U, Dore GJ, Powell E. IL28B is associated with response to chronic hepatitis C interferon-alpha and ribavirin therapy. Nat Genet. 2009;41:1100-1104. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1505] [Cited by in F6Publishing: 1482] [Article Influence: 98.8] [Reference Citation Analysis (0)] |
| 5. | Tanaka Y, Nishida N, Sugiyama M, Kurosaki M, Matsuura K, Sakamoto N, Nakagawa M, Korenaga M, Hino K, Hige S. Genome-wide association of IL28B with response to pegylated interferon-alpha and ribavirin therapy for chronic hepatitis C. Nat Genet. 2009;41:1105-1109. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1747] [Cited by in F6Publishing: 1746] [Article Influence: 116.4] [Reference Citation Analysis (0)] |
| 6. | Thomas DL, Thio CL, Martin MP, Qi Y, Ge D, O’Huigin C, Kidd J, Kidd K, Khakoo SI, Alexander G. Genetic variation in IL28B and spontaneous clearance of hepatitis C virus. Nature. 2009;461:798-801. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1667] [Cited by in F6Publishing: 1635] [Article Influence: 109.0] [Reference Citation Analysis (0)] |
| 7. | Tillmann HL, Thompson AJ, Patel K, Wiese M, Tenckhoff H, Nischalke HD, Lokhnygina Y, Kullig U, Göbel U, Capka E. A polymorphism near IL28B is associated with spontaneous clearance of acute hepatitis C virus and jaundice. Gastroenterology. 2010;139:1586-1592, 1592.e1. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 257] [Cited by in F6Publishing: 235] [Article Influence: 16.8] [Reference Citation Analysis (0)] |
| 8. | Lambotin M, Raghuraman S, Stoll-Keller F, Baumert TF, Barth H. A look behind closed doors: interaction of persistent viruses with dendritic cells. Nat Rev Microbiol. 2010;8:350-360. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 52] [Cited by in F6Publishing: 51] [Article Influence: 3.6] [Reference Citation Analysis (0)] |
| 9. | Rauch A, Kutalik Z, Descombes P, Cai T, Di Iulio J, Mueller T, Bochud M, Battegay M, Bernasconi E, Borovicka J. Genetic variation in IL28B is associated with chronic hepatitis C and treatment failure: a genome-wide association study. Gastroenterology. 2010;138:1338-1345, 1345.e1-e7. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 850] [Cited by in F6Publishing: 854] [Article Influence: 61.0] [Reference Citation Analysis (0)] |
| 10. | Eslam M, Leung R, Romero-Gomez M, Mangia A, Irving WL, Sheridan D, Spengler U, Mollison L, Cheng W, Bugianesi E. IFNL3 polymorphisms predict response to therapy in chronic hepatitis C genotype 2/3 infection. J Hepatol. 2014;61:235-241. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 33] [Cited by in F6Publishing: 34] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 11. | Noureddin M, Wright EC, Alter HJ, Clark S, Thomas E, Chen R, Zhao X, Conry-Cantilena C, Kleiner DE, Liang TJ. Association of IL28B genotype with fibrosis progression and clinical outcomes in patients with chronic hepatitis C: a longitudinal analysis. Hepatology. 2013;58:1548-1557. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 89] [Cited by in F6Publishing: 89] [Article Influence: 8.1] [Reference Citation Analysis (0)] |
| 12. | Honda M, Shirasaki T, Shimakami T, Sakai A, Horii R, Arai K, Yamashita T, Sakai Y, Yamashita T, Okada H. Hepatic interferon-stimulated genes are differentially regulated in the liver of chronic hepatitis C patients with different interleukin-28B genotypes. Hepatology. 2014;59:828-838. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 34] [Cited by in F6Publishing: 35] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 13. | Bochud PY, Bibert S, Kutalik Z, Patin E, Guergnon J, Nalpas B, Goossens N, Kuske L, Müllhaupt B, Gerlach T. IL28B alleles associated with poor hepatitis C virus (HCV) clearance protect against inflammation and fibrosis in patients infected with non-1 HCV genotypes. Hepatology. 2012;55:384-394. [PubMed] [Cited in This Article: ] |
| 14. | Randall RE, Goodbourn S. Interferons and viruses: an interplay between induction, signalling, antiviral responses and virus countermeasures. J Gen Virol. 2008;89:1-47. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1165] [Cited by in F6Publishing: 1203] [Article Influence: 75.2] [Reference Citation Analysis (0)] |
| 15. | Kotenko SV, Gallagher G, Baurin VV, Lewis-Antes A, Shen M, Shah NK, Langer JA, Sheikh F, Dickensheets H, Donnelly RP. IFN-lambdas mediate antiviral protection through a distinct class II cytokine receptor complex. Nat Immunol. 2003;4:69-77. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1416] [Cited by in F6Publishing: 1420] [Article Influence: 67.6] [Reference Citation Analysis (0)] |
| 16. | Sheppard P, Kindsvogel W, Xu W, Henderson K, Schlutsmeyer S, Whitmore TE, Kuestner R, Garrigues U, Birks C, Roraback J. IL-28, IL-29 and their class II cytokine receptor IL-28R. Nat Immunol. 2003;4:63-68. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1221] [Cited by in F6Publishing: 1159] [Article Influence: 55.2] [Reference Citation Analysis (0)] |
| 17. | Li M, Liu X, Zhou Y, Su SB. Interferon-lambdas: the modulators of antivirus, antitumor, and immune responses. J Leukoc Biol. 2009;86:23-32. [PubMed] [Cited in This Article: ] |
| 18. | Kotenko SV. IFN-λs. Curr Opin Immunol. 2011;23:583-590. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 174] [Cited by in F6Publishing: 172] [Article Influence: 13.2] [Reference Citation Analysis (0)] |
| 19. | Marcello T, Grakoui A, Barba-Spaeth G, Machlin ES, Kotenko SV, MacDonald MR, Rice CM. Interferons alpha and lambda inhibit hepatitis C virus replication with distinct signal transduction and gene regulation kinetics. Gastroenterology. 2006;131:1887-1898. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 478] [Cited by in F6Publishing: 463] [Article Influence: 25.7] [Reference Citation Analysis (0)] |
| 20. | Robek MD, Boyd BS, Chisari FV. Lambda interferon inhibits hepatitis B and C virus replication. J Virol. 2005;79:3851-3854. [PubMed] [Cited in This Article: ] |
| 21. | Zhang L, Jilg N, Shao RX, Lin W, Fusco DN, Zhao H, Goto K, Peng LF, Chen WC, Chung RT. IL28B inhibits hepatitis C virus replication through the JAK-STAT pathway. J Hepatol. 2011;55:289-298. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 104] [Cited by in F6Publishing: 108] [Article Influence: 8.3] [Reference Citation Analysis (0)] |
| 22. | Pagliaccetti NE, Eduardo R, Kleinstein SH, Mu XJ, Bandi P, Robek MD. Interleukin-29 functions cooperatively with interferon to induce antiviral gene expression and inhibit hepatitis C virus replication. J Biol Chem. 2008;283:30079-30089. [PubMed] [Cited in This Article: ] |
| 23. | Dai J, Megjugorac NJ, Gallagher GE, Yu RY, Gallagher G. IFN-lambda1 (IL-29) inhibits GATA3 expression and suppresses Th2 responses in human naive and memory T cells. Blood. 2009;113:5829-5838. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 93] [Cited by in F6Publishing: 94] [Article Influence: 6.3] [Reference Citation Analysis (0)] |
| 24. | Jordan WJ, Eskdale J, Srinivas S, Pekarek V, Kelner D, Rodia M, Gallagher G. Human interferon lambda-1 (IFN-lambda1/IL-29) modulates the Th1/Th2 response. Genes Immun. 2007;8:254-261. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 167] [Cited by in F6Publishing: 169] [Article Influence: 9.9] [Reference Citation Analysis (0)] |
| 25. | Morrow MP, Pankhong P, Laddy DJ, Schoenly KA, Yan J, Cisper N, Weiner DB. Comparative ability of IL-12 and IL-28B to regulate Treg populations and enhance adaptive cellular immunity. Blood. 2009;113:5868-5877. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 112] [Cited by in F6Publishing: 118] [Article Influence: 7.9] [Reference Citation Analysis (0)] |
| 26. | Morrow MP, Yan J, Pankhong P, Shedlock DJ, Lewis MG, Talbott K, Toporovski R, Khan AS, Sardesai NY, Weiner DB. IL-28B/IFN-lambda 3 drives granzyme B loading and significantly increases CTL killing activity in macaques. Mol Ther. 2010;18:1714-1723. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 43] [Cited by in F6Publishing: 48] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 27. | Prokunina-Olsson L, Muchmore B, Tang W, Pfeiffer RM, Park H, Dickensheets H, Hergott D, Porter-Gill P, Mumy A, Kohaar I. A variant upstream of IFNL3 (IL28B) creating a new interferon gene IFNL4 is associated with impaired clearance of hepatitis C virus. Nat Genet. 2013;45:164-171. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 701] [Cited by in F6Publishing: 728] [Article Influence: 66.2] [Reference Citation Analysis (0)] |
| 28. | Bibert S, Roger T, Calandra T, Bochud M, Cerny A, Semmo N, Duong FH, Gerlach T, Malinverni R, Moradpour D. IL28B expression depends on a novel TT/-G polymorphism which improves HCV clearance prediction. J Exp Med. 2013;210:1109-1116. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 168] [Cited by in F6Publishing: 174] [Article Influence: 15.8] [Reference Citation Analysis (0)] |
| 29. | Booth D, George J. Loss of function of the new interferon IFN-λ4 may confer protection from hepatitis C. Nat Genet. 2013;45:119-120. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 28] [Cited by in F6Publishing: 30] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 30. | Witte K, Gruetz G, Volk HD, Looman AC, Asadullah K, Sterry W, Sabat R, Wolk K. Despite IFN-lambda receptor expression, blood immune cells, but not keratinocytes or melanocytes, have an impaired response to type III interferons: implications for therapeutic applications of these cytokines. Genes Immun. 2009;10:702-714. [PubMed] [Cited in This Article: ] |
| 31. | Wolk K, Witte K, Witte E, Proesch S, Schulze-Tanzil G, Nasilowska K, Thilo J, Asadullah K, Sterry W, Volk HD. Maturing dendritic cells are an important source of IL-29 and IL-20 that may cooperatively increase the innate immunity of keratinocytes. J Leukoc Biol. 2008;83:1181-1193. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 124] [Cited by in F6Publishing: 118] [Article Influence: 7.4] [Reference Citation Analysis (0)] |
| 32. | Ank N, Iversen MB, Bartholdy C, Staeheli P, Hartmann R, Jensen UB, Dagnaes-Hansen F, Thomsen AR, Chen Z, Haugen H. An important role for type III interferon (IFN-lambda/IL-28) in TLR-induced antiviral activity. J Immunol. 2008;180:2474-2485. [PubMed] [Cited in This Article: ] |
| 33. | Donnelly RP, Kotenko SV. Interferon-lambda: a new addition to an old family. J Interferon Cytokine Res. 2010;30:555-564. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 296] [Cited by in F6Publishing: 304] [Article Influence: 23.4] [Reference Citation Analysis (0)] |
| 34. | Sommereyns C, Paul S, Staeheli P, Michiels T. IFN-lambda (IFN-lambda) is expressed in a tissue-dependent fashion and primarily acts on epithelial cells in vivo. PLoS Pathog. 2008;4:e1000017. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 578] [Cited by in F6Publishing: 610] [Article Influence: 38.1] [Reference Citation Analysis (0)] |
| 35. | Yin Z, Dai J, Deng J, Sheikh F, Natalia M, Shih T, Lewis-Antes A, Amrute SB, Garrigues U, Doyle S. Type III IFNs are produced by and stimulate human plasmacytoid dendritic cells. J Immunol. 2012;189:2735-2745. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 148] [Cited by in F6Publishing: 140] [Article Influence: 11.7] [Reference Citation Analysis (0)] |
| 36. | O'Connor KS, Ahlenstiel G, Suppiah V, Schibeci S, Ong A, Leung R, van der Poorten D, Douglas MW, Weltman MD, Stewart GJ. IFNL3 mediates interaction between innate immune cells: Implications for hepatitis C virus pathogenesis. Innate Immun. 2013;20:598-605. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 14] [Cited by in F6Publishing: 15] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 37. | Mennechet FJ, Uzé G. Interferon-lambda-treated dendritic cells specifically induce proliferation of FOXP3-expressing suppressor T cells. Blood. 2006;107:4417-4423. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 171] [Cited by in F6Publishing: 163] [Article Influence: 9.1] [Reference Citation Analysis (0)] |
| 38. | Jordan WJ, Eskdale J, Boniotto M, Rodia M, Kellner D, Gallagher G. Modulation of the human cytokine response by interferon lambda-1 (IFN-lambda1/IL-29). Genes Immun. 2007;8:13-20. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 103] [Cited by in F6Publishing: 102] [Article Influence: 6.0] [Reference Citation Analysis (0)] |
| 39. | Pekarek V, Srinivas S, Eskdale J, Gallagher G. Interferon lambda-1 (IFN-lambda1/IL-29) induces ELR(-) CXC chemokine mRNA in human peripheral blood mononuclear cells, in an IFN-gamma-independent manner. Genes Immun. 2007;8:177-180. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 55] [Cited by in F6Publishing: 56] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 40. | Chi B, Dickensheets HL, Spann KM, Alston MA, Luongo C, Dumoutier L, Huang J, Renauld JC, Kotenko SV, Roederer M. Alpha and lambda interferon together mediate suppression of CD4 T cells induced by respiratory syncytial virus. J Virol. 2006;80:5032-5040. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 90] [Cited by in F6Publishing: 91] [Article Influence: 5.1] [Reference Citation Analysis (0)] |
| 41. | Megjugorac NJ, Gallagher GE, Gallagher G. Modulation of human plasmacytoid DC function by IFN-lambda1 (IL-29). J Leukoc Biol. 2009;86:1359-1363. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 88] [Cited by in F6Publishing: 90] [Article Influence: 6.0] [Reference Citation Analysis (0)] |
| 42. | Zhang S, Kodys K, Li K, Szabo G. Human type 2 myeloid dendritic cells produce interferon-λ and amplify interferon-α in response to hepatitis C virus infection. Gastroenterology. 2013;144:414-425.e7. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 86] [Cited by in F6Publishing: 90] [Article Influence: 8.2] [Reference Citation Analysis (0)] |
| 43. | Spaan M, Janssen HL, Boonstra A. Immunology of hepatitis C virus infections. Best Pract Res Clin Gastroenterol. 2012;26:391-400. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 20] [Cited by in F6Publishing: 21] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 44. | O’Doherty U, Peng M, Gezelter S, Swiggard WJ, Betjes M, Bhardwaj N, Steinman RM. Human blood contains two subsets of dendritic cells, one immunologically mature and the other immature. Immunology. 1994;82:487-493. [PubMed] [Cited in This Article: ] |
| 45. | Tavakoli S, Mederacke I, Herzog-Hauff S, Glebe D, Grün S, Strand D, Urban S, Gehring A, Galle PR, Böcher WO. Peripheral blood dendritic cells are phenotypically and functionally intact in chronic hepatitis B virus (HBV) infection. Clin Exp Immunol. 2008;151:61-70. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 45] [Cited by in F6Publishing: 51] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 46. | Shortman K, Liu YJ. Mouse and human dendritic cell subtypes. Nat Rev Immunol. 2002;2:151-161. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1693] [Cited by in F6Publishing: 1600] [Article Influence: 72.7] [Reference Citation Analysis (0)] |
| 47. | Ziegler-Heitbrock L, Ancuta P, Crowe S, Dalod M, Grau V, Hart DN, Leenen PJ, Liu YJ, MacPherson G, Randolph GJ. Nomenclature of monocytes and dendritic cells in blood. Blood. 2010;116:e74-e80. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1589] [Cited by in F6Publishing: 1646] [Article Influence: 117.6] [Reference Citation Analysis (0)] |
| 48. | Poulin LF, Salio M, Griessinger E, Anjos-Afonso F, Craciun L, Chen JL, Keller AM, Joffre O, Zelenay S, Nye E. Characterization of human DNGR-1+ BDCA3+ leukocytes as putative equivalents of mouse CD8alpha+ dendritic cells. J Exp Med. 2010;207:1261-1271. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 512] [Cited by in F6Publishing: 543] [Article Influence: 38.8] [Reference Citation Analysis (0)] |
| 49. | Losikoff PT, Self AA, Gregory SH. Dendritic cells, regulatory T cells and the pathogenesis of chronic hepatitis C. Virulence. 2012;3:610-620. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 36] [Cited by in F6Publishing: 38] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 50. | Jongbloed SL, Kassianos AJ, McDonald KJ, Clark GJ, Ju X, Angel CE, Chen CJ, Dunbar PR, Wadley RB, Jeet V. Human CD141+ (BDCA-3)+ dendritic cells (DCs) represent a unique myeloid DC subset that cross-presents necrotic cell antigens. J Exp Med. 2010;207:1247-1260. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 760] [Cited by in F6Publishing: 801] [Article Influence: 57.2] [Reference Citation Analysis (0)] |
| 51. | Nizzoli G, Krietsch J, Weick A, Steinfelder S, Facciotti F, Gruarin P, Bianco A, Steckel B, Moro M, Crosti M. Human CD1c+ dendritic cells secrete high levels of IL-12 and potently prime cytotoxic T-cell responses. Blood. 2013;122:932-942. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 229] [Cited by in F6Publishing: 246] [Article Influence: 22.4] [Reference Citation Analysis (0)] |
| 52. | Hoeffel G, Ripoche AC, Matheoud D, Nascimbeni M, Escriou N, Lebon P, Heshmati F, Guillet JG, Gannagé M, Caillat-Zucman S. Antigen crosspresentation by human plasmacytoid dendritic cells. Immunity. 2007;27:481-492. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 200] [Cited by in F6Publishing: 211] [Article Influence: 12.4] [Reference Citation Analysis (0)] |
| 53. | Merad M, Sathe P, Helft J, Miller J, Mortha A. The dendritic cell lineage: ontogeny and function of dendritic cells and their subsets in the steady state and the inflamed setting. Annu Rev Immunol. 2013;31:563-604. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1510] [Cited by in F6Publishing: 1610] [Article Influence: 146.4] [Reference Citation Analysis (0)] |
| 54. | Zitvogel L, Terme M, Borg C, Trinchieri G. Dendritic cell-NK cell cross-talk: regulation and physiopathology. Curr Top Microbiol Immunol. 2006;298:157-174. [PubMed] [Cited in This Article: ] |
| 55. | Jego G, Palucka AK, Blanck JP, Chalouni C, Pascual V, Banchereau J. Plasmacytoid dendritic cells induce plasma cell differentiation through type I interferon and interleukin 6. Immunity. 2003;19:225-234. [PubMed] [Cited in This Article: ] |
| 56. | Ito T, Amakawa R, Inaba M, Hori T, Ota M, Nakamura K, Takebayashi M, Miyaji M, Yoshimura T, Inaba K. Plasmacytoid dendritic cells regulate Th cell responses through OX40 ligand and type I IFNs. J Immunol. 2004;172:4253-4259. [PubMed] [Cited in This Article: ] |
| 57. | Ryan EJ, O’Farrelly C. The affect of chronic hepatitis C infection on dendritic cell function: a summary of the experimental evidence. J Viral Hepat. 2011;18:601-607. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 23] [Cited by in F6Publishing: 21] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 58. | Ulsenheimer A, Gerlach JT, Jung MC, Gruener N, Wächtler M, Backmund M, Santantonio T, Schraut W, Heeg MH, Schirren CA. Plasmacytoid dendritic cells in acute and chronic hepatitis C virus infection. Hepatology. 2005;41:643-651. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 89] [Cited by in F6Publishing: 93] [Article Influence: 4.9] [Reference Citation Analysis (0)] |
| 59. | Takahashi K, Asabe S, Wieland S, Garaigorta U, Gastaminza P, Isogawa M, Chisari FV. Plasmacytoid dendritic cells sense hepatitis C virus-infected cells, produce interferon, and inhibit infection. Proc Natl Acad Sci USA. 2010;107:7431-7436. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 218] [Cited by in F6Publishing: 212] [Article Influence: 15.1] [Reference Citation Analysis (0)] |
| 60. | Wertheimer AM, Bakke A, Rosen HR. Direct enumeration and functional assessment of circulating dendritic cells in patients with liver disease. Hepatology. 2004;40:335-345. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 89] [Cited by in F6Publishing: 93] [Article Influence: 4.7] [Reference Citation Analysis (0)] |
| 61. | Cicinnati VR, Kang J, Sotiropoulos GC, Hilgard P, Frilling A, Broelsch CE, Gerken G, Beckebaum S. Altered chemotactic response of myeloid and plasmacytoid dendritic cells from patients with chronic hepatitis C: role of alpha interferon. J Gen Virol. 2008;89:1243-1253. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 22] [Cited by in F6Publishing: 22] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 62. | Nattermann J, Zimmermann H, Iwan A, von Lilienfeld-Toal M, Leifeld L, Nischalke HD, Langhans B, Sauerbruch T, Spengler U. Hepatitis C virus E2 and CD81 interaction may be associated with altered trafficking of dendritic cells in chronic hepatitis C. Hepatology. 2006;44:945-954. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 71] [Cited by in F6Publishing: 73] [Article Influence: 4.1] [Reference Citation Analysis (0)] |
| 63. | Kunitani H, Shimizu Y, Murata H, Higuchi K, Watanabe A. Phenotypic analysis of circulating and intrahepatic dendritic cell subsets in patients with chronic liver diseases. J Hepatol. 2002;36:734-741. [PubMed] [Cited in This Article: ] |
| 64. | Velazquez VM, Hon H, Ibegbu C, Knechtle SJ, Kirk AD, Grakoui A. Hepatic enrichment and activation of myeloid dendritic cells during chronic hepatitis C virus infection. Hepatology. 2012;56:2071-2081. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 31] [Cited by in F6Publishing: 32] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 65. | Yoshio S, Kanto T, Kuroda S, Matsubara T, Higashitani K, Kakita N, Ishida H, Hiramatsu N, Nagano H, Sugiyama M. Human blood dendritic cell antigen 3 (BDCA3)(+) dendritic cells are a potent producer of interferon-λ in response to hepatitis C virus. Hepatology. 2013;57:1705-1715. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 78] [Cited by in F6Publishing: 77] [Article Influence: 7.0] [Reference Citation Analysis (0)] |
| 66. | Heim MH. Innate immunity and HCV. J Hepatol. 2013;58:564-574. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 116] [Cited by in F6Publishing: 117] [Article Influence: 10.6] [Reference Citation Analysis (0)] |
| 67. | Takeuchi O, Akira S. Innate immunity to virus infection. Immunol Rev. 2009;227:75-86. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 925] [Cited by in F6Publishing: 905] [Article Influence: 60.3] [Reference Citation Analysis (0)] |
| 68. | Kawai T, Akira S. Toll-like receptor and RIG-I-like receptor signaling. Ann N Y Acad Sci. 2008;1143:1-20. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 681] [Cited by in F6Publishing: 719] [Article Influence: 44.9] [Reference Citation Analysis (0)] |
| 69. | Guo JT, Sohn JA, Zhu Q, Seeger C. Mechanism of the interferon alpha response against hepatitis C virus replicons. Virology. 2004;325:71-81. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 46] [Cited by in F6Publishing: 41] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 70. | Chen L, Borozan I, Feld J, Sun J, Tannis LL, Coltescu C, Heathcote J, Edwards AM, McGilvray ID. Hepatic gene expression discriminates responders and nonresponders in treatment of chronic hepatitis C viral infection. Gastroenterology. 2005;128:1437-1444. [PubMed] [Cited in This Article: ] |
| 71. | Asselah T, Bieche I, Narguet S, Sabbagh A, Laurendeau I, Ripault MP, Boyer N, Martinot-Peignoux M, Valla D, Vidaud M. Liver gene expression signature to predict response to pegylated interferon plus ribavirin combination therapy in patients with chronic hepatitis C. Gut. 2008;57:516-524. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 162] [Cited by in F6Publishing: 165] [Article Influence: 10.3] [Reference Citation Analysis (0)] |
| 72. | Feld JJ, Nanda S, Huang Y, Chen W, Cam M, Pusek SN, Schweigler LM, Theodore D, Zacks SL, Liang TJ. Hepatic gene expression during treatment with peginterferon and ribavirin: Identifying molecular pathways for treatment response. Hepatology. 2007;46:1548-1563. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 222] [Cited by in F6Publishing: 216] [Article Influence: 12.7] [Reference Citation Analysis (0)] |
| 73. | Thomas E, Gonzalez VD, Li Q, Modi AA, Chen W, Noureddin M, Rotman Y, Liang TJ. HCV infection induces a unique hepatic innate immune response associated with robust production of type III interferons. Gastroenterology. 2012;142:978-988. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 222] [Cited by in F6Publishing: 217] [Article Influence: 18.1] [Reference Citation Analysis (0)] |
| 74. | Rehermann B. Hepatitis C virus versus innate and adaptive immune responses: a tale of coevolution and coexistence. J Clin Invest. 2009;119:1745-1754. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 394] [Cited by in F6Publishing: 405] [Article Influence: 27.0] [Reference Citation Analysis (0)] |
| 75. | Li K, Li NL, Wei D, Pfeffer SR, Fan M, Pfeffer LM. Activation of chemokine and inflammatory cytokine response in hepatitis C virus-infected hepatocytes depends on Toll-like receptor 3 sensing of hepatitis C virus double-stranded RNA intermediates. Hepatology. 2012;55:666-675. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 140] [Cited by in F6Publishing: 137] [Article Influence: 11.4] [Reference Citation Analysis (0)] |
| 76. | Marukian S, Andrus L, Sheahan TP, Jones CT, Charles ED, Ploss A, Rice CM, Dustin LB. Hepatitis C virus induces interferon-λ and interferon-stimulated genes in primary liver cultures. Hepatology. 2011;54:1913-1923. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 147] [Cited by in F6Publishing: 154] [Article Influence: 11.8] [Reference Citation Analysis (0)] |
| 77. | Pagliaccetti NE, Robek MD. Interferon-lambda in the immune response to hepatitis B virus and hepatitis C virus. J Interferon Cytokine Res. 2010;30:585-590. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 31] [Cited by in F6Publishing: 36] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 78. | Coccia EM, Severa M, Giacomini E, Monneron D, Remoli ME, Julkunen I, Cella M, Lande R, Uzé G. Viral infection and Toll-like receptor agonists induce a differential expression of type I and lambda interferons in human plasmacytoid and monocyte-derived dendritic cells. Eur J Immunol. 2004;34:796-805. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 376] [Cited by in F6Publishing: 374] [Article Influence: 18.7] [Reference Citation Analysis (0)] |
| 79. | Mihm S, Frese M, Meier V, Wietzke-Braun P, Scharf JG, Bartenschlager R, Ramadori G. Interferon type I gene expression in chronic hepatitis C. Lab Invest. 2004;84:1148-1159. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 71] [Cited by in F6Publishing: 76] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 80. | Lauterbach H, Bathke B, Gilles S, Traidl-Hoffmann C, Luber CA, Fejer G, Freudenberg MA, Davey GM, Vremec D, Kallies A. Mouse CD8alpha+ DCs and human BDCA3+ DCs are major producers of IFN-lambda in response to poly IC. J Exp Med. 2010;207:2703-2717. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 224] [Cited by in F6Publishing: 214] [Article Influence: 15.3] [Reference Citation Analysis (0)] |
| 81. | Murata K, Sugiyama M, Kimura T, Yoshio S, Kanto T, Kirikae I, Saito H, Aoki Y, Hiramine S, Matsui T. Ex vivo induction of IFN-λ3 by a TLR7 agonist determines response to Peg-IFN/ribavirin therapy in chronic hepatitis C patients. J Gastroenterol. 2014;49:126-137. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 14] [Cited by in F6Publishing: 14] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 82. | Dill MT, Duong FH, Vogt JE, Bibert S, Bochud PY, Terracciano L, Papassotiropoulos A, Roth V, Heim MH. Interferon-induced gene expression is a stronger predictor of treatment response than IL28B genotype in patients with hepatitis C. Gastroenterology. 2011;140:1021-1031. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 208] [Cited by in F6Publishing: 206] [Article Influence: 15.8] [Reference Citation Analysis (0)] |
| 83. | Honda M, Sakai A, Yamashita T, Nakamoto Y, Mizukoshi E, Sakai Y, Yamashita T, Nakamura M, Shirasaki T, Horimoto K. Hepatic ISG expression is associated with genetic variation in interleukin 28B and the outcome of IFN therapy for chronic hepatitis C. Gastroenterology. 2010;139:499-509. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 330] [Cited by in F6Publishing: 339] [Article Influence: 24.2] [Reference Citation Analysis (0)] |
| 84. | Urban TJ, Thompson AJ, Bradrick SS, Fellay J, Schuppan D, Cronin KD, Hong L, McKenzie A, Patel K, Shianna KV. IL28B genotype is associated with differential expression of intrahepatic interferon-stimulated genes in patients with chronic hepatitis C. Hepatology. 2010;52:1888-1896. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 303] [Cited by in F6Publishing: 321] [Article Influence: 22.9] [Reference Citation Analysis (0)] |
| 85. | Harman AN, Bye CR, Nasr N, Sandgren KJ, Kim M, Mercier SK, Botting RA, Lewin SR, Cunningham AL, Cameron PU. Identification of lineage relationships and novel markers of blood and skin human dendritic cells. J Immunol. 2013;190:66-79. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 77] [Cited by in F6Publishing: 79] [Article Influence: 6.6] [Reference Citation Analysis (0)] |
| 86. | Cunningham AL, Harman A, Kim M, Nasr N, Lai J. Immunobiology of dendritic cells and the influence of HIV infection. Adv Exp Med Biol. 2013;762:1-44. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 10] [Cited by in F6Publishing: 7] [Article Influence: 0.6] [Reference Citation Analysis (0)] |