Published online Oct 28, 2014. doi: 10.3748/wjg.v20.i40.14904
Revised: May 19, 2014
Accepted: June 26, 2014
Published online: October 28, 2014
AIM: To investigate expression of microRNA (miRNA) and potential targets in chemotherapy resistant esophageal cancer cell lines.
METHODS: An in-vitro model of acquired chemotherapy resistance in esophageal adeno- (EAC) and squamous cell carcinoma (ESCC) cells was used, and microRNA expression profiles for cisplatin or 5-fluorouracil (5-FU) resistant variants vs chemotherapy sensitive controls were compared using microarray and quantitative real-time polymerase chain reaction (PCR). The expression of chemotherapy-relevant genes potentially targeted by the dysregulated microRNAs in the chemotherapy resistant variants was also evaluated.
RESULTS: Chemotherapy resistant sublines were found to have specific miRNA signatures, and these miRNA signatures were different for the cisplatin vs 5-FU resistant cells from the same tumor cell line, and also for EAC vs ESCC cells with resistance to the same specific chemotherapy agent. Amongst others, miR-27b-3p, miR-193b-3p, miR-192-5p, miR-378 a-3p, miR-125a-5p and miR-18a-3p were dysregulated, consistent with negative posttranscriptional control of KRAS, TYMS, ABCC3, CBL-B and ERBB2 expression via these miRNAs.
CONCLUSION: The current study supports the hypothesis that microRNA expression has an impact on chemotherapy resistance in esophageal cancer.
Core tip: The current study demonstrates that chemotherapy resistant esophageal adeno- and squamous cell carcinoma cells present distinct microRNA (miRNA) signatures, with a number of well known resistance relevant miRNAs differentially expressed in the derived cisplatin or 5-fluorouracil resistant cell lines. Furthermore, a number of putative target genes that are known to have an impact on chemotherapy resistance are dysregulated in the chemotherapy resistant cell lines in a direction consistent with negative posttranscriptional control of target gene expression via the respective miRNA, thereby implicating a potential mediatory effect in terms of chemotherapy resistance development.
- Citation: Hummel R, Sie C, Watson DI, Wang T, Ansar A, Michael MZ, Hoek MVD, Haier J, Hussey DJ. MicroRNA signatures in chemotherapy resistant esophageal cancer cell lines. World J Gastroenterol 2014; 20(40): 14904-14912
- URL: https://www.wjgnet.com/1007-9327/full/v20/i40/14904.htm
- DOI: https://dx.doi.org/10.3748/wjg.v20.i40.14904
Esophageal cancer is the eighth most common cancer worldwide, and the sixth most common cause of cancer related death [GLOBOCAN 2008 (IARC), Section of Cancer Information (30/1/2012)]. Despite improvements in the management of esophageal cancer, however, the outcome for individuals developing this disease remains poor. Chemotherapy and/or radiotherapy based treatment approaches are used in many patients, and recently several meta-analyses have demonstrated a survival advantage for patients undergoing either neoadjuvant chemotherapy or combined chemo-radiotherapy treatment before surgery for esophageal cancer, compared to surgery alone[1,2]. In addition, in patients not suitable for surgical resection, combined chemo-radiotherapy is followed by complete macroscopic tumor regression, when assessed by endoscopy, in up to 50% of patients, with a partial response achieved in approximately half of the remaining patients[3]. However, individuals respond to these treatments in a variable fashion, and those who respond poorly to chemotherapy probably undergo futile treatment. Identification of individuals who are unlikely to benefit before treatment starts is desirable as it would allow treatment to be tailored to the individuals most likely to benefit. It is possible that novel molecular biomarkers, which predict response to chemotherapy, might be identifiable and might be then used to tailor treatment. It is also possible that molecular biomarkers of chemotherapy response might provide novel therapeutic targets to overcome potential chemotherapy resistance.
In this context, microRNA (miRNA) biomarkers are promising candidates. It has been demonstrated that miRNA expression profiles differ between various esophageal derived tissues, correlate with prognosis and clinico-pathological features in esophageal cancer, and impact directly on tumor growth, tumor cell proliferation, and tumor invasion[4,5]. In addition, for some other cancer types there is evidence that miRNA expression has an impact on the response to chemotherapy[6], but so far there is very little data available from studies evaluating esophageal cancer in this context.
To investigate this further, we sought associations between miRNA expression and response to chemotherapy in esophageal cancer. Specifically, we assessed whether chemotherapy resistant esophageal cancer cells exhibit a specific miRNA expression pattern, and whether the expression of potential chemotherapy resistance-relevant targets of the dysregulated miRNAs is altered in chemotherapy resistant cancer cells.
The chemotherapy sensitive human adenocarcinoma (EAC) cell line OE19 and the squamous cell carcinoma (ESCC) cell line KYSE410, and cisplatin and 5-Fluorouracil (5-FU) resistant variants of both cell lines which were developed in our laboratory, were used for the current study. Resistant sublines of both sensitive cell lines were generated using a pulsatile treatment approach which included repetitive treatment of cells with constant concentrations of cisplatin or 5-FU. Briefly, KYSE 410 cells were subjected to a 4-d exposure of 2 μmol/L cisplatin or 5 μmol/L 5-FU, and OE 19 cells were exposed to 5 μmol/L cisplatin or 10 μmol/L 5-FU for 3 d. The medium was not changed during this period, thereby providing a constant exposure to the chemotherapy agent. After removal of the chemotherapy agent, cells were allowed to recover, split when approximately 70%-80% confluent, and then exposed to the next cycle of chemotherapy. All derived cell lines were shown to have significant resistance to the corresponding chemotherapy agent[7].
Cells were cultured in a humidified atmosphere containing 5% CO2 at 37 °C, using DMEM high glucose/phenol red free DMEM/F12 1:1 medium (OE-19) or 1 × RPMI 1640/phenol red free RPMI 1640 medium (KYSE410) supplemented with 10% fetal bovine serum, 1% Penicillin-Streptomycin (GIBCO® Invitrogen, Life Technologies Australia Pty Ltd., Victoria, Australia) and 2‰ Normocin™ (InvivoGen, Life Research Pty Ltd, Victoria, Australia). Resistant variants were further repeatedly exposed to the respective chemotherapeutic agent as outlined above in order to maintain the selection pressure[7].
Cells from the chemotherapy resistant variants and their original cell line were seeded into 6 replicate flasks each. Prior to harvest, resistant cell lines were allowed to grow for at least 24-48 h in chemotherapy-free medium in order to avoid an acute toxicity response to the respective chemotherapeutic agent. Then, cells were harvested at a confluency of about 80% in TRIzol® (Invitrogen) and total RNA was extracted from the cells according to the manufacturer’s protocol. RNA concentration was measured using UV spectrophotometry (NanoDrop® ND-8000 Spectrophotometer, Thermo Fisher Scientific), and quality determined by visualization of distinct 28S and 18S rRNA species via electrophoresis through a 1% agarose gel.
500 ng of total RNA was used for microarray studies. Total RNA was labeled using the FlashTag HSR Biotin RNA Labelling Kits (Genisphere LLC. Hatfield) and hybridized to microRNA Arrays (Affymetrix GeneChip miRNA Array which contained 848 miRNAs representing Sanger miRBase v11, Affymetrix Inc.) as per the Genisphere manual. Briefly, RNA was poly-A tailed, and then a proprietary biotin-labelled dendramer molecule was joined to the 3’ end using DNA ligase. Labeled samples were hybridized to the arrays at 48 °C for 16 h and then washed and stained with a Steptavidin-PE solution prior to imaging. The resultant CEL files containing the raw intensity data were background corrected and statistically analyzed using Partek Genomics Suite (Partek Inc. St Louis, United States) using the subset of human targeting probes on the array. Raw data was processed using Robust Multichip Average (RMA) background correction, quantile normalization and probe summarization. Differential miRNA expression was determined by ANOVA (analysis of variance) with the p-value adjusted using the step-up multiple test correction. The step-up method used was the Benjamini_Hochberg method for multiple test correction, which is widely used for microarray data analysis[8]. Adjusted P < 0.05 were considered to be significant. Microarray data were deposited in the Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/geo; GEO accession number GSE50224). Subsequently, the most significantly dysregulated miRNAs from the microarray experiments for each resistant cell line were selected and expression of these candidates was assessed via quantitative real-time polymerase chain reaction (PCR).
For quantitative real-time PCR validation, all samples underwent DNase-pretreatment using Ambion® TURBO DNA-free™ (Applied Biosystems) in order to remove contaminating DNA. 500 ng of RNA dissolved in 8 μL nuclease free water was incubated with 1 μL 10 × Turbo DNase Buffer and 1 μL rDNase I for 30 min at 37 °C in a thermocycler (Eppendorf Mastercycler). Samples were then incubated with 1 μL DNase Inactivation Reagent for 5 min at room temperature, and centrifuged at 10000 g for 5 min at 4 °C. DNase treated RNA was finally incubated for 10 min at 75ºC, and stored on ice for further processing.
The miScript Reverse Transcription Kit (Qiagen) was used, as it allows quantification of mature miRNA and mRNA from the same cDNA sample. 500 ng of DNase treated RNA was combined with 4 μL miScript 5x RT Buffer, 1 μL Reverse Transcriptase and 5 μL nuclease free water before incubation (protocol: 60 min at 37 °C, 5 min at 95 °C, then hold at 4 °C). For miRNA validation via quantitative real-time PCR (PCR), the miScript PCR system was used, and 5 μL of cDNA was mixed with 10 μL 2 × QuantiTect SYBR, 2 μL 10 × miScript Universal Primer, 2 μL gene specific 10 × miScript Primer Assay, and 1 μL nuclease free water. Quantitative analysis was performed using Q-Gene software. MiRNA expression data was normalized to the expression levels of RNU44, which displayed comparable expression across the different groups (data not shown).
To evaluate the functional relevance of the miRNAs that were identified to be dysregulated in the development of chemotherapy resistance, we next performed gene expression analyses of the mRNA levels for putative targets of these miRNAs. In this context, potential gene targets of the dysregulated miRNAs were identified from the literature and bioinformatics tools (DIANA-mirPath[9]), and we focused on genes that were known to be relevant to cellular resistance to chemotherapy (cisplatin resistant EAC: ZEB1/ZEB 2[10], Bcl-2[11], DUSP16[12], KRAS[13], MAP4K4[14]; 5-FU resistant EAC: KRAS[13], CDKN1b[15], TYMS[16], ABCC3[17], CBL-B[18]; 5-FU resistant ESCC: ERBB2/ERBB3[19], MUC1[20], MAP4K4[14], KRAS[13]). To determine the mRNA levels, the same cDNA that was used for the determination of miRNA expression levels was used (see section “DNase-pretreatment, reverse transcription and quantitative real-time PCR (qPCR) for miRNA validation”). TaqMan® Assays (Applied Biosystems) were used as follows: 3 μL of cDNA was mixed with 10 μL TaqMan® Gene Expression Master Mix (× 2), 1 μL of the respective TaqMan® Primer (× 20) and 6 μL nuclease free water. All samples were assayed in triplicate reactions using a Rotorgene 6000 thermocycler (Corbett Life Science). Quantitative analysis was performed using Q-Gene software. mRNA expression data was normalized to 18S rRNA, which displayed comparable expression across the different groups (data not shown).
Expression data for miRNAs/mRNAs were expressed as means of normalized expression with standard deviation. These data were assessed for statistical significance using Student’s t-test for equal and unequal variances, as appropriate, based on Levene’s test for homogeneity of variances. P≤ 0.05 was considered to be statistically significant. Analyses were performed using SPSS 17.0 for Windows (SPSS, Chicago, IL).
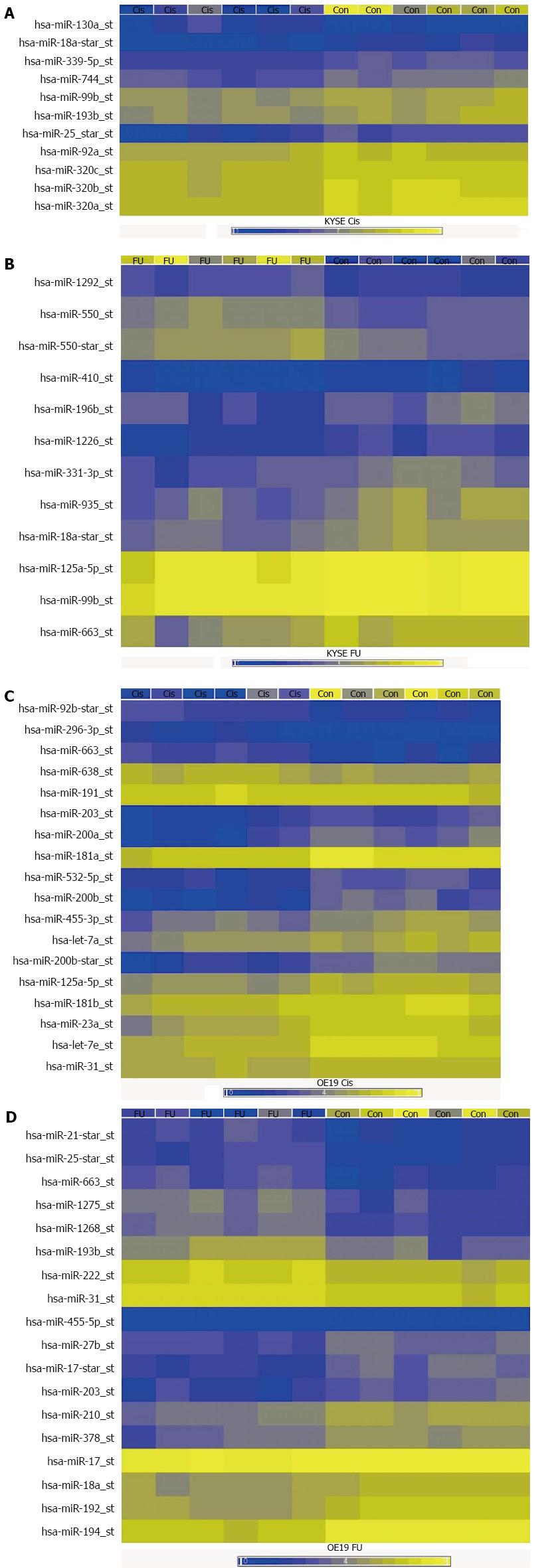
The results of the miRNA microarray experiments are summarized in Figure 1 and Table 1. A large number of miRNAs that were dysregulated in the different chemotherapy resistant cell lines were identified, compared to the chemotherapy sensitive controls. In cisplatin resistant EAC cells, 18 miRNAs were significantly dysregulated compared to controls (13 downregulated, 5 upregulated). In cisplatin resistant ESCC cells, 8 miRNAs were significantly dysregulated (all 8 downregulated). In 5-FU resistant EAC cells, 18 miRNAs were significantly dysregulated (10 downregulated, 8 upregulated). In 5-FU resistant ESCC cells, 5 miRNAs were significantly dysregulated (3 downregulated, 2 upregulated).
| EAC | ESCC | |||||
| miRNA | Stepup P-value | Fold-change | miRNA | Stepup P-value | Fold-change | |
| Dysregulated microRNAs in cisplatin-resistant variants | miR-181a-5p | 0.00141 | -1.78 | miR-320a | 0.002 | -1.75 |
| miR-125a-5p | 0.00169 | -2.23 | miR-320b | 0.002 | -1.76 | |
| let-7e-5p | 0.00170 | -2.21 | miR-320c | 0.002 | -1.66 | |
| miR-181b-5p | 0.00170 | -1.68 | miR-744-5p | 0.023 | -1.90 | |
| miR-200b-3p | 0.00170 | -5.38 | miR-99b-5p | 0.023 | -1.70 | |
| miR-31-5p | 0.00188 | -1.57 | miR-92a-3p | 0.030 | -1.53 | |
| miR-200b-5p | 0.00278 | -5.11 | miR-25-5p | 0.041 | -2.52 | |
| miR-455-3p | 0.00510 | -2.78 | miR-339-5p | 0.047 | -1.60 | |
| miR-191-5p | 0.00934 | 1.30 | miR-18a-3p | 0.056 | -2.42 | |
| miR-638 | 0.00934 | 1.74 | miR-193b-3p | 0.062 | -1.73 | |
| miR-92b-5p | 0.01344 | 1.79 | miR-130a-3p | 0.077 | 2.28 | |
| miR-23a-3p | 0.01799 | -2.31 | ||||
| miR-200a-3p | 0.03240 | -5.05 | ||||
| miR-296-3p | 0.04236 | 1.66 | ||||
| let-7a-5p | 0.04236 | -1.87 | ||||
| miR-203a | 0.04236 | -2.84 | ||||
| miR-663a | 0.04480 | 2.40 | ||||
| miR-532-5p | 0.04913 | -2.44 | ||||
| Dysregulated microRNAs in 5-FU-resistant variants | miR-31-5p | 0.00031 | 1.84 | miR-550 a-5p | 0.001 | 3.23 |
| miR-192-5p | 0.00151 | -2.57 | miR-935 | 0.008 | -3.69 | |
| miR-194-5p | 0.00151 | -2.55 | miR-550 a-3p | 0.037 | 2.65 | |
| miR-378 a-3p | 0.00277 | -3.17 | miR-99b-5p | 0.045 | -1.70 | |
| miR-222-3p | 0.00277 | 2.06 | miR-18a-3p | 0.047 | -2.71 | |
| miR-210-3p | 0.00289 | -2.90 | miR-1226-3p | 0.072 | -2.45 | |
| miR-18a-3p | 0.00456 | -1.84 | miR-1292-5p | 0.072 | 1.77 | |
| miR-21-3p | 0.00556 | 2.53 | miR-125a-5p | 0.127 | -1.92 | |
| miR-1268a | 0.00880 | 2.95 | miR-410-3p | 0.233 | -1.46 | |
| miR-25-5p | 0.01189 | 2.21 | miR-331-3p | 0.240 | -2.21 | |
| miR-27b-3p | 0.01797 | -2.12 | miR-663a | 0.240 | -2.61 | |
| miR-17-5p | 0.01928 | -1.35 | miR-196b-5p | 0.251 | -2.58 | |
| miR-193b-3p | 0.01961 | 3.78 | ||||
| miR-17-3p | 0.03118 | -3.03 | ||||
| miR-455-5p | 0.03143 | -1.40 | ||||
| miR-1275 | 0.03143 | 3.29 | ||||
| miR-203a | 0.03456 | -2.85 | ||||
| miR-663a | 0.04074 | 2.39 | ||||
Quantitative real-time PCR validation of the microarray experiments validated significant up- and downregulation for 11 miRNAs in cisplatin resistant EAC cells, 9 miRNAs in 5-FU resistant EAC cells, 1 miRNA in cisplatin resistant ESCC cells and 6 miRNAs in 5-FU resistant ESCC cells (Table 2). More miRNAs were downregulated in resistant cells compared to sensitive controls (19 downregulated vs 8 upregulated miRNAs). Two miRNAs were found to have altered expression in both cisplatin and 5-FU resistant cells: miR-31 and miR-125a-5p. Interestingly, miR-31-5p was downregulated in cisplatin resistant EAC but upregulated in 5-FU resistant EAC, whereas miR-125a-5p expression was suppressed in both, cisplatin resistant EAC and 5-FU resistant ESCC variants.
| Cisplatin resistant EAC | Cisplatin resistant ESCC | 5-FU resistant EAC | 5-FU resistant ESCC | |
| Significantly dysregulated miRNAs in resistant variants | miR-455-3p (-3.06) | miR-130a-3p (1.70) | miR-378a-3p (-3.58) | miR-935 (-2.29) |
| miR-200b-3p (-2.46) | miR-192-5p (-2.32) | miR-550a-3p (2.21) | ||
| let-7e-5p (-2.44) | miR-210-3p (-2.19) | miR-125a-5p (-2.20) | ||
| miR-181b-5p (-1.88) | miR-194-5p (-2.01) | miR-1226-3p (-2.12) | ||
| miR-125a-5p (-1.79) | miR-222-3p (1.85) | miR-99b-5p (-1.88) | ||
| miR-181a-5p (-1.63) | miR-17-3p (-1.83) | miR-18a-3p (-1.75) | ||
| miR-200b-5p (-1.63) | miR-193b-3p (1.54) | |||
| miR-31-5p (-1.58) | miR-31-5p (1.40) | |||
| miR-200a-3p (-1.57) | miR-27b-3p (1.36) | |||
| miR-638 (1.55) | ||||
| miR-191-5p (1.42) |
The expression of potential target genes for the dysregulated miRNAs was examined for 19 putative target candidates. Significant up- or downregulation of 9 of these 19 targets were found in the respective cell lines (5-FU resistant EAC: KRAS, TYMS, ABCC3 and CBL-B; cisplatin resistant EAC: BCL-2 and MAP4K4; 5-FU resistant ESCC: ERBB2, MUC1 and KRAS). Comparison between target gene expression and miRNA expression revealed that 6 targets (KRAS, TYMS, ABCC3 and CBL-B in 5-FU resistant EAC; ERBB2 and KRAS in 5-FU resistant ESCC) were regulated in a direction consistent with negative post-transcriptional control of target gene expression via the respective miRNA (Table 3).
| MicroRNA | Target | ||||
| Dysregulated miRNA | Up-/down regulation | mRNA target | Up-/down regulation | X-fold (P-value) | |
| 5-FU resistant EAC | miR-192-5p | ↓ | TYMS | ↑ | +2.30 (< 0.001) |
| miR-378a-3p | ↓ | CBL-B | ↑ | +1.93 (0.003) | |
| miR-192-5p | ↓ | ABCC3 | ↑ | +1.30 (0.008) | |
| miR-27b-3p | ↑ | KRAS | ↓ | -1.24 (0.036) | |
| miR-193b-3p | ↑ | KRAS | ↓ | -1.24 (0.036) | |
| 5-FU resistant ESCC | miR-18a-3p | ↓ | KRAS | ↑ | +1.60 (0.036) |
| miR-125a-5p | ↓ | ERBB2 | ↑ | +1.40 (0.026) | |
miRNAs represent a new class of epigenetic regulators of global gene expression, and their expression appears to influence chemotherapy sensitivity and resistance[6]. In the current study, miRNA microarrays were used to evaluate global miRNA expression profiling within an in-vitro model of acquired chemotherapy resistance in esophageal EAC and ESCC cells, and then quantitative real-time PCR was used to validate the findings. This demonstrated specific miRNA signatures for each chemotherapy resistant subline, and that the miRNA signatures are different for cisplatin vs 5-FU resistant cells from the same tumor cell line, and also for EAC vs ESCC cells with resistance to the same specific chemotherapy agent. The latter observation of differences in miRNA expression between EAC vs ESCC cell lines is consistent with the different tumor biology for EAC and ESCC. Furthermore, our study also identified dysregulated expression of the miRNAs’ potential target genes [namely KRAS, TYMS, ABCC3 and CBL-B (5-FU resistant EAC) respectively ERBB2 and KRAS (5-FU resistant ESCC)] in a direction consistent with negative posttranscriptional control of target gene expression via miR-27b-3p, miR-193b-3p, miR-192-5p, miR-378 a-3p, miR-125a-5p and miR-18a-3p.
These findings are consistent with other studies that suggest that these genes have an impact on chemotherapy resistance in cancer. For example, overexpression of TYMS has been linked to resistance to TYMS-targeted chemotherapy agents such as 5-FU and methotrexate[16], and ABCC3 is associated with chemotherapy resistance[17]. In addition, overexpression of Cbl-b has been shown to downregulate both p-Akt and P-gp, and partially reverse resistance to chemotherapeutic agents in gastric cancer[18]. KRAS also has an impact on sensitivity to chemotherapy in colorectal cancer[13], and ERBB2 affects chemotherapy resistance in breast cancer cells[19].
There is more than one possible explanation for dysregulation of miRNA expression in (cisplatin and 5-FU) resistant cell lines, and we can only speculate on this, based on currently known paradigms. For example, epigenetic changes such as DNA methylation or histone modification, that might occur during acquisition of chemotherapy resistance could directly impact upon the level of transcription of miRNA genes. The resulting changes in miRNA expression might then be important for further driving acquisition of a highly chemotherapy resistant phenotype. However, recent reports also suggest that many of the identified miRNAs in our study have been shown to have an impact on chemotherapy resistance in other cancers. For example, members of the miR-200 family, the miR-181 family, the let-7 family, or single miRNAs such as miR-130a-3p, miR-221-3p/miR-222-3p and miR-31-5p have been reported to be differentially expressed in various chemotherapy resistant cancer cell lines, or to have an impact on sensitivity to anticancer treatment including chemotherapy and radiation[6]. The miRNA candidates in our study, for which we identified potential target genes, revealed a similar pattern, with miR-27b-3p, miR-193b-3p/miR-193b-5p, miR-192-5p, miR-378 a-3p or miR-125a-5p associated with resistance towards multiple chemotherapeutic agents in a number of other tumor entities, including gastric[21] and colorectal cancer[22].
Data on esophageal cancer are still very rare, but the results from our study and other studies addressing this issue suggest that chemotherapy or radiotherapy resistance in esophageal cancer is also modulated by miRNAs[23]. For example, Imanaka et al[24] found 10 miRNAs to be significantly dysregulated between nine ESCC cell lines with varying sensitivity towards cisplatin. However, as the authors used a model that compares miRNA expression between several different cell lines, it remains unclear whether the differences in miRNA expression are caused by variations in chemotherapy resistance or phenotypic differences between different cells originating from different human individuals. Data from other studies suggest that modulation of the expression of single miRNAs such as miR-27a-3p, miR-31-5p, miR-141-3p, miR-148a-3p, miR-200c-3p and miR-296-5p affect sensitivity towards various chemotherapeutic agents or radiation in esophageal cancer[7,24-28].
There are some limitations to the current study. Firstly, cell line experiments do not always yield results that can be directly translated to the clinical setting, as tumor tissues from patients often show considerable heterogeneity. Hence, the miRNAs identified in our study may not be relevant to all esophageal EACs or ESCCs in the clinical setting, and could be specific to the particular cell lines tested. This highlights the need for further confirmation in experiments using human tumor samples. However, most of the miRNAs identified in our study have been reported in other tumor types, in both human tumor samples and cell culture settings, and this does suggest that the data from the current study is relevant.
Secondly, the current study was limited to only two esophageal cancer cell lines, and their resistant sublines, and confirmation of our results in other (sensitive and resistant) cell lines would provide further support to the findings.
Finally, a thorough validation of potential gene targets has not been undertaken at this stage, and we have not conducted functional analyses to confirm the role of miRNA in chemoresistance.
However, the primary aims of the current study were to investigate (1) if chemotherapy resistant esophageal cancer cell lines exhibit specific miRNA expression patterns, and (2) if the expression of potential resistance-relevant target genes of the dysregulated miRNAs is altered in the resistant cell lines. We were able to answer these questions with the current study. However, future experiments exploring the downstream pathways in more detail will be needed and should help to understand how miRNAs might have an impact on chemotherapy resistance.
In conclusion, the current study demonstrated that chemotherapy resistant esophageal EAC and ESCC cells present distinct miRNA signatures, with a number of well known resistance relevant miRNAs differentially expressed in the derived cisplatin or 5-FU resistant cell lines. Furthermore, a number of putative target genes that are known to have an impact on chemotherapy resistance were dysregulated in the chemotherapy resistant cell lines in a direction consistent with negative posttranscriptional control of target gene expression via the respective miRNA, thereby implicating a potential mediatory effect in terms of chemotherapy resistance development. This data presents initial basic research results pertinent to this topic, although the data remains limited by its in-vitro character. Nevertheless, the results from this study do support undertaking further in vivo studies that address whether miRNAs can be used as novel biomarkers for chemotherapy response prediction, or perhaps even evaluate modulation of miRNAs as novel therapies for overcoming chemotherapy resistance in esophageal cancer.
Neoadjuvant (radio-) chemotherapy plays an important role in the treatment of patients with advanced esophageal cancer. However, individuals respond to these treatments in a variable fashion, and those who respond poorly to chemotherapy probably undergo futile treatment. Identification of individuals who are unlikely to benefit before treatment starts is desirable as it would allow treatment to be tailored to the individuals most likely to benefit. So far, no reliable biomarkers are available for prediction of response to neoadjuvant chemotherapy in esophageal cancer patients. MicroRNAs might be promising candidates in this context, as there is some first evidence that microRNA expression has an impact on the response to chemotherapy in a number of other malignancies.
MicroRNAs are important regulators of global gene expression. To date, it is accepted that microRNAs play a crucial role in many physiological and pathological processes, such as in the initiation and progression of cancer. Most importantly, microRNAs have been demonstrated to impact on sensitivity towards various chemotherapeutic drugs including cisplatin and 5-FU resistant cells (5-FU) amongst others in a variety of cancers by regulating several resistance-relevant intracellular pathways that control for example apoptosis, cell survival, DNA-repair and others.
By using an in-vitro model of acquired chemotherapy resistance in esophageal adeno- (EAC) and squamous cell carcinoma (ESCC) cells, the current study demonstrated that chemotherapy resistant esophageal EAC and ESCC cells present distinct miRNA signatures, with a number of well known resistance relevant miRNAs differentially expressed in the derived cisplatin or 5-FU resistant cell lines. Furthermore, a number of putative target genes that are known to have an impact on chemotherapy resistance were dysregulated in the chemotherapy resistant cell lines in a direction consistent with negative posttranscriptional control of target gene expression via the respective miRNA, thereby implicating a potential mediatory effect in terms of chemotherapy resistance development. In detail, miR-27b-3p, miR-193b-3p, miR-192-5p, miR-378 a-3p, miR-125a-5p and miR-18a-3p were dysregulated amongst others, consistent with negative posttranscriptional control of the expression of their potential targets KRAS, TYMS, ABCC3, CBL-B and ERBB2.
The results from this study do support undertaking further in vivo studies that address whether miRNAs can be used as novel biomarkers for chemotherapy response prediction, or perhaps even evaluate modulation of miRNAs as novel therapies for overcoming chemotherapy resistance in esophageal cancer.
Chemotherapy resistance is a lack of response of cancer cells to drug-induced tumor growth inhibition. MicroRNAs are small non-coding RNA-molecules that control global gene expression on a post-transcriptional level.
The current study supports the hypothesis that microRNA expression has an impact on chemotherapy resistance in esophageal cancer. This provides a basis for further experimental and clinical investigations which could better tailor chemotherapy to individuals most likely to benefit, or identify and modulate chemotherapy resistance to achieve a better clinical response.
P- Reviewer: Fujita T, Yang MH, Zhao BS S- Editor: Qi Y L- Editor: A E- Editor: Zhang DN
| 1. | Gebski V, Burmeister B, Smithers BM, Foo K, Zalcberg J, Simes J. Survival benefits from neoadjuvant chemoradiotherapy or chemotherapy in oesophageal carcinoma: a meta-analysis. Lancet Oncol. 2007;8:226-234. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 898] [Cited by in F6Publishing: 874] [Article Influence: 51.4] [Reference Citation Analysis (0)] |
| 2. | Sjoquist KM, Burmeister BH, Smithers BM, Zalcberg JR, Simes RJ, Barbour A, Gebski V. Survival after neoadjuvant chemotherapy or chemoradiotherapy for resectable oesophageal carcinoma: an updated meta-analysis. Lancet Oncol. 2011;12:681-692. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1141] [Cited by in F6Publishing: 1203] [Article Influence: 92.5] [Reference Citation Analysis (0)] |
| 3. | Pike GK, Bessell JR, Mathew G, Watson DI, Mitchell PC, Jamieson GG. Changes in fibrinogen levels in patients undergoing open and laparoscopic Nissen fundoplication. Aust N Z J Surg. 1996;66:94-96. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 7] [Cited by in F6Publishing: 7] [Article Influence: 0.3] [Reference Citation Analysis (0)] |
| 4. | Mayne GC, Hussey DJ, Watson DI. MicroRNAs and esophageal cancer--implications for pathogenesis and therapy. Curr Pharm Des. 2013;19:1211-1226. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1] [Cited by in F6Publishing: 1] [Article Influence: 0.1] [Reference Citation Analysis (0)] |
| 5. | Hiyoshi Y, Kamohara H, Karashima R, Sato N, Imamura Y, Nagai Y, Yoshida N, Toyama E, Hayashi N, Watanabe M. MicroRNA-21 regulates the proliferation and invasion in esophageal squamous cell carcinoma. Clin Cancer Res. 2009;15:1915-1922. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 207] [Cited by in F6Publishing: 220] [Article Influence: 14.7] [Reference Citation Analysis (0)] |
| 6. | Hummel R, Hussey DJ, Haier J. MicroRNAs: predictors and modifiers of chemo- and radiotherapy in different tumour types. Eur J Cancer. 2010;46:298-311. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 259] [Cited by in F6Publishing: 243] [Article Influence: 17.4] [Reference Citation Analysis (0)] |
| 7. | Hummel R, Watson DI, Smith C, Kist J, Michael MZ, Haier J, Hussey DJ. Mir-148a improves response to chemotherapy in sensitive and resistant oesophageal adenocarcinoma and squamous cell carcinoma cells. J Gastrointest Surg. 2011;15:429-438. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 70] [Cited by in F6Publishing: 78] [Article Influence: 6.0] [Reference Citation Analysis (0)] |
| 8. | Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. JRSS Series B. 1995;57:289-300. [Cited in This Article: ] |
| 9. | Papadopoulos GL, Alexiou P, Maragkakis M, Reczko M, Hatzigeorgiou AG. DIANA-mirPath: Integrating human and mouse microRNAs in pathways. Bioinformatics. 2009;25:1991-1993. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 228] [Cited by in F6Publishing: 241] [Article Influence: 16.1] [Reference Citation Analysis (0)] |
| 10. | Krasnapolski MA, Todaro LB, de Kier Joffé EB. Is the epithelial-to-mesenchymal transition clinically relevant for the cancer patient? Curr Pharm Biotechnol. 2011;12:1891-1899. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 20] [Cited by in F6Publishing: 21] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 11. | Davids MS, Letai A. Targeting the B-cell lymphoma/leukemia 2 family in cancer. J Clin Oncol. 2012;30:3127-3135. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 210] [Cited by in F6Publishing: 208] [Article Influence: 17.3] [Reference Citation Analysis (0)] |
| 12. | Sabine VS, Sims AH, Macaskill EJ, Renshaw L, Thomas JS, Dixon JM, Bartlett JM. Gene expression profiling of response to mTOR inhibitor everolimus in pre-operatively treated post-menopausal women with oestrogen receptor-positive breast cancer. Breast Cancer Res Treat. 2010;122:419-428. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 33] [Cited by in F6Publishing: 38] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 13. | de Bruijn MT, Raats DA, Hoogwater FJ, van Houdt WJ, Cameron K, Medema JP, Borel Rinkes IH, Kranenburg O. Oncogenic KRAS sensitises colorectal tumour cells to chemotherapy by p53-dependent induction of Noxa. Br J Cancer. 2010;102:1254-1264. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 21] [Cited by in F6Publishing: 21] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 14. | López-Guerra M, Trigueros-Motos L, Molina-Arcas M, Villamor N, Casado FJ, Montserrat E, Campo E, Colomer D, Pastor-Anglada M. Identification of TIGAR in the equilibrative nucleoside transporter 2-mediated response to fludarabine in chronic lymphocytic leukemia cells. Haematologica. 2008;93:1843-1851. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 16] [Cited by in F6Publishing: 16] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 15. | St Croix B, Kerbel RS. Cell adhesion and drug resistance in cancer. Curr Opin Oncol. 1997;9:549-556. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 81] [Cited by in F6Publishing: 91] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 16. | Marsh S. Thymidylate synthase pharmacogenetics. Invest New Drugs. 2005;23:533-537. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 82] [Cited by in F6Publishing: 70] [Article Influence: 3.9] [Reference Citation Analysis (0)] |
| 17. | Zhou SF, Wang LL, Di YM, Xue CC, Duan W, Li CG, Li Y. Substrates and inhibitors of human multidrug resistance associated proteins and the implications in drug development. Curr Med Chem. 2008;15:1981-2039. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 259] [Cited by in F6Publishing: 243] [Article Influence: 15.2] [Reference Citation Analysis (0)] |
| 18. | Zhang Y, Qu X, Hu X, Yang X, Hou K, Teng Y, Zhang J, Sada K, Liu Y. Reversal of P-glycoprotein-mediated multi-drug resistance by the E3 ubiquitin ligase Cbl-b in human gastric adenocarcinoma cells. J Pathol. 2009;218:248-255. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 37] [Cited by in F6Publishing: 40] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 19. | Chen X, Yeung TK, Wang Z. Enhanced drug resistance in cells coexpressing ErbB2 with EGF receptor or ErbB3. Biochem Biophys Res Commun. 2000;277:757-763. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 93] [Cited by in F6Publishing: 101] [Article Influence: 4.2] [Reference Citation Analysis (0)] |
| 20. | Ren J, Agata N, Chen D, Li Y, Yu WH, Huang L, Raina D, Chen W, Kharbanda S, Kufe D. Human MUC1 carcinoma-associated protein confers resistance to genotoxic anticancer agents. Cancer Cell. 2004;5:163-175. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 253] [Cited by in F6Publishing: 269] [Article Influence: 13.5] [Reference Citation Analysis (0)] |
| 21. | Nishida N, Mimori K, Fabbri M, Yokobori T, Sudo T, Tanaka F, Shibata K, Ishii H, Doki Y, Mori M. MicroRNA-125a-5p is an independent prognostic factor in gastric cancer and inhibits the proliferation of human gastric cancer cells in combination with trastuzumab. Clin Cancer Res. 2011;17:2725-2733. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 167] [Cited by in F6Publishing: 199] [Article Influence: 15.3] [Reference Citation Analysis (0)] |
| 22. | Boni V, Bitarte N, Cristobal I, Zarate R, Rodriguez J, Maiello E, Garcia-Foncillas J, Bandres E. miR-192/miR-215 influence 5-fluorouracil resistance through cell cycle-mediated mechanisms complementary to its post-transcriptional thymidilate synthase regulation. Mol Cancer Ther. 2010;9:2265-2275. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 121] [Cited by in F6Publishing: 133] [Article Influence: 9.5] [Reference Citation Analysis (0)] |
| 23. | Fang Y, Fang D, Hu J. MicroRNA and its roles in esophageal cancer. Med Sci Monit. 2012;18:RA22-RA30. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 21] [Cited by in F6Publishing: 27] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 24. | Imanaka Y, Tsuchiya S, Sato F, Shimada Y, Shimizu K, Tsujimoto G. MicroRNA-141 confers resistance to cisplatin-induced apoptosis by targeting YAP1 in human esophageal squamous cell carcinoma. J Hum Genet. 2011;56:270-276. [PubMed] [DOI] [Cited in This Article: ] [Cited by in F6Publishing: 1] [Reference Citation Analysis (0)] |
| 25. | Zhang H, Li M, Han Y, Hong L, Gong T, Sun L, Zheng X. Down-regulation of miR-27a might reverse multidrug resistance of esophageal squamous cell carcinoma. Dig Dis Sci. 2010;55:2545-2551. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 99] [Cited by in F6Publishing: 105] [Article Influence: 7.5] [Reference Citation Analysis (0)] |
| 26. | Lynam-Lennon N, Reynolds JV, Marignol L, Sheils OM, Pidgeon GP, Maher SG. MicroRNA-31 modulates tumour sensitivity to radiation in oesophageal adenocarcinoma. J Mol Med (Berl). 2012;90:1449-1458. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 79] [Cited by in F6Publishing: 84] [Article Influence: 7.0] [Reference Citation Analysis (0)] |
| 27. | Hamano R, Miyata H, Yamasaki M, Kurokawa Y, Hara J, Moon JH, Nakajima K, Takiguchi S, Fujiwara Y, Mori M. Overexpression of miR-200c induces chemoresistance in esophageal cancers mediated through activation of the Akt signaling pathway. Clin Cancer Res. 2011;17:3029-3038. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 175] [Cited by in F6Publishing: 197] [Article Influence: 15.2] [Reference Citation Analysis (0)] |
| 28. | Hong L, Han Y, Zhang H, Li M, Gong T, Sun L, Wu K, Zhao Q, Fan D. The prognostic and chemotherapeutic value of miR-296 in esophageal squamous cell carcinoma. Ann Surg. 2010;251:1056-1063. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 117] [Cited by in F6Publishing: 123] [Article Influence: 8.8] [Reference Citation Analysis (0)] |