Published online Sep 14, 2014. doi: 10.3748/wjg.v20.i34.11991
Revised: January 29, 2014
Accepted: April 8, 2014
Published online: September 14, 2014
Despite a decline in the overall incidence of gastric cancer (GC), the disease remains the second most common cause of cancer-related death worldwide and is thus a significant global health problem. The best means of improving the survival of GC patients is to screen for and treat early lesions. However, GC is often diagnosed at an advanced stage and is associated with a poor prognosis. Current diagnostic and therapeutic strategies have not been successful in decreasing the global burden of the disease; therefore, the identification of reliable biomarkers for an early diagnosis, predictive markers of recurrence and survival and markers of drug sensitivity and/or resistance is urgently needed. The initiation and progression of GC depends not only on genetic alterations but also epigenetic changes, such as DNA methylation and histone modification. Aberrant DNA methylation is the most well-defined epigenetic change in human cancers and is associated with inappropriate gene silencing. Therefore, an increasing number of genes methylated at the promoter region have been targeted as possible biomarkers for different purposes, including early detection, classification, the assessment of the tumor prognosis, the development of therapeutic strategies and patient follow-up. This review article summarizes the current understanding and recent evidence regarding DNA methylation markers in GC with a focus on the clinical potential of these markers.
Core tip: This article summarizes the current understanding and recent evidence regarding DNA methylation markers in gastric cancer (GC) and includes our previous works. Current diagnostic and therapeutic tools of GC have not been successful in decreasing the global burden of this disease; however, it is promising that the early diagnosis and careful selection of patient subsets prior to initiating chemotherapy is a key factor for improving the outcomes of patients with GC. Methylation biomarkers would be useful for different purposes, including early detection, classification, assessment of the tumor prognosis, the development of therapeutic strategies and patient follow-up.
- Citation: Nakamura J, Tanaka T, Kitajima Y, Noshiro H, Miyazaki K. Methylation-mediated gene silencing as biomarkers of gastric cancer: A review. World J Gastroenterol 2014; 20(34): 11991-12006
- URL: https://www.wjgnet.com/1007-9327/full/v20/i34/11991.htm
- DOI: https://dx.doi.org/10.3748/wjg.v20.i34.11991
Although the incidence of gastric cancer (GC) has declined on a yearly basis, it remains the fourth most common cancer and the second leading cause of cancer-related death worldwide[1]. Recent progress in early diagnosis, surgical techniques and perioperative management have improved patient satisfaction and outcomes; however, GC remains a major clinical challenge due to its high prevalence, poor prognosis and limited treatment options. The case fatality rate of GC is higher than that of other common malignancies, such as colorectal, breast and prostate cancer[2]. Because the prognosis of patients with GC depends on the tumor stage at diagnosis and the selection of an appropriate treatment strategy, the identification of novel biomarkers for early diagnosis and predictive markers of drug sensitivity is urgently needed.
Accumulating evidence suggests that cancer is caused by both epigenetic and genetic abnormalities[3]. DNA methylation, a major epigenetic modification that is strongly involved in the control of gene expression, is an early landmark event in carcinogenesis[4]. DNA methylation is a covalent chemical modification catalyzed by enzymes of the DNA methyltransferase family that mediate the addition of a methyl (CH3) group to the fifth carbon of the cytosine. DNA methylation usually occurs within the context of 5’-CG-3’ (CpG dinucleotide) clustered regions known as CpG islands (CGIs), which are frequently found proximal to the promoters of housekeeping genes[5]. The occurrence of methylation within the CGI of a gene promoter is associated with a compact chromatin structure and is linked to the transcriptional silencing of the affiliated gene, particularly tumor suppressor genes[6]. Numerous studies have reported that many tumor suppressor genes, which play key roles in functions pertaining to cancer prevention (DNA repair, cell adhesion, cell cycle control and apoptosis), are silenced by the hypermethylation of their promoters during carcinogenesis[7]. Therefore, it is reasonable to hypothesize that the DNA methylation status of certain genes serves as a useful biomarker for predicting tumor behavior. Furthermore, DNA methylation biomarkers offer several advantages over genetic and serum markers[8,9]. First, the incidence of aberrant DNA methylation of specific CGIs is higher than that observed in genetic abnormalities[9,10]. Second, the aberrant DNA methylation observed in cancer can be sensitively detected using a simple technique, methylation-specific PCR. Third, aberrant DNA methylation appears to occur in early-stage tumors, causing the loss- and/or gain-of-function of key processes and signaling properties[4].
In this review, we summarize the current status and recent evidence regarding DNA methylation biomarkers in GC and assess the clinical potential of these biomarkers for use in risk assessment, early diagnosis and the evaluation of treatment and prognosis in patients with GC.
The methylation of gene promoters is now recognized to be one of the primary mechanisms used to inactivate tumor-related genes (particularly tumor suppressor genes) as well as genetic alterations. Since the first article by Fang et al[11] in 1996 describing the DNA hypomethylation of c-myc and c-Ha-ras in GC, numerous studies have been published on the involvement of DNA methylation in GC. To date, the aberrant DNA methylation of more than 100 genes, such as tumor suppressor genes (including E-cadherin, RASSF1A, p16, GSTP1, SOCS1, SFRP1 and PTEN) has been reported in GC[12-17]. A recent meta-analysis[18] reviewed 143 case-control studies reporting the methylation frequency of 142 individual genes. Consequently, a total of 70 genes were found to be significantly hypermethylated in cancer tissue compared with those observed in normal tissue obtained from GC subjects. Table 1 shows a list of genes that are commonly methylated in GC compared with those in corresponding normal gastric tissue[14,19-34].
| Gene | Frequency (%) | Assay | Function | Ref. |
| ITGA4 | 96.0 | Q-MSP | Other | [19] |
| ZIC1 | 94.6 | MSP | Transcriptional regulation | [20] |
| PRDM5 | 88.0 | MSP | Cell cycle | [21] |
| PCDH10 | 82.0 | MSP | Apoptosis | [22] |
| TFPI2 | 80.9 | MSP | Other | [23] |
| RUNX3 | 75.2 | Q-MSP | Transcriptional regulation | [24] |
| SPINT2 | 75.0 | MSP | Cell growth/differentiation | [25] |
| BTG4 | 73.7 | MSP | Other | [26] |
| SFRP2 | 73.3 | Q-MSP | Apoptosis | [27] |
| hMLH1 | 72.9 | MSP | DNA repair | [28] |
| DKK-3 | 67.6 | MSP | Wnt pathway | [29] |
| TCF4 | 67.0 | Pyrosequencing | Cell cycle | [30] |
| GRIK2 | 66.6 | Q-MSP | Cell adhesion/invasion/migration | [31] |
| RARβ | 65.8 | MSP | Retinoic acid pathway | [32] |
| CHFR | 65.0 | Q-MSP | Other | [33] |
| BNIP3 | 65.0 | Q-MSP | Apoptosis | [33] |
| RASSF1A | 61.8 | MSP | Ras pathway | [14] |
| LRP1B | 61.0 | Q-MSP | Other | [34] |
| SFRP5 | 56.0 | Q-MSP | Wnt pathway | [19] |
E-cadherin is one of the most important tumor suppressor genes in GC, and its inactivation is thought to contribute to tumor progression via subsequent increases in proliferation, invasion and metastasis[35-37]. E-cadherin is silenced by CpG methylation in various cancers, including GC and particularly undifferentiated GC, at the early stage[12,13]. However, E-cadherin methylation is frequently observed in both neoplastic and the corresponding non-neoplastic gastric mucosa. Age-related E-cadherin methylation is commonly present in the gastric mucosa starting from approximately 45 years of age[38]. However, RASSF1A, a member of the Ras association domain family, is a tumor suppressor gene that plays a critical role in cell cycle regulation, apoptosis and microtubule stability by regulating the Ras signaling pathway[39]. Mutations of RASSF1A are uncommon, whereas silencing by promoter methylation is frequent in cancers, including GC[40,41]. In contrast, such methylation occurs in only a small proportion of non-neoplastic gastric epithelia[42]. Moreover, RASSF1A methylation is closely associated with the TNM stage and a poor prognosis in GC patients[43]. Therefore, RASSF1A represents a potential diagnostic and therapeutic target in GC.
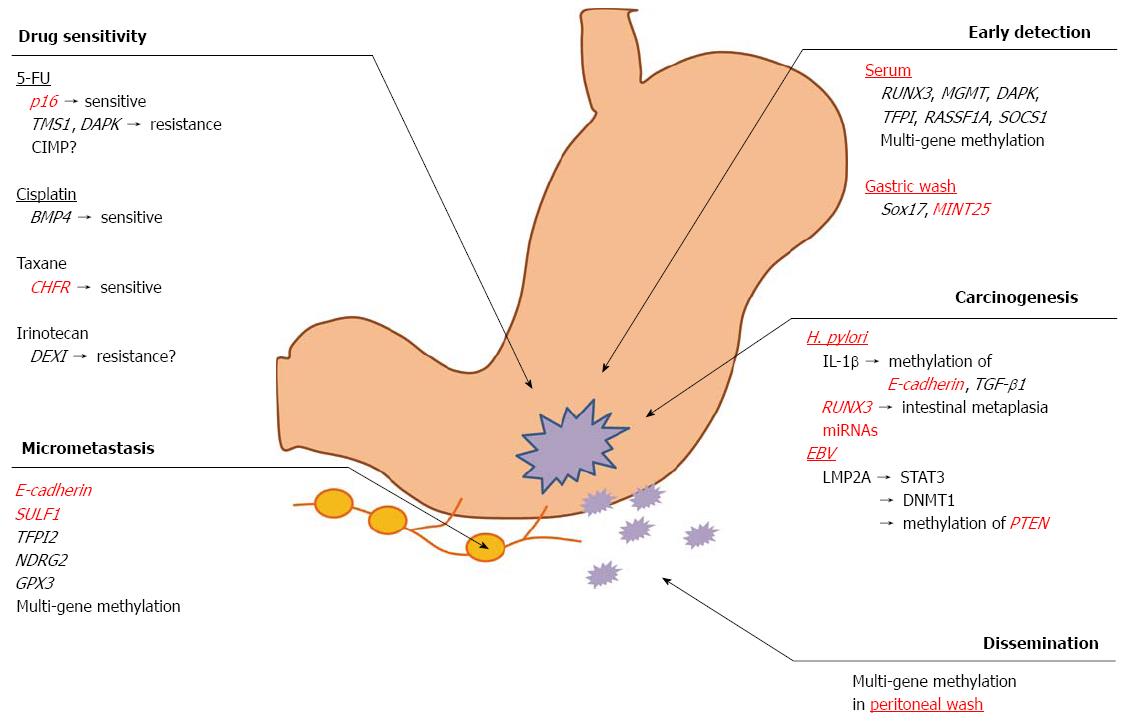
It has been reported that a widely known pathogen, Helicobacter pylori (H. pylori), is involved in the development of GC[44]. H. pylori is a Gram-negative spiral-shaped bacterium that is present in the stomach of approximately half of the world’s population[45,46]. Numerous prospective studies have shown that H. pylori infection plays an essential role in gastric carcinogenesis[47], and the mechanisms underlying gastric carcinogenesis due to H. pylori-induced DNA methylation have been clarified. Maekita et al[48] collected tissue samples of gastric mucosa from 154 healthy volunteers and 72 patients with differentiated-type GC via endoscopy and evaluated the methylation levels in seven CGIs among eight lesions. The data indicated that H. pylori infection potently and temporarily induces the methylation of multiple CGIs to various degrees and that the methylation levels in specific CGIs in noncancerous gastric mucosa are associated with the risk of GC in H. pylori-negative individuals. Our previous study also demonstrated that H. pylori infection contributes to the loss of RUNX3 expression via promoter methylation in GC[49]. In RUNX3-deficient mouse, some gastric epithelial cells differentiated into intestinal type cells, suggesting that the loss of RUNX3 expression triggered precancerous intestinal metaplasia (IM), which possibly leads to cancer in the stomach[50]. This speculation is supported by independent evidence recently published by Lu et al[24]. Furthermore, regardless of the status of H. pylori infection, the number of methylated genes in IM was significantly higher than that found in chronic gastritis without IM[51]. However, recent studies have intensively investigated the role of miRNA methylation in GC. Ando et al[52] demonstrated that gastric mucosa infected with H. pylori exhibits significantly higher methylation levels of three miRNAs (miR-124a-1, miR-124a-2 and miR-124a-3) than that without H. pylori infection among healthy volunteers and that the noncancerous gastric mucosa of GC patients exhibits higher methylation levels than the gastric mucosa of healthy volunteers among H. pylori-negative individuals. Therefore, the methylation-induced silencing of miRNA genes, as well as protein-coding genes, may contribute to the formation of field defects for GC. With respect to the correlation between the methylation level in the gastric mucosa and risk of GC, Nakajima et al[53] showed that the methylation levels in the gastric mucosa are significantly increased in patients with a single GC and even more prominently increased in patients with multiple GCs among H. pylori-negative individuals. In contrast, the methylation levels in H. pylori-positive individuals were increased to various degrees. Moreover, it has previously been shown that the development of inflammation triggered by H. pylori infection is pivotal for aberrant methylation and that the expression of inflammation-related genes, such as IL-1β, Nos2 and TNF in the stomach is associated with the induction of DNA methylation[54,55]. IL-1β directly induces the promoter methylation of E-cadherin, an important extracellular matrix component involved in the maintenance of epithelial stability[56]. Namely, the H. pylori-induced methylation of E-cadherin promoter is mediated through IL-1β. Furthermore, IL-1β is an important mediator of H. pylori-induced TGF-β1 methylation[57-59]. As previously described, basic and clinical studies have demonstrated that H. pylori infection is strongly correlated with aberrant methylation in GC. Meanwhile, it has become increasingly clear that the eradication of H. pylori significantly reduces gene methylation[60,61]. The removal of the aberrant DNA methylation induced by H. pylori in pre-cancerous lesions would be a novel approach to preventing cancer. Niwa et al[62] showed that 5-aza-dC treatment prevents the development of H. pylori-induced GC using a Mongolian gerbil model. Therefore, the removal and/or suppression of H. pylori-induced aberrant DNA methylation may prevent H. pylori-associated cancers.
Epstein-barr virus (EBV) causes infectious mononucleosis on initial infection, and more than 90% of adult patients become EBV carriers[63]. It is well known that EBV is associated with several malignancies, such as Burkitt lymphoma[64] and nasopharyngeal carcinoma[65]. EBV-positive GC was first discovered in 1990[66], and the incidence of EBV-positive GC ranges from 7% to 15% regardless of race or region[67,68]. EBV infection occurs at an early stage of carcinogenesis and plays an important role in cancer development. Furthermore, previous studies have reported that aberrant promoter methylation is more frequently involved in EBV-positive GC than EBV-negative GC[69-72]. DNA methylation of the promoter region in the APC, p16, MINT1, TP73 and HOXA10 genes[19,69,73] has been specifically observed in patients with EBV-positive GC. Moreover, one recent study demonstrated that the frequency of the methylation of seven genes (TP73, BLU, FSD1, BCL7A, MARK1, SCRN1 and NKX3.1) is significantly higher in EBV-positive GC patients than in EBV-negative GC patients[74]. Regarding the molecular mechanisms underlying host DNA methylation during the early stages of EBV infection in gastric epithelium, human DNA methyltransferases (DNMTs) have received attention as methylation drivers during EBV infection. It has been reported that LMP2A induces the phosphorylation of STAT3, which activates DNMT1 transcription and causes a loss of the PTEN expression via the methylation of the PTEN promoter in EBV-associated GC[75]. Zhao et al[76] also showed that the induction of promoter methylation by EBV is regulated by the upregulation of DNMT3b by LMP2A. Furthermore, a recent large-scale analysis performed by Matsusaka et al[77] assessed the DNA methylation profiling of GC using the Infinium Human Methylation27 BeadChip (Infinium, Illumina, San Diego, CA, United States). The authors classified GC into three epigenotypes (EBV−/low methylation, EBV−/high methylation and EBV+/high methylation) according to the pattern of DNA methylation. EBV-positive GCs exhibited distinct and markedly high levels of methylation, while the CXXC4, TIMP2 and PLXND1 genes were specifically methylated in the EBV-positive epigenotype. MLH1 was preferentially methylated in the EBV−/high methylation epigenotype; however, no methylation was detected in the EBV+/high methylation epigenotype. Namely, the authors identified the specific genotype associated with EBV infection and proved that the epigenetic alteration observed in EBV-positive GC is directly caused by EBV infection.
Less than 25% of GC cases are diagnosed at an early stage, and the 5-year survival rate is approximately 20%-25% worldwide[78]. However, the survival rate improves to over 60% if the disease is detected at an early stage[78], emphasizing the importance of making an early diagnosis of GC. Bodily fluids, particularly blood, contain various molecules that originate from other tissues and organs. These molecules can signal the presence of cancer and therefore are potential cancer biomarkers. First-generation tumor markers, such as CEA, CA19-9 and CA72-4, are associated with the development of GC; however, none of these markers are available for the purpose of screening and early detection of GC. Nucleic acids, which can be easily amplified using the PCR technique, represent obvious potential targets for biomarker development. The detection of circulating tumor DNA released from apoptotic or necrotic tumor cells or actively secreted from proliferating cells was first reported approximately three decades ago[79]. Circulating tumor DNA carries not only tumor-specific genetic information but also epigenetic marks, particularly DNA methylation. The use of DNA methylation tests in GC patients would facilitate the development of novel biomarkers for the early detection and diagnosis of GC.
Abbaszadegan et al[80] evaluated the methylation of p16, one of the most frequently methylated genes in GC, in cancer tissues and the corresponding serum obtained from GC patients. Aberrant methylation of the p16 promoter was detected in 44.2% of the GC tissues (23/52), whereas all normal gastric mucosa samples (n = 50) were unmethylated. Furthermore, among the patients with p16 methylation, 60.9% (14/23) also exhibited methylation in their corresponding serum samples. Therefore, the detection of DNA methylation in the serum is a potential biomarker for the early detection of GC. Kanyama et al[81] also demonstrated that the aberrant DNA methylation of p16 was observed in six of 23 serum samples obtained from GC patients (26.1%), regardless of the tumor stage and/or clinicopathological features, thus suggesting that p16 methylation is also a potential biomarker for the early detection of GC. Additionally, other methylation markers in the serum, such as RUNX3, MGMT, DAPK, TFPI, RASSF1A and SOCS1, were reported to be useful for the early detection of GC[82-87]. Leung et al[88] investigated multiple gene methylation in the serum DNA of GC patients, focusing on the incidence of promoter hypermethylation in 10 tumor-related genes (APC, E-cadherin, GSTP1, hMLH1, MGMT, p15, p16, SOCS1, TIMP3 and TGF-beta RII). Among these 10 genes, APC (17%), E-cadherin (13%), hMLH1 (41%) and TIMP3 (17%) were significantly methylated in the GC patients compared with that observed in the healthy volunteers. Furthermore, methylation was detected in the serum of 33 of 60 (55%) patients in at least one of these four genes. These results suggest that the detection of DNA methylation in the serum has diagnostic value in GC patients.
Another attempt regarding the early detection of GC was made in which methylation markers in gastric washes were investigated. Because many mucosal cells can be found in stomach juice, the detection of molecular markers in gastric washes is a possible noninvasive approach to screening for GC. Oishi et al[89] identified Sox17 methylation to be a candidate biomarker for detecting GC using a genome-wide DNA methylation analysis of gastric wash samples. Moreover, Watanabe et al[90] suggested that MINT25 is a sensitive and specific marker for screening GC. The authors initially assessed 24 tissue samples and identified six methylated genes (MINT25, RORA, GDNF, ADAM23, PRDM5 and MLF1). The methylation levels of these six genes significantly increased according to the progression from non-neoplastic gastric mucosa to normal adjacent tissue to dysplasia and finally to early GC. The power of these six genes was validated in 153 different populations using gastric washes. Consequently, MINT25 methylation exhibited the best sensitivity (90%) and specificity (96%) in terms of tumor detection in the gastric washes.
The postoperative recurrence of GC usually occurs in the peritoneum, lymph nodes (LNs) or liver[91]. Peritoneal metastasis is the most frequent event in recurrent GCs[91], and 50%-60% of GC patients with serosal invasion following curative resection eventually develop peritoneal metastasis[92]. Peritoneal metastasis exhibits resistance to various chemotherapeutic agents, causes massive ascites and occasionally results in intestinal obstruction. The average length of survival following the development of peritoneal metastasis is 4.9 mo[93]. Therefore, the ability to predict peritoneal metastasis is essential for selecting the treatment strategy before metastatic nodules in the peritoneum become large. We previously reported that DNA methylation is a possible marker for detecting cancer cells in the peritoneal washes (PW) of GC patients[33]. In that study, the DNA methylation of six genes, including BNIP3, CHFR, CYP1B1, MINT25, SFRP2 and RASSF2, was analyzed in 107 PW specimens obtained from GC patients who underwent surgery. The patients were classified into the following three groups: group A (n = 42), patients with a depth of cancer invasion at the muscularis propria (MP) or less than the MP; group B (n = 45), a depth of cancer invasion beyond the MP; and group C (n = 20), histologically diagnosed peritoneal metastasis or cytologically detected cancer cells in the peritoneal cavity. The methylation status of the six genes significantly differed between the three groups (group A, 0%-5%; group B, 0%-15%; group C, 15%-45%; P < 0.01). Furthermore, three of nine patients in group B in whom methylation was detected in PW in at least one of these six genes experienced peritoneal recurrence after surgery. However, only one of 36 patients without gene methylation experienced peritoneal recurrence (P < 0.05). Yu et al[94] also showed that the presence of methylated E-cadherin in PW predicts a poor prognosis in GC patients. Conducting methylation analyses and cytological examinations of PW is therefore useful for predicting the risk of recurrence in patients with GC.
LN metastasis is one of the most important factors in predicting recurrence in patients with GC[95]. The presence of micrometastasis, which cannot be detected using conventional histological methods, is significantly correlated with the postoperative prognosis of GC patients[96-98]. The detection of micrometastasis in LNs is therefore very important for making an accurate classification of the cancer stage and selecting the appropriate treatment strategy. Several studies have reported RT-PCR analyses to detect a positive expression of CEA, CK19 or CK20 mRNA in 5.3%-23.8% of LNs that are free from histological metastasis in patients with GC[99-101]. However, accumulating evidence suggests that gene methylation, such as that of E-cadherin[102], SULF1[103], TFPI2[86], NDRG2[104] and GPX3[105], is a useful biomarker for detecting LN metastasis in GC patients. Yi Kim et al[102] investigated the expression of E-cadherin in resected specimens obtained from 60 patients with early GC and correlated the findings with the metastatic potential of the tumors. Their data suggested that the methylation of E-cadherin is involved in LN metastasis in patients with GC. Furthermore, Hur et al[103] examined the expression of SULF1 and SULF2 in a large cohort of 450 GC tissues. The GC tissues exhibited a conspicuously higher expression of SULF1 (P = 0.0002) regulated by the promoter hypomethylation compared with that observed in the normal mucosa. Additionally, SULF1 is an independent prognostic (P = 0.0123) and LN metastasis predictive factor (P = 0.0003) in patients with GC. Our previous study[106] also evaluated the methylation of multiple genes to detect the micrometastasis of GC. Methylation analyses are useful for accurately staging tumors and selecting optimal candidates for chemotherapy.
In addition to several chemotherapeutic agents, 5-fluorouracil (5-FU) is widely used as a key drug in chemotherapy for advanced GC. 5-FU is converted into fluorodeoxyuridine monophosphate, which inhibits the activity of thymidylate synthase (TS), an enzyme required for DNA synthesis[107]. Several enzymes involved in the metabolic pathway of 5-FU, including TS, dihydropyrimidine dehydrogenase, thymidine phosphorylase (TP) and orotate phosphoribosyltransferase, have been intensively examined as candidate biomarkers for the efficacy of 5-FU therapy. Our previous work also focused on the correlation between the response to 5-FU and the expression of the above enzymes[108]. However, the predictive power of these molecules with respect to the efficacy of 5-FU remains controversial[109]. Therefore, it is necessary to establish new predictive tools for selecting patients who will benefit from treatment with this conventional cytotoxic drug.
We previously demonstrated that the methylation of p16 predicts survival benefits from 5-FU in patients with GC[110]. p16 proteins, a product of the CDKN2/MTS1 gene, are G1/S-specific cell cycle inhibitors[111]. p16 silencing induced via DNA methylation accelerates the cell cycle and increases the cell number in the S-phase. These conditions allow cancer cells to easily absorb 5-FU into S-phase cells, resulting in the enhancement of drug efficacy. Another in vitro study showed that p16 dysfunction is correlated with increased sensitivity to 5-FU in patients with brain tumors[112]. Furthermore, Kato et al[113] showed that the methylation of two proapoptotic genes, TMS1 (target of methylation-induced silencing; also called ‘‘ASC’’) and DAPK (death-associated protein kinase), is associated with resistance to 5-FU and a poor prognosis in GC patients. The expression of ASCL2 (achaete scute-like 2), a cancer stem cell (CSC) marker in colon cancer[114], is also mediated via epigenetic mechanisms, and the upregulation of ASCL2 by promoter demethylation in GC is associated with resistance to 5-FU[115]. CSCs are relatively resistant to commonly used cancer therapies, such as radiation and chemotherapy. Therefore, the development of reliable CTC biomarkers and CSC-targeted therapies is crucial for improving survival in cancer patients.
Multigene methylation has also been investigated as another approach for developing predictive biomarkers for a response to 5-FU. The prognostic relevance of the CpG island methylator phenotype (CIMP) following the administration of standard therapy or a 5-FU-based chemotherapy regimen has been reported in different types of cancers, including GC[70,116-118]. Although no direct correlations between the CIMP status and chemosensitivity and/or resistance have thus far been demonstrated in patients with GC, the detection of the CIMP-positive phenotype can be used to independently predict better survival after 5-FU-based chemotherapy in patients with stage III colorectal cancer[116]. Moreover, the CIMP status in GC has been shown to be associated with microsatellite instability and clinicopathological features, such as tumor location, histology and the TNM stage[70,117]. Further investigations should clarify the role of the CIMP status in the chemosensitivity and/or resistance of GC.
Cisplatin is a platinum-containing compound that is commonly used in various cancers, including GC. The use of cisplatin-based regimens, such as ECF (epirubicin/cisplatin/fluorouracil) and DCF (docetaxel/cisplatin/fluorouracil), has improved the survival of GC patients[119]. Cisplatin forms intrastrand and interstrand cross-links between purines, and these platinum-DNA adducts activate several signaling pathways, resulting in double-stranded DNA breaks and the impairment of replication and gene expression[120]. The initial approach to identifying biomarkers of cisplatin sensitivity focused on the genes involved in the drug’s mechanisms of action. ERCC1 (excision repair cross-complementation group 1) is a major component of the nucleotide excision repair (NER) complex that acts as the rate-limiting enzyme in the NER pathway[121]. De Dosso et al[122] showed that the overall survival of GC patients who receive cisplatin-based adjuvant chemotherapy following curative resection is longer among patients who exhibit a negative expression of ERCC1 proteins. A recent meta-analysis also indicated a significant association between polymorphisms of the NER genes (ERCC1 and ERCC2) and the response to platinum-based chemotherapy[123]. In GC patients, a high expression of NER (ERCC1, ERCC2, GSTP1, XRCC1) and DNA damage proteins (TP53, GADD45A) is negatively associated with a response to cisplatin[124,125].
Despite the intensive investigation of genetic markers for platinum agents (as described above), little information regarding the epigenetic variability affecting the clinical outcomes of platinum-based therapy is available in the setting of GC. One recent in vitro study evaluated the methylation profiles of 20 GC cell lines and identified BMP4 (bone morphogenetic protein 4) to be an epigenetically regulated gene that is highly expressed in cisplatin-resistant cells[126]. BMP4 encodes a secreted protein belonging to the TGFβ superfamily. BMP4 binds to BMP type I/II receptors, thereby activating a signaling cascade that results in the phosphorylation of SMAD1/5/8. BMP4 induces the epithelial mesenchymal transition via the SMAD pathway in several cancers[127,128] and may also be involved in the development of IM of the esophagus and stomach[129]. Other epigenetic markers for CDDP have been frequently assessed in patients with ovarian and lung cancers[130-132]. Stefansson et al[131] recently showed that BRCA1 silencing induced by promoter hypermethylation predicts enhanced sensitivity to platinum in ovarian and breast cancer patients. Tumor suppressor BRCA1 proteins as well as BRCA2 proteins play major roles in DNA double-strand break repair via homologous recombination[133]. BRCA1 and BRCA2 are frequently mutated in patients with familial breast and ovarian cancer, and BRCA1/2-deficient cancers often exhibit a better response to DNA cross-linking agents, such as platinum[133]. Furthermore, Wang et al[134] demonstrated that the methylation of 14-3-3 sigma predicts survival benefits from cisplatin plus gemcitabine treatment in lung cancer patients. BRCA1 and 14-3-3 sigma silencing induced by promoter methylation is also observed in GC[69,135]. Further attempts to identify novel methylation biomarkers of the efficacy of cisplatin therapy in GC patients are needed.
Paclitaxel and docetaxel, both of which are microtubule-stabilizing agents that block cell division by interfering with the function of mitotic spindles by inhibiting microtubule dynamics[136,137], are commonly used in second-line chemotherapy regimens in GC patients who exhibit resistance to 5-FU-based chemotherapy in Japan[138]. Checkpoint with forkhead-associated domain and ring finger (CHFR) has been intensively investigated as a candidate biomarker of the response to microtubule inhibitors in various cancers, including GC[139-141]. CHFR functions as a mitotic checkpoint by detecting mitotic stress induced by microtubule inhibitors, such as paclitaxel, and, under such conditions, induces cell cycle arrest in the G2 phase to repair damaged DNA and is consequently resistant to taxane[142]. However, cells with a CHFR gene inactivated by aberrant hypermethylation cannot detect DNA damage and proceed to mitosis with subsequent cell death due to mitotic catastrophe, i.e., these cells demonstrate a high sensitivity to taxane. We previously demonstrated that the methylation of CHFR is correlated with the response to treatment with paclitaxel in both GC cell lines and patients[143].
Stone et al[144] demonstrated that, in addition to CHFR methylation, BCL-2 expression is associated with DNA methylation in human breast cancer and that BCL-2 hypermethylation is a potential biomarker of sensitivity to antimitotic chemotherapy, including that with paclitaxel and docetaxel. The promoter hypermethylation of RASSF1A as well as BCL-2 was also shown to be an important factor modulating the efficacy of docetaxel-based chemotherapy in patients with breast cancer[145]. Furthermore, a recent in vitro study indicated that TGFβI (transforming growth factor-beta-inducible gene-h3) is frequently methylated and associated with paclitaxel resistance in patients with ovarian cancer[146]. However, to our knowledge, no methylation markers for the efficacy of paclitaxel and/or docetaxel have thus far been reported in GC patients (except for CHFR methylation).
Irinotecan (CPT-11) is a semisynthetic derivative of camptothecin (CPT), which interferes with DNA replication and cell division through its potent interaction with the enzyme topoisomerase 1 (Topo 1)[147]. Both CPT-11 and CPT are Topo1 inhibitors. CPT-11 is primarily used in second-line or subsequent chemotherapy in the treatment of GC.
Several mechanisms have been proposed for resistance to CPT, such as variable levels of the enzymes involved in the conversion of CPT-11, reduced cellular accumulation due to active drug efflux, a reduced expression of Topo 1, alteration in the structure of Topo 1, alterations in the cellular response to CPT-Topo 1-DNA complex formation and the activation of NF-kappaB[148,149]. To identify the epigenetic predictive markers for the efficacy of CPT-11, we previously analyzed the gene methylation of CHFR, p16, RUNX3, E-cadherin, MGMT, hMLH1, ABCG2, UGT1A1 and BNIP3 in 27 colorectal cancer patients who were postoperatively treated with S-1 plus CPT-11 combined therapy. Among these candidate genes, we identified BNIP3 gene methylation as a possible marker for predicting a poor response to S-1 plus CPT-11 therapy in patients with colorectal cancer[150]. Another study also demonstrated that metastatic colorectal cancer patients with BNIP3 methylation exhibit resistance to first-line CPT-11 chemotherapy compared with those without such methylation[151]. Furthermore, two independent studies suggested that the methylation of the UGT1A1 gene in colorectal cancer patients is an important mechanism of UGT1A1 gene silencing and is associated with CPT-11 resistance[152,153]. However, no reliable markers of the sensitivity and/or resistance to CPT-11 have thus far been identified in GC, including BNIP3 and UGT1A1 methylation. Recently, Miyaki et al[154] focused on the DNA methylation profiles of colorectal cancer and GC and identified a novel gene, DEXI (glucocorticoid-induced protein coding gene), that is frequently methylated in colorectal cancer and GC patients using the MS-AFLP (methylation-sensitive amplified fragment-length polymorphism) technique[155]. DEXI methylation results in a poor response to CPT-11-based chemotherapy, suggesting that DEXI is a potent therapeutic target and an epigenetic biomarker for the selection of patients who will benefit from CPT-11-based chemotherapy.
Two recent phase III RCTs demonstrated the clinical efficacy and safety of antibody therapy in patients with advanced GC. The ToGA study investigated the additional benefits of trastuzumab in combination with chemotherapy as a first-line treatment of human epidermal growth factor receptor 2 (HER2)-positive advanced GC. The median overall survival was 13.8 mo in the trastuzumab plus chemotherapy group and 11.1 mo in the chemotherapy alone group (HR = 0.74; 95%CI: 0.60-0.91; P = 0.0046)[156]. The ToGA trial also demonstrated that patients who express a higher level of HER2 receive the greatest benefits from trastuzumab. Although the efficacy of trastuzumab appears to be predictable based on the expression level of HER2, the molecular mechanisms underlying trastuzumab resistance in GC patients are unknown. Preclinical studies have indicated that increased signaling via the PI3K/AKT pathway contributes to trastuzumab resistance in patients with breast cancer[157,158]. Moreover, several reports have shown that DNA methylation profiles can be used to predict the breast cancer subtype and support the prognosis determination and therapeutic stratification of patients with breast cancer[159-161]. Terada et al[162] evaluated the methylation levels in the promoter CGIs of 11 genes in 63 human breast cancer samples and concluded that frequent methylation exhibits a strong association with HER2 amplification in patients with breast cancer. Based on this evidence, increasing the understanding of epigenetic profiles in the setting of GC may enable physicians to classify the intrinsic subtypes of GC and predict the response to trastuzumab in GC patients.
The AVAGAST trial, which was designed to investigate the impact of an anti-vascular endothelial growth factor (VEGF) monoclonal antibody (bevacizumab) in combination with fluoropyrimidine-platinum as first-line therapy for advanced GC showed significant improvements in progression-free survival and the response rate compared with that observed in the placebo group (HR = 0.80, P = 0.0037; 46.0% vs 37.4%, P = 0.0315, respectively)[163]. Additional analyses of biomarkers[164], including VEGF-A, neuropilin-1 and VEGF receptors-1 and -2 (VEGFR-1 and VEGFR-2), showed that the baseline plasma VEGF-A levels and tumor neuropilin-1 expression are potential predictors of bevacizumab efficacy. The patients with a high plasma VEGF-A level exhibited a trend toward improved overall survival (HR = 0.72; 95%CI: 0.57-0.93) compared with the patients with a low VEGF-A level (HR = 1.01; 95%CI: 0.77-1.31; P = 0.07). Furthermore, the potential predictive power of the plasma VEGF-A level identified in the AVAGAST trial is supported by two other independent biomarker analyses performed in patients with metastatic breast cancer (the AVADO study)[165] and pancreatic cancer (the AViTA study)[166]. However, the levels of VEGFRs failed to predict the efficacy of bevacizumab in the AVAGAST study, although the VEGFR-2 expression in the AVADO study and the VEGFR-1 expression in the AViTA study were significantly associated with bevacizumab efficacy[167,168]. In a previous study, the methylation of the VEGF gene was not observed in most cancer cells, whereas the promoter hypermethylation of the VEGFR-1 gene (Flt1) and VEGFR-2 gene (KDR) were widely detected in various cancer cells, including those of GC[169]. A recent in vitro study by Kim et al[170] also suggested that the efficacy of various VEGF-target drugs, including anti-VEGF monoclonal antibodies, is influenced by the epigenetic alteration of VEGFRs.
Among the several molecular targeted therapies for GC to date, only trastuzumab has succeeded in significantly increasing the overall survival in patients with HER2-overexpressing GC. Other agents, including bevacizumab, have failed to achieve efficacy in increasing survival rates over those obtained with standard chemotherapy. It is promising that the careful selection of patient subsets is a key factor for improving GC outcomes. Therefore, further investigations to identify reliable predictive biomarkers of drug efficacy, including methylation profiling, are needed.
The most common tool for predicting patient prognosis of GC is the TNM classification. However, some GCs exhibit different behavior from that predicted by the TNM classification; hence, the accumulation of further information and the identification of indicators, such as molecular biomarkers, is necessary to precisely predict the prognosis. Recently, numerous studies have investigated the molecular basis of GC, and an association between aberrant DNA methylation and the prognosis of GC patients has been reported[171-174]. A large number of genes, including p16, E-cadherin, MGMT, RASSF1, RUNX3, etc., have been shown to be suppressed by CGI hypermethylation[175]. Among these genes, the promoter hypermethylation of E-cadherin[176] (http://link.springer.com/article/10.1007/s10120-012-0209-7/fulltext.html - CR32) and MGMT[177,178] (http://link.springer.com/article/10.1007/s10120-012-0209-7/fulltext.html - CR34) is associated with worse outcomes after surgery in GC patients. Additionally, Wanajo et al[172] identified the methylation of CACNA2D3 (calcium channel voltage-dependent alpha 2/delta subunit 3) to be a strong indicator of poor prognosis among patients with advanced GC. Furthermore, Kim et al[179] suggested that the expression of galectin-7 is critically regulated by DNA hypermethylation and may play a role as a prognostic marker of GC. Our recent work also demonstrated that trefoil factor 1 (TFF1) expression is silenced by DNA methylation and is associated with tumor invasion and poor survival in GC patients[180]. TFF1 is considered to be a tumor suppressor gene in GC. We assessed the immunohistochemical expression of TFF1 in 182 GC patients and examined whether the level of TFF1 is associated with clinicopathological factors and/or patient survival. Consequently, a low expression of TFF1 was found to be independently associated with a poor survival in 108 GC patients treated with surgery alone. Furthermore, bisulfite sequencing demonstrated that TFF1 expression was strongly mediated by DNA methylation in both the GC cells and tissues. Although most previous studies evaluated DNA methylation in GC tissues, epigenetic information obtained from blood samples may also be an important prognostic biomarker in GC patients. It has been reported that patients with hypermethylated insulin-like growth factor 2 (IGF2) in blood leukocyte DNA exhibit a significantly better survival rate than those with hypomethylated IGF2[181].
As described above, many previous studies of methylation in GC have focused on the prognostic significance of single methylated genes. However, the LINE-1 methylation level is regarded to be a surrogate marker of global DNA methylation, and LINE-1 hypomethylation is strongly associated with a poor outcome in several types of human cancers[182,183] (http://link.springer.com/article/10.1007/s10120-012-0209-7/fulltext.html - CR14). Shigaki et al[184] reported for the first time that the genome-wide DNA hypomethylation status measured according to the LINE-1 level is independently associated with poor survival among patients with GC. Furthermore, regarding the prognostic value of CIMP status, it has been reported that CIMP-high is associated with better overall survival, although this parameter is not an independent prognostic factor in patients with resected GC[117]. In another study, patients with a CIMP-negative status demonstrated significantly worse survival than patients with a CIMP-high or CIMP-low status among 78 primary GC series in Japan[70]. In contrast, two investigators reported that hypermethylation at 14 CGI loci or more is closely associated with a poor clinical outcome and was found to be an independent prognostic factor in 196 cases of GC in South Korea[185,186]. Moreover, another report demonstrated no significant differences in survival between the CIMP-positive group and the CIMP-negative group[187]. The CIMP status prognostic value is therefore controversial in the setting of GC. A large-scale study is needed to validate the association between CIMP status and the prognosis of patients with GC.
Although DNA methylation at the fifth position of cytosine (5-mC) is a key epigenetic marker associated with the prognosis of GC, 5-mC is converted to 5-hydroxymethylcytosine (5-hmC) by the ten-eleven translocation family of DNA hydroxylases. In recent years, the effects of 5-hmC on the characteristics of cancer have been widely recognized. However, the significance of 5-hmC for the prognosis of human cancers, including GC, remains largely unknown. Yang et al[188] indicated that a decreased 5-hmC level is a strong and independent poor prognostic factor in patients with GC. The direct or consequential methylation of microRNAs may also influence the prognosis of GC[189]. Prompt and reliable methods for the analysis of the molecular basis of GC have recently been developed, and the impact of epigenetic modification on the prognosis of GC patients has been increasingly revealed. Increasing understanding of epigenetic molecular profiles will hopefully be successfully applied to predict and improve the prognosis of GC.
The field of cancer epigenetics has significantly expanded over the last decade, and technological advances have dramatically accelerated investigation in this area of cancer research. Aberrant DNA methylation results in the misregulation of cellular processes, such as proliferation, transformation and anti-apoptotic effects, all of which promote cancer progression. However, DNA methylation has great potential to provide valuable information for understanding the malignant behavior of GC (Figure 1). Further investigations of the DNA methylation status, which regulates cancer initiation, proliferation, invasion, metastasis and drug resistance, will aid in designing strategies for earlier detection and better therapeutic decision making in the setting of GC.
P- Reviewer: Chi SG, Hiyama T, Imaeda H, Liu XF S- Editor: Ma YJ L- Editor: A E- Editor: Liu XM
| 1. | Ferlay J, Shin HR, Bray F, Forman D, Mathers C, Parkin DM. Estimates of worldwide burden of cancer in 2008: GLOBOCAN 2008. Int J Cancer. 2010;127:2893-2917. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 11128] [Cited by in F6Publishing: 11614] [Article Influence: 893.4] [Reference Citation Analysis (4)] |
| 2. | Nagini S. Carcinoma of the stomach: A review of epidemiology, pathogenesis, molecular genetics and chemoprevention. World J Gastrointest Oncol. 2012;4:156-169. [PubMed] [DOI] [Cited in This Article: ] [Cited by in CrossRef: 303] [Cited by in F6Publishing: 311] [Article Influence: 25.9] [Reference Citation Analysis (0)] |
| 3. | Baylin SB, Jones PA. A decade of exploring the cancer epigenome - biological and translational implications. Nat Rev Cancer. 2011;11:726-734. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 2049] [Cited by in F6Publishing: 1973] [Article Influence: 151.8] [Reference Citation Analysis (0)] |
| 4. | Miyamoto K, Ushijima T. Diagnostic and therapeutic applications of epigenetics. Jpn J Clin Oncol. 2005;35:293-301. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 89] [Cited by in F6Publishing: 100] [Article Influence: 5.3] [Reference Citation Analysis (0)] |
| 5. | Gardiner-Garden M, Frommer M. CpG islands in vertebrate genomes. J Mol Biol. 1987;196:261-282. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 2342] [Cited by in F6Publishing: 2283] [Article Influence: 61.7] [Reference Citation Analysis (0)] |
| 6. | Baylin SB, Ohm JE. Epigenetic gene silencing in cancer - a mechanism for early oncogenic pathway addiction? Nat Rev Cancer. 2006;6:107-116. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1182] [Cited by in F6Publishing: 1155] [Article Influence: 64.2] [Reference Citation Analysis (0)] |
| 7. | Shames DS, Minna JD, Gazdar AF. DNA methylation in health, disease, and cancer. Curr Mol Med. 2007;7:85-102. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 145] [Cited by in F6Publishing: 131] [Article Influence: 7.7] [Reference Citation Analysis (0)] |
| 8. | Laird PW. The power and the promise of DNA methylation markers. Nat Rev Cancer. 2003;3:253-266. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1143] [Cited by in F6Publishing: 1083] [Article Influence: 51.6] [Reference Citation Analysis (0)] |
| 9. | Schuebel KE, Chen W, Cope L, Glöckner SC, Suzuki H, Yi JM, Chan TA, Van Neste L, Van Criekinge W, van den Bosch S. Comparing the DNA hypermethylome with gene mutations in human colorectal cancer. PLoS Genet. 2007;3:1709-1723. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 248] [Cited by in F6Publishing: 280] [Article Influence: 16.5] [Reference Citation Analysis (0)] |
| 10. | Sjöblom T, Jones S, Wood LD, Parsons DW, Lin J, Barber TD, Mandelker D, Leary RJ, Ptak J, Silliman N. The consensus coding sequences of human breast and colorectal cancers. Science. 2006;314:268-274. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 2562] [Cited by in F6Publishing: 2468] [Article Influence: 137.1] [Reference Citation Analysis (0)] |
| 11. | Fang J, Zhu S, Xiao S, Shi Y, Jiang S, Zhou X, Qian L. Alterations of level of total genomic DNA methylation and pattern of c-myc, c-Ha-ras oncogene methylation in human gastric carcinogenesis. Chin Med J (Engl). 1996;109:787-791. [PubMed] [Cited in This Article: ] |
| 12. | Tamura G, Yin J, Wang S, Fleisher AS, Zou T, Abraham JM, Kong D, Smolinski KN, Wilson KT, James SP. E-Cadherin gene promoter hypermethylation in primary human gastric carcinomas. J Natl Cancer Inst. 2000;92:569-573. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 254] [Cited by in F6Publishing: 253] [Article Influence: 10.5] [Reference Citation Analysis (0)] |
| 13. | Tamura G, Sato K, Akiyama S, Tsuchiya T, Endoh Y, Usuba O, Kimura W, Nishizuka S, Motoyama T. Molecular characterization of undifferentiated-type gastric carcinoma. Lab Invest. 2001;81:593-598. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 57] [Cited by in F6Publishing: 58] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 14. | Ksiaa F, Ziadi S, Amara K, Korbi S, Trimeche M. Biological significance of promoter hypermethylation of tumor-related genes in patients with gastric carcinoma. Clin Chim Acta. 2009;404:128-133. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 56] [Cited by in F6Publishing: 62] [Article Influence: 4.1] [Reference Citation Analysis (0)] |
| 15. | Oshimo Y, Kuraoka K, Nakayama H, Kitadai Y, Yoshida K, Chayama K, Yasui W. Epigenetic inactivation of SOCS-1 by CpG island hypermethylation in human gastric carcinoma. Int J Cancer. 2004;112:1003-1009. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 82] [Cited by in F6Publishing: 90] [Article Influence: 4.5] [Reference Citation Analysis (0)] |
| 16. | Nojima M, Suzuki H, Toyota M, Watanabe Y, Maruyama R, Sasaki S, Sasaki Y, Mita H, Nishikawa N, Yamaguchi K. Frequent epigenetic inactivation of SFRP genes and constitutive activation of Wnt signaling in gastric cancer. Oncogene. 2007;26:4699-4713. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 169] [Cited by in F6Publishing: 176] [Article Influence: 10.4] [Reference Citation Analysis (0)] |
| 17. | Kang YH, Lee HS, Kim WH. Promoter methylation and silencing of PTEN in gastric carcinoma. Lab Invest. 2002;82:285-291. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 159] [Cited by in F6Publishing: 175] [Article Influence: 8.0] [Reference Citation Analysis (0)] |
| 18. | Sapari NS, Loh M, Vaithilingam A, Soong R. Clinical potential of DNA methylation in gastric cancer: a meta-analysis. PLoS One. 2012;7:e36275. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 64] [Cited by in F6Publishing: 74] [Article Influence: 6.2] [Reference Citation Analysis (0)] |
| 19. | Kang GH, Lee S, Cho NY, Gandamihardja T, Long TI, Weisenberger DJ, Campan M, Laird PW. DNA methylation profiles of gastric carcinoma characterized by quantitative DNA methylation analysis. Lab Invest. 2008;88:161-170. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 126] [Cited by in F6Publishing: 134] [Article Influence: 8.4] [Reference Citation Analysis (0)] |
| 20. | Wang LJ, Jin HC, Wang X, Lam EK, Zhang JB, Liu X, Chan FK, Si JM, Sung JJ. ZIC1 is downregulated through promoter hypermethylation in gastric cancer. Biochem Biophys Res Commun. 2009;379:959-963. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 39] [Cited by in F6Publishing: 46] [Article Influence: 3.1] [Reference Citation Analysis (0)] |
| 21. | Shu XS, Geng H, Li L, Ying J, Ma C, Wang Y, Poon FF, Wang X, Ying Y, Yeo W. The epigenetic modifier PRDM5 functions as a tumor suppressor through modulating WNT/β-catenin signaling and is frequently silenced in multiple tumors. PLoS One. 2011;6:e27346. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 54] [Cited by in F6Publishing: 54] [Article Influence: 4.2] [Reference Citation Analysis (0)] |
| 22. | Yu J, Cheng YY, Tao Q, Cheung KF, Lam CN, Geng H, Tian LW, Wong YP, Tong JH, Ying JM. Methylation of protocadherin 10, a novel tumor suppressor, is associated with poor prognosis in patients with gastric cancer. Gastroenterology. 2009;136:640-651.e1. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 146] [Cited by in F6Publishing: 164] [Article Influence: 10.9] [Reference Citation Analysis (0)] |
| 23. | Jee CD, Kim MA, Jung EJ, Kim J, Kim WH. Identification of genes epigenetically silenced by CpG methylation in human gastric carcinoma. Eur J Cancer. 2009;45:1282-1293. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 64] [Cited by in F6Publishing: 74] [Article Influence: 4.9] [Reference Citation Analysis (0)] |
| 24. | Lu XX, Yu JL, Ying LS, Han J, Wang S, Yu QM, Wang XB, Fang XH, Ling ZQ. Stepwise cumulation of RUNX3 methylation mediated by Helicobacter pylori infection contributes to gastric carcinoma progression. Cancer. 2012;118:5507-5517. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 53] [Cited by in F6Publishing: 63] [Article Influence: 5.3] [Reference Citation Analysis (0)] |
| 25. | Jones PA, Baylin SB. The epigenomics of cancer. Cell. 2007;128:683-692. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 3299] [Cited by in F6Publishing: 3278] [Article Influence: 192.8] [Reference Citation Analysis (0)] |
| 26. | Dong W, Tu S, Xie J, Sun P, Wu Y, Wang L. Frequent promoter hypermethylation and transcriptional downregulation of BTG4 gene in gastric cancer. Biochem Biophys Res Commun. 2009;387:132-138. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 17] [Cited by in F6Publishing: 18] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 27. | Cheng YY, Yu J, Wong YP, Man EP, To KF, Jin VX, Li J, Tao Q, Sung JJ, Chan FK. Frequent epigenetic inactivation of secreted frizzled-related protein 2 (SFRP2) by promoter methylation in human gastric cancer. Br J Cancer. 2007;97:895-901. [PubMed] [Cited in This Article: ] |
| 28. | Wani M, Afroze D, Makhdoomi M, Hamid I, Wani B, Bhat G, Wani R, Wani K. Promoter methylation status of DNA repair gene (hMLH1) in gastric carcinoma patients of the Kashmir valley. Asian Pac J Cancer Prev. 2012;13:4177-4181. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 15] [Cited by in F6Publishing: 19] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 29. | Yu J, Tao Q, Cheng YY, Lee KY, Ng SS, Cheung KF, Tian L, Rha SY, Neumann U, Röcken C. Promoter methylation of the Wnt/beta-catenin signaling antagonist Dkk-3 is associated with poor survival in gastric cancer. Cancer. 2009;115:49-60. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 96] [Cited by in F6Publishing: 110] [Article Influence: 7.3] [Reference Citation Analysis (0)] |
| 30. | Joo JK, Kim SH, Kim HG, Kim DY, Ryu SY, Lee KH, Lee JH. CpG methylation of transcription factor 4 in gastric carcinoma. Ann Surg Oncol. 2010;17:3344-3353. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 11] [Cited by in F6Publishing: 11] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 31. | Wu CS, Lu YJ, Li HP, Hsueh C, Lu CY, Leu YW, Liu HP, Lin KH, Hui-Ming Huang T, Chang YS. Glutamate receptor, ionotropic, kainate 2 silencing by DNA hypermethylation possesses tumor suppressor function in gastric cancer. Int J Cancer. 2010;126:2542-2552. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 22] [Cited by in F6Publishing: 28] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 32. | Ben Ayed-Guerfali D, Benhaj K, Khabir A, Abid M, Bayrouti MI, Sellami-Boudawara T, Gargouri A, Mokdad-Gargouri R. Hypermethylation of tumor-related genes in Tunisian patients with gastric carcinoma: clinical and biological significance. J Surg Oncol. 2011;103:687-694. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 22] [Cited by in F6Publishing: 27] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 33. | Hiraki M, Kitajima Y, Koga Y, Tanaka T, Nakamura J, Hashiguchi K, Noshiro H, Miyazaki K. Aberrant gene methylation is a biomarker for the detection of cancer cells in peritoneal wash samples from advanced gastric cancer patients. Ann Surg Oncol. 2011;18:3013-3019. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 33] [Cited by in F6Publishing: 35] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 34. | Lu YJ, Wu CS, Li HP, Liu HP, Lu CY, Leu YW, Wang CS, Chen LC, Lin KH, Chang YS. Aberrant methylation impairs low density lipoprotein receptor-related protein 1B tumor suppressor function in gastric cancer. Genes Chromosomes Cancer. 2010;49:412-424. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 11] [Cited by in F6Publishing: 29] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 35. | Christofori G, Semb H. The role of the cell-adhesion molecule E-cadherin as a tumour-suppressor gene. Trends Biochem Sci. 1999;24:73-76. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 525] [Cited by in F6Publishing: 531] [Article Influence: 21.2] [Reference Citation Analysis (0)] |
| 36. | Chan AO. E-cadherin in gastric cancer. World J Gastroenterol. 2006;12:199-203. [PubMed] [Cited in This Article: ] |
| 37. | Tamura G. Alterations of tumor suppressor and tumor-related genes in the development and progression of gastric cancer. World J Gastroenterol. 2006;12:192-198. [PubMed] [Cited in This Article: ] |
| 38. | Waki T, Tamura G, Tsuchiya T, Sato K, Nishizuka S, Motoyama T. Promoter methylation status of E-cadherin, hMLH1, and p16 genes in nonneoplastic gastric epithelia. Am J Pathol. 2002;161:399-403. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 112] [Cited by in F6Publishing: 119] [Article Influence: 5.4] [Reference Citation Analysis (0)] |
| 39. | Dammann R, Li C, Yoon JH, Chin PL, Bates S, Pfeifer GP. Epigenetic inactivation of a RAS association domain family protein from the lung tumour suppressor locus 3p21.3. Nat Genet. 2000;25:315-319. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 814] [Cited by in F6Publishing: 847] [Article Influence: 35.3] [Reference Citation Analysis (0)] |
| 40. | Byun DS, Lee MG, Chae KS, Ryu BG, Chi SG. Frequent epigenetic inactivation of RASSF1A by aberrant promoter hypermethylation in human gastric adenocarcinoma. Cancer Res. 2001;61:7034-7038. [PubMed] [Cited in This Article: ] |
| 41. | Dammann R, Schagdarsurengin U, Strunnikova M, Rastetter M, Seidel C, Liu L, Tommasi S, Pfeifer GP. Epigenetic inactivation of the Ras-association domain family 1 (RASSF1A) gene and its function in human carcinogenesis. Histol Histopathol. 2003;18:665-677. [PubMed] [Cited in This Article: ] |
| 42. | To KF, Leung WK, Lee TL, Yu J, Tong JH, Chan MW, Ng EK, Chung SC, Sung JJ. Promoter hypermethylation of tumor-related genes in gastric intestinal metaplasia of patients with and without gastric cancer. Int J Cancer. 2002;102:623-628. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 98] [Cited by in F6Publishing: 104] [Article Influence: 4.7] [Reference Citation Analysis (0)] |
| 43. | Yao D, Shi J, Shi B, Wang N, Liu W, Zhang G, Ji M, Xu L, He N, Hou P. Quantitative assessment of gene methylation and their impact on clinical outcome in gastric cancer. Clin Chim Acta. 2012;413:787-794. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 33] [Cited by in F6Publishing: 31] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 44. | Marshall BJ, Warren JR. Unidentified curved bacilli in the stomach of patients with gastritis and peptic ulceration. Lancet. 1984;1:1311-1315. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 3302] [Cited by in F6Publishing: 3116] [Article Influence: 77.9] [Reference Citation Analysis (0)] |
| 45. | Nomura A, Stemmermann GN, Chyou PH, Kato I, Perez-Perez GI, Blaser MJ. Helicobacter pylori infection and gastric carcinoma among Japanese Americans in Hawaii. N Engl J Med. 1991;325:1132-1136. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1302] [Cited by in F6Publishing: 1203] [Article Influence: 36.5] [Reference Citation Analysis (0)] |
| 46. | Parsonnet J, Friedman GD, Vandersteen DP, Chang Y, Vogelman JH, Orentreich N, Sibley RK. Helicobacter pylori infection and the risk of gastric carcinoma. N Engl J Med. 1991;325:1127-1131. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 2805] [Cited by in F6Publishing: 2665] [Article Influence: 80.8] [Reference Citation Analysis (0)] |
| 47. | Uemura N, Okamoto S, Yamamoto S, Matsumura N, Yamaguchi S, Yamakido M, Taniyama K, Sasaki N, Schlemper RJ. Helicobacter pylori infection and the development of gastric cancer. N Engl J Med. 2001;345:784-789. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 3126] [Cited by in F6Publishing: 3020] [Article Influence: 131.3] [Reference Citation Analysis (0)] |
| 48. | Maekita T, Nakazawa K, Mihara M, Nakajima T, Yanaoka K, Iguchi M, Arii K, Kaneda A, Tsukamoto T, Tatematsu M. High levels of aberrant DNA methylation in Helicobacter pylori-infected gastric mucosae and its possible association with gastric cancer risk. Clin Cancer Res. 2006;12:989-995. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 474] [Cited by in F6Publishing: 460] [Article Influence: 25.6] [Reference Citation Analysis (0)] |
| 49. | Kitajima Y, Ohtaka K, Mitsuno M, Tanaka M, Sato S, Nakafusa Y, Miyazaki K. Helicobacter pylori infection is an independent risk factor for Runx3 methylation in gastric cancer. Oncol Rep. 2008;19:197-202. [PubMed] [Cited in This Article: ] |
| 50. | Fukamachi H, Ito K, Ito Y. Runx3-/- gastric epithelial cells differentiate into intestinal type cells. Biochem Biophys Res Commun. 2004;321:58-64. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 40] [Cited by in F6Publishing: 43] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 51. | Park SY, Yoo EJ, Cho NY, Kim N, Kang GH. Comparison of CpG island hypermethylation and repetitive DNA hypomethylation in premalignant stages of gastric cancer, stratified for Helicobacter pylori infection. J Pathol. 2009;219:410-416. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 86] [Cited by in F6Publishing: 93] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 52. | Ando T, Yoshida T, Enomoto S, Asada K, Tatematsu M, Ichinose M, Sugiyama T, Ushijima T. DNA methylation of microRNA genes in gastric mucosae of gastric cancer patients: its possible involvement in the formation of epigenetic field defect. Int J Cancer. 2009;124:2367-2374. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 211] [Cited by in F6Publishing: 215] [Article Influence: 14.3] [Reference Citation Analysis (0)] |
| 53. | Nakajima T, Maekita T, Oda I, Gotoda T, Yamamoto S, Umemura S, Ichinose M, Sugimura T, Ushijima T, Saito D. Higher methylation levels in gastric mucosae significantly correlate with higher risk of gastric cancers. Cancer Epidemiol Biomarkers Prev. 2006;15:2317-2321. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 134] [Cited by in F6Publishing: 130] [Article Influence: 7.6] [Reference Citation Analysis (0)] |
| 54. | Niwa T, Tsukamoto T, Toyoda T, Mori A, Tanaka H, Maekita T, Ichinose M, Tatematsu M, Ushijima T. Inflammatory processes triggered by Helicobacter pylori infection cause aberrant DNA methylation in gastric epithelial cells. Cancer Res. 2010;70:1430-1440. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 289] [Cited by in F6Publishing: 305] [Article Influence: 21.8] [Reference Citation Analysis (0)] |
| 55. | Hur K, Niwa T, Toyoda T, Tsukamoto T, Tatematsu M, Yang HK, Ushijima T. Insufficient role of cell proliferation in aberrant DNA methylation induction and involvement of specific types of inflammation. Carcinogenesis. 2011;32:35-41. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 65] [Cited by in F6Publishing: 72] [Article Influence: 5.1] [Reference Citation Analysis (0)] |
| 56. | Qian X, Huang C, Cho CH, Hui WM, Rashid A, Chan AO. E-cadherin promoter hypermethylation induced by interleukin-1beta treatment or H. pylori infection in human gastric cancer cell lines. Cancer Lett. 2008;263:107-113. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 39] [Cited by in F6Publishing: 42] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 57. | Wang YQ, Li YM, Li X, Liu T, Liu XK, Zhang JQ, Guo JW, Guo LY, Qiao L. Hypermethylation of TGF-β1 gene promoter in gastric cancer. World J Gastroenterol. 2013;19:5557-5564. [PubMed] [DOI] [Cited in This Article: ] [Cited by in CrossRef: 18] [Cited by in F6Publishing: 16] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 58. | Wang M, Furuta T, Takashima M, Futami H, Shirai N, Hanai H, Kaneko E. Relation between interleukin-1beta messenger RNA in gastric fundic mucosa and gastric juice pH in patients infected with Helicobacter pylori. J Gastroenterol. 1999;34 Suppl 11:10-17. [PubMed] [Cited in This Article: ] |
| 59. | Yamaoka Y, Kita M, Kodama T, Sawai N, Kashima K, Imanishi J. Induction of various cytokines and development of severe mucosal inflammation by cagA gene positive Helicobacter pylori strains. Gut. 1997;41:442-451. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 265] [Cited by in F6Publishing: 292] [Article Influence: 10.8] [Reference Citation Analysis (0)] |
| 60. | Leung WK, Yu J, Ng EK, To KF, Ma PK, Lee TL, Go MY, Chung SC, Sung JJ. Concurrent hypermethylation of multiple tumor-related genes in gastric carcinoma and adjacent normal tissues. Cancer. 2001;91:2294-2301. [PubMed] [Cited in This Article: ] |
| 61. | Chan AO, Peng JZ, Lam SK, Lai KC, Yuen MF, Cheung HK, Kwong YL, Rashid A, Chan CK, Wong BC. Eradication of Helicobacter pylori infection reverses E-cadherin promoter hypermethylation. Gut. 2006;55:463-468. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 133] [Cited by in F6Publishing: 132] [Article Influence: 7.3] [Reference Citation Analysis (0)] |
| 62. | Niwa T, Toyoda T, Tsukamoto T, Mori A, Tatematsu M, Ushijima T. Prevention of Helicobacter pylori-induced gastric cancers in gerbils by a DNA demethylating agent. Cancer Prev Res (Phila). 2013;6:263-270. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 50] [Cited by in F6Publishing: 55] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 63. | Niedobitek G, Agathanggelou A, Steven N, Young LS. Epstein-Barr virus (EBV) in infectious mononucleosis: detection of the virus in tonsillar B lymphocytes but not in desquamated oropharyngeal epithelial cells. Mol Pathol. 2000;53:37-42. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 52] [Cited by in F6Publishing: 56] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 64. | Epstein MA, Barr YM. Cultivation in vitro of human lymphoblasts from burkitt’s malignant lymphoma. Lancet. 1964;1:252-253. [PubMed] [Cited in This Article: ] |
| 65. | zur Hausen H, Schulte-Holthausen H, Klein G, Henle W, Henle G, Clifford P, Santesson L. EBV DNA in biopsies of Burkitt tumours and anaplastic carcinomas of the nasopharynx. Nature. 1970;228:1056-1058. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 925] [Cited by in F6Publishing: 986] [Article Influence: 18.3] [Reference Citation Analysis (0)] |
| 66. | Burke AP, Yen TS, Shekitka KM, Sobin LH. Lymphoepithelial carcinoma of the stomach with Epstein-Barr virus demonstrated by polymerase chain reaction. Mod Pathol. 1990;3:377-380. [PubMed] [Cited in This Article: ] |
| 67. | Tokunaga M, Land CE, Uemura Y, Tokudome T, Tanaka S, Sato E. Epstein-Barr virus in gastric carcinoma. Am J Pathol. 1993;143:1250-1254. [PubMed] [Cited in This Article: ] |
| 68. | Shibata D, Weiss LM. Epstein-Barr virus-associated gastric adenocarcinoma. Am J Pathol. 1992;140:769-774. [PubMed] [Cited in This Article: ] |
| 69. | Kang GH, Lee S, Kim WH, Lee HW, Kim JC, Rhyu MG, Ro JY. Epstein-barr virus-positive gastric carcinoma demonstrates frequent aberrant methylation of multiple genes and constitutes CpG island methylator phenotype-positive gastric carcinoma. Am J Pathol. 2002;160:787-794. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 247] [Cited by in F6Publishing: 262] [Article Influence: 11.9] [Reference Citation Analysis (0)] |
| 70. | Kusano M, Toyota M, Suzuki H, Akino K, Aoki F, Fujita M, Hosokawa M, Shinomura Y, Imai K, Tokino T. Genetic, epigenetic, and clinicopathologic features of gastric carcinomas with the CpG island methylator phenotype and an association with Epstein-Barr virus. Cancer. 2006;106:1467-1479. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 132] [Cited by in F6Publishing: 134] [Article Influence: 7.4] [Reference Citation Analysis (0)] |
| 71. | Chang MS, Uozaki H, Chong JM, Ushiku T, Sakuma K, Ishikawa S, Hino R, Barua RR, Iwasaki Y, Arai K. CpG island methylation status in gastric carcinoma with and without infection of Epstein-Barr virus. Clin Cancer Res. 2006;12:2995-3002. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 142] [Cited by in F6Publishing: 154] [Article Influence: 8.6] [Reference Citation Analysis (0)] |
| 72. | Kaneda A, Matsusaka K, Aburatani H, Fukayama M. Epstein-Barr virus infection as an epigenetic driver of tumorigenesis. Cancer Res. 2012;72:3445-3450. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 128] [Cited by in F6Publishing: 138] [Article Influence: 11.5] [Reference Citation Analysis (0)] |
| 73. | Ushiku T, Chong JM, Uozaki H, Hino R, Chang MS, Sudo M, Rani BR, Sakuma K, Nagai H, Fukayama M. p73 gene promoter methylation in Epstein-Barr virus-associated gastric carcinoma. Int J Cancer. 2007;120:60-66. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 65] [Cited by in F6Publishing: 71] [Article Influence: 4.2] [Reference Citation Analysis (0)] |
| 74. | Okada T, Nakamura M, Nishikawa J, Sakai K, Zhang Y, Saito M, Morishige A, Oga A, Sasaki K, Suehiro Y. Identification of genes specifically methylated in Epstein-Barr virus-associated gastric carcinomas. Cancer Sci. 2013;104:1309-1314. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 26] [Cited by in F6Publishing: 30] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 75. | Hino R, Uozaki H, Murakami N, Ushiku T, Shinozaki A, Ishikawa S, Morikawa T, Nakaya T, Sakatani T, Takada K. Activation of DNA methyltransferase 1 by EBV latent membrane protein 2A leads to promoter hypermethylation of PTEN gene in gastric carcinoma. Cancer Res. 2009;69:2766-2774. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 258] [Cited by in F6Publishing: 272] [Article Influence: 18.1] [Reference Citation Analysis (0)] |
| 76. | Zhao J, Liang Q, Cheung KF, Kang W, Lung RW, Tong JH, To KF, Sung JJ, Yu J. Genome-wide identification of Epstein-Barr virus-driven promoter methylation profiles of human genes in gastric cancer cells. Cancer. 2013;119:304-312. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 109] [Cited by in F6Publishing: 108] [Article Influence: 9.8] [Reference Citation Analysis (0)] |
| 77. | Matsusaka K, Kaneda A, Nagae G, Ushiku T, Kikuchi Y, Hino R, Uozaki H, Seto Y, Takada K, Aburatani H. Classification of Epstein-Barr virus-positive gastric cancers by definition of DNA methylation epigenotypes. Cancer Res. 2011;71:7187-7197. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 159] [Cited by in F6Publishing: 178] [Article Influence: 13.7] [Reference Citation Analysis (0)] |
| 78. | Garcia M, Jemal A. Global Cancer Facts and Figures 2011. Atlanta, GA: American Cancer Society 2011; . [Cited in This Article: ] |
| 79. | Shapiro B, Chakrabarty M, Cohn EM, Leon SA. Determination of circulating DNA levels in patients with benign or malignant gastrointestinal disease. Cancer. 1983;51:2116-2120. [PubMed] [Cited in This Article: ] |
| 80. | Abbaszadegan MR, Moaven O, Sima HR, Ghafarzadegan K, A’rabi A, Forghani MN, Raziee HR, Mashhadinejad A, Jafarzadeh M, Esmaili-Shandiz E. p16 promoter hypermethylation: a useful serum marker for early detection of gastric cancer. World J Gastroenterol. 2008;14:2055-2060. [PubMed] [DOI] [Cited in This Article: ] [Cited by in CrossRef: 60] [Cited by in F6Publishing: 69] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 81. | Kanyama Y, Hibi K, Nakayama H, Kodera Y, Ito K, Akiyama S, Nakao A. Detection of p16 promoter hypermethylation in serum of gastric cancer patients. Cancer Sci. 2003;94:418-420. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 33] [Cited by in F6Publishing: 38] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 82. | Zou XP, Zhang B, Zhang XQ, Chen M, Cao J, Liu WJ. Promoter hypermethylation of multiple genes in early gastric adenocarcinoma and precancerous lesions. Hum Pathol. 2009;40:1534-1542. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 52] [Cited by in F6Publishing: 61] [Article Influence: 4.1] [Reference Citation Analysis (0)] |
| 83. | Zheng Y, Zhang Y, Huang X, Chen L. Analysis of the RUNX3 gene methylation in serum DNA from esophagus squamous cell carcinoma, gastric and colorectal adenocarcinoma patients. Hepatogastroenterology. 2011;58:2007-2011. [PubMed] [DOI] [Cited in This Article: ] [Cited by in F6Publishing: 14] [Reference Citation Analysis (0)] |
| 84. | Sakakura C, Hamada T, Miyagawa K, Nishio M, Miyashita A, Nagata H, Ida H, Yazumi S, Otsuji E, Chiba T. Quantitative analysis of tumor-derived methylated RUNX3 sequences in the serum of gastric cancer patients. Anticancer Res. 2009;29:2619-2625. [PubMed] [Cited in This Article: ] |
| 85. | Wang YC, Yu ZH, Liu C, Xu LZ, Yu W, Lu J, Zhu RM, Li GL, Xia XY, Wei XW. Detection of RASSF1A promoter hypermethylation in serum from gastric and colorectal adenocarcinoma patients. World J Gastroenterol. 2008;14:3074-3080. [PubMed] [DOI] [Cited in This Article: ] [Cited by in CrossRef: 61] [Cited by in F6Publishing: 65] [Article Influence: 4.1] [Reference Citation Analysis (0)] |
| 86. | Hibi K, Goto T, Shirahata A, Saito M, Kigawa G, Nemoto H, Sanada Y. Detection of TFPI2 methylation in the serum of gastric cancer patients. Anticancer Res. 2011;31:3835-3838. [PubMed] [Cited in This Article: ] |
| 87. | Chan MW, Chu ES, To KF, Leung WK. Quantitative detection of methylated SOCS-1, a tumor suppressor gene, by a modified protocol of quantitative real time methylation-specific PCR using SYBR green and its use in early gastric cancer detection. Biotechnol Lett. 2004;26:1289-1293. [PubMed] [Cited in This Article: ] |
| 88. | Leung WK, To KF, Chu ES, Chan MW, Bai AH, Ng EK, Chan FK, Sung JJ. Potential diagnostic and prognostic values of detecting promoter hypermethylation in the serum of patients with gastric cancer. Br J Cancer. 2005;92:2190-2194. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 72] [Cited by in F6Publishing: 80] [Article Influence: 4.2] [Reference Citation Analysis (0)] |
| 89. | Oishi Y, Watanabe Y, Yoshida Y, Sato Y, Hiraishi T, Oikawa R, Maehata T, Suzuki H, Toyota M, Niwa H. Hypermethylation of Sox17 gene is useful as a molecular diagnostic application in early gastric cancer. Tumour Biol. 2012;33:383-393. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 34] [Cited by in F6Publishing: 40] [Article Influence: 3.1] [Reference Citation Analysis (0)] |
| 90. | Watanabe Y, Kim HS, Castoro RJ, Chung W, Estecio MR, Kondo K, Guo Y, Ahmed SS, Toyota M, Itoh F. Sensitive and specific detection of early gastric cancer with DNA methylation analysis of gastric washes. Gastroenterology. 2009;136:2149-2158. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 100] [Cited by in F6Publishing: 97] [Article Influence: 6.5] [Reference Citation Analysis (0)] |
| 91. | Yoo CH, Noh SH, Shin DW, Choi SH, Min JS. Recurrence following curative resection for gastric carcinoma. Br J Surg. 2000;87:236-242. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 523] [Cited by in F6Publishing: 531] [Article Influence: 22.1] [Reference Citation Analysis (0)] |
| 92. | Broll R, Weschta M, Windhoevel U, Berndt S, Schwandner O, Roblick U, Schiedeck TH, Schimmelpenning H, Bruch HP, Duchrow M. Prognostic significance of free gastrointestinal tumor cells in peritoneal lavage detected by immunocytochemistry and polymerase chain reaction. Langenbecks Arch Surg. 2001;386:285-292. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 22] [Cited by in F6Publishing: 23] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 93. | Lee CC, Lo SS, Wu CW, Shen KH, Li AF, Hsieh MC, Lui WY. Peritoneal recurrence of gastric adenocarcinoma after curative resection. Hepatogastroenterology. 2003;50:1720-1722. [PubMed] [Cited in This Article: ] |
| 94. | Yu QM, Wang XB, Luo J, Wang S, Fang XH, Yu JL, Ling ZQ. CDH1 methylation in preoperative peritoneal washes is an independent prognostic factor for gastric cancer. J Surg Oncol. 2012;106:765-771. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 27] [Cited by in F6Publishing: 28] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 95. | Ikeda Y, Saku M, Kishihara F, Maehara Y. Effective follow-up for recurrence or a second primary cancer in patients with early gastric cancer. Br J Surg. 2005;92:235-239. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 63] [Cited by in F6Publishing: 69] [Article Influence: 3.6] [Reference Citation Analysis (0)] |
| 96. | Doekhie FS, Mesker WE, van Krieken JH, Kok NF, Hartgrink HH, Kranenbarg EK, Putter H, Kuppen PJ, Tanke HJ, Tollenaar RA. Clinical relevance of occult tumor cells in lymph nodes from gastric cancer patients. Am J Surg Pathol. 2005;29:1135-1144. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 26] [Cited by in F6Publishing: 28] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 97. | Yonemura Y, Endo Y, Hayashi I, Kawamura T, Yun HY, Bandou E. Proliferative activity of micrometastases in the lymph nodes of patients with gastric cancer. Br J Surg. 2007;94:731-736. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 47] [Cited by in F6Publishing: 42] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 98. | Kim JH, Park JM, Jung CW, Park SS, Kim SJ, Mok YJ, Kim CS, Chae YS, Bae JW. The significances of lymph node micrometastasis and its correlation with E-cadherin expression in pT1-T3N0 gastric adenocarcinoma. J Surg Oncol. 2008;97:125-130. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 51] [Cited by in F6Publishing: 52] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 99. | Kubota K, Nakanishi H, Hiki N, Shimizu N, Tsuji E, Yamaguchi H, Mafune K, Tange T, Tatematsu M, Kaminishi M. Quantitative detection of micrometastases in the lymph nodes of gastric cancer patients with real-time RT-PCR: a comparative study with immunohistochemistry. Int J Cancer. 2003;105:136-143. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 47] [Cited by in F6Publishing: 52] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 100. | Ikeda S, Funakoshi N, Usui S, Takiguchi N, Hiranuma S, Shibata T. Prognostic significance of gastric cancer metastasis in second-tier lymph nodes detected on reverse transcriptase-polymerase chain reaction and immunohistochemistry. Pathol Int. 2008;58:45-50. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 6] [Cited by in F6Publishing: 6] [Article Influence: 0.4] [Reference Citation Analysis (0)] |
| 101. | Arigami T, Natsugoe S, Uenosono Y, Yanagita S, Ehi K, Arima H, Mataki Y, Nakajo A, Ishigami S, Aikou T. Vascular endothelial growth factor-C and -D expression correlates with lymph node micrometastasis in pN0 early gastric cancer. J Surg Oncol. 2009;99:148-153. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 24] [Cited by in F6Publishing: 27] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 102. | Yi Kim D, Kyoon Joo J, Kyu Park Y, Yeob Ryu S, Soo Kim H, Kyun Noh B, Hwa Lee K, Hyuk Lee J. E-cadherin expression in early gastric carcinoma and correlation with lymph node metastasis. J Surg Oncol. 2007;96:429-435. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 18] [Cited by in F6Publishing: 19] [Article Influence: 1.1] [Reference Citation Analysis (0)] |
| 103. | Hur K, Han TS, Jung EJ, Yu J, Lee HJ, Kim WH, Goel A, Yang HK. Up-regulated expression of sulfatases (SULF1 and SULF2) as prognostic and metastasis predictive markers in human gastric cancer. J Pathol. 2012;228:88-98. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 42] [Cited by in F6Publishing: 45] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 104. | Chang X, Li Z, Ma J, Deng P, Zhang S, Zhi Y, Chen J, Dai D. DNA methylation of NDRG2 in gastric cancer and its clinical significance. Dig Dis Sci. 2013;58:715-723. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 26] [Cited by in F6Publishing: 26] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 105. | Peng DF, Hu TL, Schneider BG, Chen Z, Xu ZK, El-Rifai W. Silencing of glutathione peroxidase 3 through DNA hypermethylation is associated with lymph node metastasis in gastric carcinomas. PLoS One. 2012;7:e46214. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 40] [Cited by in F6Publishing: 46] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 106. | Hiraki M, Kitajima Y, Sato S, Mitsuno M, Koga Y, Nakamura J, Hashiguchi K, Noshiro H, Miyazaki K. Aberrant gene methylation in the lymph nodes provides a possible marker for diagnosing micrometastasis in gastric cancer. Ann Surg Oncol. 2010;17:1177-1186. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 23] [Cited by in F6Publishing: 28] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 107. | Longley DB, Harkin DP, Johnston PG. 5-fluorouracil: mechanisms of action and clinical strategies. Nat Rev Cancer. 2003;3:330-338. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 3255] [Cited by in F6Publishing: 3302] [Article Influence: 157.2] [Reference Citation Analysis (0)] |
| 108. | Hashiguchi K, Kitajima Y, Kai K, Hiraki M, Nakamura J, Tokunaga O, Noshiro H, Miyazaki K. A quantitative evaluation of the determinant proteins for S-1 responsiveness in a biopsy specimen assists in patient selection to neoadjuvant therapy in cases of advanced gastric cancer. Int J Oncol. 2010;37:257-264. [PubMed] [Cited in This Article: ] |
| 109. | Ichikawa W. Prediction of clinical outcome of fluoropyrimidine-based chemotherapy for gastric cancer patients, in terms of the 5-fluorouracil metabolic pathway. Gastric Cancer. 2006;9:145-155. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 43] [Cited by in F6Publishing: 42] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 110. | Mitsuno M, Kitajima Y, Ide T, Ohtaka K, Tanaka M, Satoh S, Miyazaki K. Aberrant methylation of p16 predicts candidates for 5-fluorouracil-based adjuvant therapy in gastric cancer patients. J Gastroenterol. 2007;42:866-873. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 32] [Cited by in F6Publishing: 35] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 111. | Serrano M, Hannon GJ, Beach D. A new regulatory motif in cell-cycle control causing specific inhibition of cyclin D/CDK4. Nature. 1993;366:704-707. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 2515] [Cited by in F6Publishing: 2493] [Article Influence: 80.4] [Reference Citation Analysis (0)] |
| 112. | Iwadate Y, Mochizuki S, Fujimoto S, Namba H, Sakiyama S, Tagawa M, Yamaura A. Alteration of CDKN2/p16 in human astrocytic tumors is related with increased susceptibility to antimetabolite anticancer agents. Int J Oncol. 2000;17:501-505. [PubMed] [Cited in This Article: ] |
| 113. | Kato K, Iida S, Uetake H, Takagi Y, Yamashita T, Inokuchi M, Yamada H, Kojima K, Sugihara K. Methylated TMS1 and DAPK genes predict prognosis and response to chemotherapy in gastric cancer. Int J Cancer. 2008;122:603-608. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 49] [Cited by in F6Publishing: 51] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 114. | van der Flier LG, van Gijn ME, Hatzis P, Kujala P, Haegebarth A, Stange DE, Begthel H, van den Born M, Guryev V, Oving I. Transcription factor achaete scute-like 2 controls intestinal stem cell fate. Cell. 2009;136:903-912. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 536] [Cited by in F6Publishing: 535] [Article Influence: 35.7] [Reference Citation Analysis (0)] |
| 115. | Kwon OH, Park JL, Baek SJ, Noh SM, Song KS, Kim SY, Kim YS. Aberrant upregulation of ASCL2 by promoter demethylation promotes the growth and resistance to 5-fluorouracil of gastric cancer cells. Cancer Sci. 2013;104:391-397. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 23] [Cited by in F6Publishing: 27] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 116. | Van Rijnsoever M, Elsaleh H, Joseph D, McCaul K, Iacopetta B. CpG island methylator phenotype is an independent predictor of survival benefit from 5-fluorouracil in stage III colorectal cancer. Clin Cancer Res. 2003;9:2898-2903. [PubMed] [Cited in This Article: ] |
| 117. | An C, Choi IS, Yao JC, Worah S, Xie K, Mansfield PF, Ajani JA, Rashid A, Hamilton SR, Wu TT. Prognostic significance of CpG island methylator phenotype and microsatellite instability in gastric carcinoma. Clin Cancer Res. 2005;11:656-663. [PubMed] [Cited in This Article: ] |
| 118. | Brock MV, Gou M, Akiyama Y, Muller A, Wu TT, Montgomery E, Deasel M, Germonpré P, Rubinson L, Heitmiller RF. Prognostic importance of promoter hypermethylation of multiple genes in esophageal adenocarcinoma. Clin Cancer Res. 2003;9:2912-2919. [PubMed] [Cited in This Article: ] |
| 119. | Ilson DH. Docetaxel, cisplatin, and fluorouracil in gastric cancer: does the punishment fit the crime? J Clin Oncol. 2007;25:3188-3190. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 42] [Cited by in F6Publishing: 47] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 120. | Siddik ZH. Cisplatin: mode of cytotoxic action and molecular basis of resistance. Oncogene. 2003;22:7265-7279. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 2302] [Cited by in F6Publishing: 2429] [Article Influence: 115.7] [Reference Citation Analysis (1)] |
| 121. | McHugh PJ, Spanswick VJ, Hartley JA. Repair of DNA interstrand crosslinks: molecular mechanisms and clinical relevance. Lancet Oncol. 2001;2:483-490. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 280] [Cited by in F6Publishing: 294] [Article Influence: 12.8] [Reference Citation Analysis (0)] |
| 122. | De Dosso S, Zanellato E, Nucifora M, Boldorini R, Sonzogni A, Biffi R, Fazio N, Bucci E, Beretta O, Crippa S. ERCC1 predicts outcome in patients with gastric cancer treated with adjuvant cisplatin-based chemotherapy. Cancer Chemother Pharmacol. 2013;72:159-165. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 31] [Cited by in F6Publishing: 28] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 123. | Yin M, Yan J, Martinez-Balibrea E, Graziano F, Lenz HJ, Kim HJ, Robert J, Im SA, Wang WS, Etienne-Grimaldi MC. ERCC1 and ERCC2 polymorphisms predict clinical outcomes of oxaliplatin-based chemotherapies in gastric and colorectal cancer: a systemic review and meta-analysis. Clin Cancer Res. 2011;17:1632-1640. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 111] [Cited by in F6Publishing: 114] [Article Influence: 8.8] [Reference Citation Analysis (0)] |
| 124. | Fareed KR, Kaye P, Soomro IN, Ilyas M, Martin S, Parsons SL, Madhusudan S. Biomarkers of response to therapy in oesophago-gastric cancer. Gut. 2009;58:127-143. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 60] [Cited by in F6Publishing: 67] [Article Influence: 4.5] [Reference Citation Analysis (0)] |
| 125. | Bataille F, Rümmele P, Dietmaier W, Gaag D, Klebl F, Reichle A, Wild P, Hofstädter F, Hartmann A. Alterations in p53 predict response to preoperative high dose chemotherapy in patients with gastric cancer. Mol Pathol. 2003;56:286-292. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 29] [Cited by in F6Publishing: 32] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 126. | Ivanova T, Zouridis H, Wu Y, Cheng LL, Tan IB, Gopalakrishnan V, Ooi CH, Lee J, Qin L, Wu J. Integrated epigenomics identifies BMP4 as a modulator of cisplatin sensitivity in gastric cancer. Gut. 2013;62:22-33. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 75] [Cited by in F6Publishing: 80] [Article Influence: 7.3] [Reference Citation Analysis (0)] |
| 127. | Thériault BL, Shepherd TG, Mujoomdar ML, Nachtigal MW. BMP4 induces EMT and Rho GTPase activation in human ovarian cancer cells. Carcinogenesis. 2007;28:1153-1162. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 118] [Cited by in F6Publishing: 122] [Article Influence: 7.2] [Reference Citation Analysis (0)] |
| 128. | Hamada S, Satoh K, Hirota M, Kimura K, Kanno A, Masamune A, Shimosegawa T. Bone morphogenetic protein 4 induces epithelial-mesenchymal transition through MSX2 induction on pancreatic cancer cell line. J Cell Physiol. 2007;213:768-774. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 68] [Cited by in F6Publishing: 76] [Article Influence: 4.5] [Reference Citation Analysis (0)] |
| 129. | Barros R, Pereira B, Duluc I, Azevedo M, Mendes N, Camilo V, Jacobs RJ, Paulo P, Santos-Silva F, van Seuningen I. Key elements of the BMP/SMAD pathway co-localize with CDX2 in intestinal metaplasia and regulate CDX2 expression in human gastric cell lines. J Pathol. 2008;215:411-420. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 45] [Cited by in F6Publishing: 51] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 130. | Lee PS, Teaberry VS, Bland AE, Huang Z, Whitaker RS, Baba T, Fujii S, Secord AA, Berchuck A, Murphy SK. Elevated MAL expression is accompanied by promoter hypomethylation and platinum resistance in epithelial ovarian cancer. Int J Cancer. 2010;126:1378-1389. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 6] [Cited by in F6Publishing: 26] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 131. | Stefansson OA, Villanueva A, Vidal A, Martí L, Esteller M. BRCA1 epigenetic inactivation predicts sensitivity to platinum-based chemotherapy in breast and ovarian cancer. Epigenetics. 2012;7:1225-1229. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 95] [Cited by in F6Publishing: 89] [Article Influence: 7.4] [Reference Citation Analysis (0)] |
| 132. | Ramirez JL, Rosell R, Taron M, Sanchez-Ronco M, Alberola V, de Las Peñas R, Sanchez JM, Moran T, Camps C, Massuti B. 14-3-3sigma methylation in pretreatment serum circulating DNA of cisplatin-plus-gemcitabine-treated advanced non-small-cell lung cancer patients predicts survival: The Spanish Lung Cancer Group. J Clin Oncol. 2005;23:9105-9112. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 97] [Cited by in F6Publishing: 105] [Article Influence: 5.8] [Reference Citation Analysis (0)] |
| 133. | Roy R, Chun J, Powell SN. BRCA1 and BRCA2: different roles in a common pathway of genome protection. Nat Rev Cancer. 2012;12:68-78. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 898] [Cited by in F6Publishing: 949] [Article Influence: 73.0] [Reference Citation Analysis (0)] |
| 134. | Wang W, Figg WD. Secondary BRCA1 and BRCA2 alterations and acquired chemoresistance. Cancer Biol Ther. 2008;7:1004-1005. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 28] [Cited by in F6Publishing: 29] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 135. | Bernal C, Vargas M, Ossandón F, Santibáñez E, Urrutia J, Luengo V, Zavala LF, Backhouse C, Palma M, Argandoña J. DNA methylation profile in diffuse type gastric cancer: evidence for hypermethylation of the BRCA1 promoter region in early-onset gastric carcinogenesis. Biol Res. 2008;41:303-315. [PubMed] [Cited in This Article: ] |
| 136. | Jordan MA, Wilson L. Microtubules and actin filaments: dynamic targets for cancer chemotherapy. Curr Opin Cell Biol. 1998;10:123-130. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 453] [Cited by in F6Publishing: 431] [Article Influence: 16.6] [Reference Citation Analysis (0)] |
| 137. | Dumontet C, Sikic BI. Mechanisms of action of and resistance to antitubulin agents: microtubule dynamics, drug transport, and cell death. J Clin Oncol. 1999;17:1061-1070. [PubMed] [Cited in This Article: ] |
| 138. | Kinoshita T, Nashimoto A, Yamamura Y, Okamura T, Sasako M, Sakamoto J, Kojima H, Hiratsuka M, Arai K, Sairenji M. Feasibility study of adjuvant chemotherapy with S-1 (TS-1; tegafur, gimeracil, oteracil potassium) for gastric cancer. Gastric Cancer. 2004;7:104-109. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 29] [Cited by in F6Publishing: 32] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 139. | Satoh A, Toyota M, Itoh F, Sasaki Y, Suzuki H, Ogi K, Kikuchi T, Mita H, Yamashita T, Kojima T. Epigenetic inactivation of CHFR and sensitivity to microtubule inhibitors in gastric cancer. Cancer Res. 2003;63:8606-8613. [PubMed] [Cited in This Article: ] |
| 140. | Yanokura M, Banno K, Kawaguchi M, Hirao N, Hirasawa A, Susumu N, Tsukazaki K, Aoki D. Relationship of aberrant DNA hypermethylation of CHFR with sensitivity to taxanes in endometrial cancer. Oncol Rep. 2007;17:41-48. [PubMed] [Cited in This Article: ] |
| 141. | Pillai RN, Brodie SA, Sica GL, Shaojin Y, Li G, Nickleach DC, Yuan L, Varma VA, Bonta D, Herman JG. CHFR protein expression predicts outcomes to taxane-based first line therapy in metastatic NSCLC. Clin Cancer Res. 2013;19:1603-1611. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 19] [Cited by in F6Publishing: 21] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 142. | Scolnick DM, Halazonetis TD. Chfr defines a mitotic stress checkpoint that delays entry into metaphase. Nature. 2000;406:430-435. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 277] [Cited by in F6Publishing: 292] [Article Influence: 12.2] [Reference Citation Analysis (0)] |
| 143. | Koga Y, Kitajima Y, Miyoshi A, Sato K, Sato S, Miyazaki K. The significance of aberrant CHFR methylation for clinical response to microtubule inhibitors in gastric cancer. J Gastroenterol. 2006;41:133-139. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 44] [Cited by in F6Publishing: 50] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 144. | Stone A, Cowley MJ, Valdes-Mora F, McCloy RA, Sergio CM, Gallego-Ortega D, Caldon CE, Ormandy CJ, Biankin AV, Gee JM. BCL-2 hypermethylation is a potential biomarker of sensitivity to antimitotic chemotherapy in endocrine-resistant breast cancer. Mol Cancer Ther. 2013;12:1874-1885. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 33] [Cited by in F6Publishing: 37] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 145. | Gil EY, Jo UH, Jeong H, Whang YM, Woo OH, Cho KR, Seo JH, Kim A, Lee ES, Koh I. Promoter methylation of RASSF1A modulates the effect of the microtubule-targeting agent docetaxel in breast cancer. Int J Oncol. 2012;41:611-620. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 13] [Cited by in F6Publishing: 16] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 146. | Wang N, Zhang H, Yao Q, Wang Y, Dai S, Yang X. TGFBI promoter hypermethylation correlating with paclitaxel chemoresistance in ovarian cancer. J Exp Clin Cancer Res. 2012;31:6. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 44] [Cited by in F6Publishing: 44] [Article Influence: 3.7] [Reference Citation Analysis (0)] |
| 147. | Liu LF, Desai SD, Li TK, Mao Y, Sun M, Sim SP. Mechanism of action of camptothecin. Ann N Y Acad Sci. 2000;922:1-10. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 321] [Cited by in F6Publishing: 339] [Article Influence: 14.7] [Reference Citation Analysis (0)] |
| 148. | Xu Y, Villalona-Calero MA. Irinotecan: mechanisms of tumor resistance and novel strategies for modulating its activity. Ann Oncol. 2002;13:1841-1851. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 247] [Cited by in F6Publishing: 264] [Article Influence: 12.0] [Reference Citation Analysis (0)] |
| 149. | Huang TT, Wuerzberger-Davis SM, Seufzer BJ, Shumway SD, Kurama T, Boothman DA, Miyamoto S. NF-kappaB activation by camptothecin. A linkage between nuclear DNA damage and cytoplasmic signaling events. J Biol Chem. 2000;275:9501-9509. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 121] [Cited by in F6Publishing: 122] [Article Influence: 5.1] [Reference Citation Analysis (0)] |
| 150. | Hiraki M, Kitajima Y, Nakafusa Y, Nakamura J, Hashiguchi K, Sumi K, Noshiro H, Miyazaki K. CpG island methylation of BNIP3 predicts resistance against S-1/CPT-11 combined therapy in colorectal cancer patients. Oncol Rep. 2010;23:191-197. [PubMed] [Cited in This Article: ] |
| 151. | Shimizu S, Iida S, Ishiguro M, Uetake H, Ishikawa T, Takagi Y, Kobayashi H, Higuchi T, Enomoto M, Mogushi K. Methylated BNIP3 gene in colorectal cancer prognosis. Oncol Lett. 2010;1:865-872. [PubMed] [Cited in This Article: ] |
| 152. | Gagnon JF, Bernard O, Villeneuve L, Têtu B, Guillemette C. Irinotecan inactivation is modulated by epigenetic silencing of UGT1A1 in colon cancer. Clin Cancer Res. 2006;12:1850-1858. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 67] [Cited by in F6Publishing: 72] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 153. | Xie FW, Peng Y, Chen X, Chen X, Li J, Wang W, Yu Z, Ouyang X. Relationship between the expression of CES2, UGT1A1, and GUSB in colorectal cancer tissues and aberrant methylation. Neoplasma. 2014;61:99-109. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 6] [Cited by in F6Publishing: 6] [Article Influence: 0.6] [Reference Citation Analysis (0)] |
| 154. | Miyaki Y, Suzuki K, Koizumi K, Kato T, Saito M, Kamiyama H, Maeda T, Shibata K, Shiya N, Konishi F. Identification of a potent epigenetic biomarker for resistance to camptothecin and poor outcome to irinotecan-based chemotherapy in colon cancer. Int J Oncol. 2012;40:217-226. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 4] [Cited by in F6Publishing: 9] [Article Influence: 0.7] [Reference Citation Analysis (0)] |
| 155. | Yamamoto F, Yamamoto M, Soto JL, Kojima E, Wang EN, Perucho M, Sekiya T, Yamanaka H. Notl-Msell methylation-sensitive amplied fragment length polymorhism for DNA methylation analysis of human cancers. Electrophoresis. 2001;22:1946-1956. [PubMed] [Cited in This Article: ] |
| 156. | Bang YJ, Van Cutsem E, Feyereislova A, Chung HC, Shen L, Sawaki A, Lordick F, Ohtsu A, Omuro Y, Satoh T. Trastuzumab in combination with chemotherapy versus chemotherapy alone for treatment of HER2-positive advanced gastric or gastro-oesophageal junction cancer (ToGA): a phase 3, open-label, randomised controlled trial. Lancet. 2010;376:687-697. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 4615] [Cited by in F6Publishing: 4831] [Article Influence: 345.1] [Reference Citation Analysis (1)] |
| 157. | Nagata Y, Lan KH, Zhou X, Tan M, Esteva FJ, Sahin AA, Klos KS, Li P, Monia BP, Nguyen NT. PTEN activation contributes to tumor inhibition by trastuzumab, and loss of PTEN predicts trastuzumab resistance in patients. Cancer Cell. 2004;6:117-127. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1362] [Cited by in F6Publishing: 1329] [Article Influence: 66.5] [Reference Citation Analysis (0)] |
| 158. | Berns K, Horlings HM, Hennessy BT, Madiredjo M, Hijmans EM, Beelen K, Linn SC, Gonzalez-Angulo AM, Stemke-Hale K, Hauptmann M. A functional genetic approach identifies the PI3K pathway as a major determinant of trastuzumab resistance in breast cancer. Cancer Cell. 2007;12:395-402. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1213] [Cited by in F6Publishing: 1207] [Article Influence: 71.0] [Reference Citation Analysis (0)] |
| 159. | Bediaga NG, Acha-Sagredo A, Guerra I, Viguri A, Albaina C, Ruiz Diaz I, Rezola R, Alberdi MJ, Dopazo J, Montaner D. DNA methylation epigenotypes in breast cancer molecular subtypes. Breast Cancer Res. 2010;12:R77. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 139] [Cited by in F6Publishing: 143] [Article Influence: 10.2] [Reference Citation Analysis (0)] |
| 160. | Holm K, Hegardt C, Staaf J, Vallon-Christersson J, Jönsson G, Olsson H, Borg A, Ringnér M. Molecular subtypes of breast cancer are associated with characteristic DNA methylation patterns. Breast Cancer Res. 2010;12:R36. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 239] [Cited by in F6Publishing: 211] [Article Influence: 15.1] [Reference Citation Analysis (0)] |
| 161. | Lee JS, Fackler MJ, Lee JH, Choi C, Park MH, Yoon JH, Zhang Z, Sukumar S. Basal-like breast cancer displays distinct patterns of promoter methylation. Cancer Biol Ther. 2010;9:1017-1024. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 28] [Cited by in F6Publishing: 30] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 162. | Terada K, Okochi-Takada E, Akashi-Tanaka S, Miyamoto K, Taniyama K, Tsuda H, Asada K, Kaminishi M, Ushijima T. Association between frequent CpG island methylation and HER2 amplification in human breast cancers. Carcinogenesis. 2009;30:466-471. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 23] [Cited by in F6Publishing: 23] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 163. | Ohtsu A, Shah MA, Van Cutsem E, Rha SY, Sawaki A, Park SR, Lim HY, Yamada Y, Wu J, Langer B. Bevacizumab in combination with chemotherapy as first-line therapy in advanced gastric cancer: a randomized, double-blind, placebo-controlled phase III study. J Clin Oncol. 2011;29:3968-3976. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 795] [Cited by in F6Publishing: 887] [Article Influence: 68.2] [Reference Citation Analysis (0)] |
| 164. | Van Cutsem E, de Haas S, Kang YK, Ohtsu A, Tebbutt NC, Ming Xu J, Peng Yong W, Langer B, Delmar P, Scherer SJ. Bevacizumab in combination with chemotherapy as first-line therapy in advanced gastric cancer: a biomarker evaluation from the AVAGAST randomized phase III trial. J Clin Oncol. 2012;30:2119-2127. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 355] [Cited by in F6Publishing: 385] [Article Influence: 32.1] [Reference Citation Analysis (0)] |
| 165. | Pivot X, Schneeweiss A, Verma S, Thomssen C, Passos-Coelho JL, Benedetti G, Ciruelos E, von Moos R, Chang HT, Duenne AA. Efficacy and safety of bevacizumab in combination with docetaxel for the first-line treatment of elderly patients with locally recurrent or metastatic breast cancer: results from AVADO. Eur J Cancer. 2011;47:2387-2395. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 72] [Cited by in F6Publishing: 77] [Article Influence: 5.9] [Reference Citation Analysis (0)] |
| 166. | Van Cutsem E, Vervenne WL, Bennouna J, Humblet Y, Gill S, Van Laethem JL, Verslype C, Scheithauer W, Shang A, Cosaert J. Phase III trial of bevacizumab in combination with gemcitabine and erlotinib in patients with metastatic pancreatic cancer. J Clin Oncol. 2009;27:2231-2237. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 469] [Cited by in F6Publishing: 478] [Article Influence: 31.9] [Reference Citation Analysis (0)] |
| 167. | Miles DW, de Haas SL, Dirix LY, Romieu G, Chan A, Pivot X, Tomczak P, Provencher L, Cortés J, Delmar PR. Biomarker results from the AVADO phase 3 trial of first-line bevacizumab plus docetaxel for HER2-negative metastatic breast cancer. Br J Cancer. 2013;108:1052-1060. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 120] [Cited by in F6Publishing: 117] [Article Influence: 10.6] [Reference Citation Analysis (0)] |
| 168. | Lambrechts D, Claes B, Delmar P, Reumers J, Mazzone M, Yesilyurt BT, Devlieger R, Verslype C, Tejpar S, Wildiers H. VEGF pathway genetic variants as biomarkers of treatment outcome with bevacizumab: an analysis of data from the AViTA and AVOREN randomised trials. Lancet Oncol. 2012;13:724-733. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 152] [Cited by in F6Publishing: 141] [Article Influence: 11.8] [Reference Citation Analysis (0)] |
| 169. | Kim JY, Hwang JH, Zhou W, Shin J, Noh SM, Song IS, Kim JY, Lee SH, Kim J. The expression of VEGF receptor genes is concurrently influenced by epigenetic gene silencing of the genes and VEGF activation. Epigenetics. 2009;4:313-321. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 36] [Cited by in F6Publishing: 36] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 170. | Kim J, Hwang J, Jeong H, Song HJ, Shin J, Hur G, Park YW, Lee SH, Kim J. Promoter methylation status of VEGF receptor genes: a possible epigenetic biomarker to anticipate the efficacy of intracellular-acting VEGF-targeted drugs in cancer cells. Epigenetics. 2012;7:191-200. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 22] [Cited by in F6Publishing: 24] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 171. | Vo QN, Geradts J, Boudreau DA, Bravo JC, Schneider BG. CDKN2A promoter methylation in gastric adenocarcinomas: clinical variables. Hum Pathol. 2002;33:1200-1204. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 20] [Cited by in F6Publishing: 20] [Article Influence: 0.9] [Reference Citation Analysis (0)] |
| 172. | Wanajo A, Sasaki A, Nagasaki H, Shimada S, Otsubo T, Owaki S, Shimizu Y, Eishi Y, Kojima K, Nakajima Y. Methylation of the calcium channel-related gene, CACNA2D3, is frequent and a poor prognostic factor in gastric cancer. Gastroenterology. 2008;135:580-590. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 70] [Cited by in F6Publishing: 69] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 173. | Hu C, Lin F, Zhu G, Xue X, Ding Y, Zhao Z, Zhang L, Shen X. Abnormal hypermethylation of promoter region downregulates chemokine CXC ligand 14 expression in gastric cancer. Int J Oncol. 2013;43:1487-1494. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 22] [Cited by in F6Publishing: 22] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 174. | Ling ZQ, Lv P, Lu XX, Yu JL, Han J, Ying LS, Zhu X, Zhu WY, Fang XH, Wang S. Circulating Methylated XAF1 DNA Indicates Poor Prognosis for Gastric Cancer. PLoS One. 2013;8:e67195. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 49] [Cited by in F6Publishing: 51] [Article Influence: 4.6] [Reference Citation Analysis (0)] |
| 175. | Kim TY, Jong HS, Jung Y, Kim TY, Kang GH, Bang YJ. DNA hypermethylation in gastric cancer. Aliment Pharmacol Ther. 2004;20 Suppl 1:131-142. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 29] [Cited by in F6Publishing: 34] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 176. | Graziano F, Arduini F, Ruzzo A, Bearzi I, Humar B, More H, Silva R, Muretto P, Guilford P, Testa E. Prognostic analysis of E-cadherin gene promoter hypermethylation in patients with surgically resected, node-positive, diffuse gastric cancer. Clin Cancer Res. 2004;10:2784-2789. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 67] [Cited by in F6Publishing: 68] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 177. | Park TJ, Han SU, Cho YK, Paik WK, Kim YB, Lim IK. Methylation of O(6)-methylguanine-DNA methyltransferase gene is associated significantly with K-ras mutation, lymph node invasion, tumor staging, and disease free survival in patients with gastric carcinoma. Cancer. 2001;92:2760-2768. [PubMed] [Cited in This Article: ] |
| 178. | Bae SI, Lee HS, Kim SH, Kim WH. Inactivation of O6-methylguanine-DNA methyltransferase by promoter CpG island hypermethylation in gastric cancers. Br J Cancer. 2002;86:1888-1892. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 40] [Cited by in F6Publishing: 48] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 179. | Kim SJ, Hwang JA, Ro JY, Lee YS, Chun KH. Galectin-7 is epigenetically-regulated tumor suppressor in gastric cancer. Oncotarget. 2013;4:1461-1471. [PubMed] [Cited in This Article: ] |
| 180. | Tanaka T, Nakamura J, Kitajima Y, Kai K, Miyake S, Hiraki M, Ide T, Koga Y, Noshiro H. Loss of trefoil factor 1 is regulated by DNA methylation and is an independent predictive factor for poor survival in advanced gastric cancer. Int J Oncol. 2013;42:894-902. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 18] [Cited by in F6Publishing: 17] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 181. | Yuasa Y, Nagasaki H, Oze I, Akiyama Y, Yoshida S, Shitara K, Ito S, Hosono S, Watanabe M, Ito H. Insulin-like growth factor 2 hypomethylation of blood leukocyte DNA is associated with gastric cancer risk. Int J Cancer. 2012;131:2596-2603. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 24] [Cited by in F6Publishing: 25] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 182. | Ogino S, Nosho K, Kirkner GJ, Kawasaki T, Chan AT, Schernhammer ES, Giovannucci EL, Fuchs CS. A cohort study of tumoral LINE-1 hypomethylation and prognosis in colon cancer. J Natl Cancer Inst. 2008;100:1734-1738. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 276] [Cited by in F6Publishing: 289] [Article Influence: 18.1] [Reference Citation Analysis (0)] |
| 183. | Ohka F, Natsume A, Motomura K, Kishida Y, Kondo Y, Abe T, Nakasu Y, Namba H, Wakai K, Fukui T. The global DNA methylation surrogate LINE-1 methylation is correlated with MGMT promoter methylation and is a better prognostic factor for glioma. PLoS One. 2011;6:e23332. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 75] [Cited by in F6Publishing: 84] [Article Influence: 6.5] [Reference Citation Analysis (0)] |
| 184. | Shigaki H, Baba Y, Watanabe M, Murata A, Iwagami S, Miyake K, Ishimoto T, Iwatsuki M, Baba H. LINE-1 hypomethylation in gastric cancer, detected by bisulfite pyrosequencing, is associated with poor prognosis. Gastric Cancer. 2013;16:480-487. [PubMed] [Cited in This Article: ] |
| 185. | Park SY, Kook MC, Kim YW, Cho NY, Jung N, Kwon HJ, Kim TY, Kang GH. CpG island hypermethylator phenotype in gastric carcinoma and its clinicopathological features. Virchows Arch. 2010;457:415-422. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 32] [Cited by in F6Publishing: 35] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 186. | Chen HY, Zhu BH, Zhang CH, Yang DJ, Peng JJ, Chen JH, Liu FK, He YL. High CpG island methylator phenotype is associated with lymph node metastasis and prognosis in gastric cancer. Cancer Sci. 2012;103:73-79. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 38] [Cited by in F6Publishing: 43] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 187. | Liu JB, Wu XM, Cai J, Zhang JY, Zhang JL, Zhou SH, Shi MX, Qiang FL. CpG island methylator phenotype and Helicobacter pylori infection associated with gastric cancer. World J Gastroenterol. 2012;18:5129-5134. [PubMed] [DOI] [Cited in This Article: ] [Cited by in CrossRef: 30] [Cited by in F6Publishing: 27] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 188. | Yang Q, Wu K, Ji M, Jin W, He N, Shi B, Hou P. Decreased 5-hydroxymethylcytosine (5-hmC) is an independent poor prognostic factor in gastric cancer patients. J Biomed Nanotechnol. 2013;9:1607-1616. [PubMed] [Cited in This Article: ] |
| 189. | Tang H, Deng M, Tang Y, Xie X, Guo J, Kong Y, Ye F, Su Q, Xie X. miR-200b and miR-200c as prognostic factors and mediators of gastric cancer cell progression. Clin Cancer Res. 2013;19:5602-5612. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 133] [Cited by in F6Publishing: 132] [Article Influence: 12.0] [Reference Citation Analysis (0)] |