Published online Aug 28, 2014. doi: 10.3748/wjg.v20.i32.11340
Revised: February 24, 2014
Accepted: May 23, 2014
Published online: August 28, 2014
AIM: To investigate microRNA-133a (miR-133a) expression in colorectal cancer (CRC) and its relationship with tumorigenesis and disease prognosis.
METHODS: Quantitative real-time polymerase chain reaction was used to measure levels of miR-133a in tumor samples and adjacent non-cancerous tissues from 169 patients undergoing radical resection for CRC. The associations between miR-133a expression and patient age, sex, as well as clinicopathologic parameters, such as tumor size, differentiation, location, invasion depth, metastasis, tumor-node-metastasis (TNM) stage and overall patient survival, were analyzed by Mann-Whitney U and Kruskal-Wallis tests. The Kaplan-Meier method and Cox proportional hazards regression analyses were performed to estimate the prognostic factors for patient survival prediction.
RESULTS: The expression of miR-133a was significantly downregulated in CRC tissues compared with adjacent non-cancerous tissues (P < 0.05). This reduction was associated with the depth of the local invasion, poor differentiation, lymph node metastasis and advanced disease (P < 0.05). Moreover, Kaplan-Meier analysis demonstrated that patients with low miR-133a expression had poorer overall survival (OS) than those with high miR-133a expression (P < 0.001). Univariate analysis revealed statistically significant correlations between OS and miR-133a level, tumor local invasion, lymph node metastasis and TNM stage (P < 0.001). Furthermore, miR-133a levels and TNM stage were independently associated with OS (HR = 0.590, 95%CI: 0.350-0.995, P < 0.05; and HR = 6.111, 95%CI: 1.029-36.278, P < 0.05, respectively).
CONCLUSION: The downregulation of miR-133a may play an important role in the progression of CRC and can be used as an independent factor to determine CRC prognosis.
Core tip: In the present study, the level of microRNA-133a (miR-133a) was found to be downregulated in colorectal cancer (CRC) tissues. The altered expression of miR-133a was significantly associated with malignant behavior, including tumor cell differentiation, local invasion, lymph node metastasis and tumor-node-metastasis stage. Multivariate analysis suggested that low expression of miR-133a is an independent prognostic factor for CRC. Furthermore, the data suggest that miR-133a may play a critical role in CRC progression, and thus may serve as a potential therapeutic target.
- Citation: Wang LL, Du LT, Li J, Liu YM, Qu AL, Yang YM, Zhang X, Zheng GX, Wang CX. Decreased expression of miR-133a correlates with poor prognosis in colorectal cancer patients. World J Gastroenterol 2014; 20(32): 11340-11346
- URL: https://www.wjgnet.com/1007-9327/full/v20/i32/11340.htm
- DOI: https://dx.doi.org/10.3748/wjg.v20.i32.11340
Colorectal cancer (CRC) is the second most common malignancy in females, and third most common in males worldwide, with over 1.2 million new cases and an estimated 608700 deaths in 2008 alone[1]. The five-year survival rate of CRC ranges from 90% for stage I patients to 8% for metastatic cases, and nearly 25% of patients with stage II may relapse or develop metastases[2]. Biomarkers such as carcinoembryonic antigen have been recommended for the prediction of CRC prognosis and postoperative surveillance in advanced disease[3], however, they show a limited sensitivity of only 30%-40% for early diagnosis and prognosis. Therefore, novel and reliable prognostic CRC biomarkers are urgently needed.
MicroRNAs (miRNAs) are a novel class of small endogenous non-coding RNAs (17-28 nucleotides) that are involved in the initiation and progression of tumors by the dysregulation of oncogenes and tumor suppressor genes. A growing amount of evidence demonstrates the valuable role of miRNAs in tumor diagnosis, progression and therapy response, which has led both researchers and clinicians to focus on the identification of novel miRNAs. Ma et al[4] carried out a comprehensive systematic review of miRNA expression in CRC and identified several up- and downregulated miRNAs as candidate biomarkers. Among those identified, the ectopic expression of miR-133a was associated with various human malignancies, including lung squamous cell carcinoma, breast cancer, renal cell carcinoma, prostate cancer and bladder urothelial carcinoma[5-9]. In addition, a correlation between miR-133a expression and the carcinogenesis of CRC has also been reported[10,11]. Recently, Wang et al[12] demonstrated that miR-133a affects CRC cell motility and represses tumor growth and metastasis by targeting Lin11, Isl-1 and Mec-3 and SRC homology 3 protein 1, and inhibiting the mitogen-activated protein kinase (MAPK) pathway. However, another study showed that miR-133a served as a gene promoter for brain metastasis[13]. Thus, further analyses are needed to clarify the role of miR-133a in CRC prognosis based on clinicopathologic stage. In the present study, miR-133a expression levels in CRC were examined, and the clinicopathologic significance and potential prognostic value for CRC were assessed.
A total of 182 CRC patients who underwent radical resection for CRC in the Department of General Surgery, Qilu Hospital of Shandong University between June 2005 and December 2007 were recruited for this study. Of these subjects, seven were excluded because of incomplete follow-up data and six with distant metastases were excluded for the reason of non-statistical significance. The remaining 169 patients had not received preoperative adjuvant therapy and were deemed eligible for the study. All patient data were obtained from clinical and pathologic records, including age, sex, tumor size and depth, lymph node metastasis and distant metastasis. The postoperative pathologic staging of each subject was determined according to the 7th edition of the Union for International Cancer Control tumor-node-metastasis (TNM) staging system for CRC. The resected tumor tissues and paired adjacent non-cancerous tissues (at least 5 cm away from the tumor margin) were immediately collected, frozen in liquid nitrogen and stored at -80 °C. This study was approved by the ethics committee of the Qilu Hospital of Shandong University and written informed consent was obtained from each patient or legal representative.
The patients were followed every 3 to 6 mo after the operation. The clinical end point of this study was death or the end of the study period (January 2013) with a median follow-up period of 63 mo (range: 10-77 mo). Overall survival (OS) was defined as the period from surgery to death. All data including physical examination, laboratory results and computed tomography findings were collected from hospital records or by patient interviews.
The human colon cancer cell line Caco-2 was kindly provided by Shuo Chen (Department of Gastroenterology, Qilu Hospital, Shandong University, China). Cells were cultured in Dulbecco’s modified Eagle’s medium (Hyclone, Logan, UT, United States), supplemented with 10% fetal bovine serum (Gibco, Carlsbad, CA, United States) and maintained at 37 °C with 5% CO2.
Total RNA was isolated from tissue or cells using TRIzol® reagent (Invitrogen, Carlsbad, CA, United States) following the manufacturer’s instructions and the concentration was determined using a NanoDrop 2000 (Thermo Fisher Scientific, Waltham, MA, United States). The cDNA was synthesized as follows: (1) 1 μg of isolated RNA was incubated at 65 °C for 5 min with 1 μL specific reverse transcription primer (RiboBio, Guangzhou, China), 1 μL U6 reverse transcription primer, and 1 μL dNTP in a 12 μL total reaction volume; (2) 4 μL of 5 × first-strand buffer, 2 μL of DTT, and 1 μL of RNase inhibitor were added and the reaction was incubated at 37 °C for 2 min; (3) 1 μL of MMLV reverse transcriptase was added and the final reaction was incubated at 37 °C for 50 min, followed by 70 °C for 5 min. For quantitative real-time polymerase chain reaction (qRT-PCR), the amplification protocol was carried out in the ABI PRISM 7500 Sequence Detection System (Applied Biosystems Inc., Waltham, MA, United States) as follows: initiation at 95 °C for 1 min, followed by 45 cycles of 95 °C for 15 s, 60 °C for 30 s, 72 °C for 45 s, followed by a dissociation protocol. The U6 small nuclear RNA was used as a reference gene to normalize RNA concentrations, and the relative expression of miR-133a in Caco-2 cells was used as a calibrator by the comparative threshold cycle (Ct) method (2-ΔΔCt). Triplicate quantification tests were performed and the average was calculated for each sample.
All statistical analyses were performed using SPSS 17.0 statistical software (IBM Corporation, Chicago, IL, United States). The median concentrations of miR-133a were compared among different groups using a Mann-Whitney U test or Kruskal-Wallis test. OS curves were calculated using the Kaplan-Meier method and the statistical differences between subgroups were compared by the log-rank test. The Cox proportional hazards regression model was employed for univariate and multivariate analyses to estimate the prognostic factors for survival prediction. Data are expressed as median and interquartile range. P < 0.05 was considered statistically significant.
To reveal the role of miR-133a in CRC, qRT-PCR was performed to measure miR-133a levels in 169 pairs of CRC tissues and adjacent non-cancerous tissues. Median miR-133a levels were significantly lower in CRC tissues compared with matched non-cancerous tissues [133.6 (33.73-508.2) vs 804.8 (298.64-1727.5), P < 0.05] (Figure 1A). In addition, the miR-133a expression was found to be decreased at least 2-fold compared with adjacent non-cancerous tissue in 55.6% (94/169) of cases (Figure 1B).
The relationship between miR-133a expression and clinicopathologic parameters was evaluated. As shown in Table 1, the level of miR-133a in CRC was strongly correlated with tumor differentiation, local invasion, lymph node metastasis and clinical TNM stage (P < 0.05 for all). However, there were no significant associations between miR-133a expression and other clinical features including sex, age, tumor location and tumor size. Taken together, these observations indicate that miR-133a expression is downregulated in CRC and is associated with the disease progression.
| Characteristic | Number of cases (n = 175) | miR-133a expressionmedian (IQR) | P value |
| Sex | 0.727 | ||
| Male | 96 | 127.40 (34.93-535.51) | |
| Female | 73 | 135.30 (31.90-435.10) | |
| Age (yr) | 0.800 | ||
| ≤ Median | 85 | 173.72 (39.43-603.81) | |
| > Median | 84 | 109.90 (27.18-311.64) | |
| Tumor location | 0.484 | ||
| Colon | 79 | 135.30 (27.31-468.70) | |
| Rectum | 90 | 132.35 (43.73-525.97) | |
| Differentiation | 0.004 | ||
| Well | 18 | 560.88 (98.18-1027.33) | |
| Moderate | 130 | 134.45 (35.88-434.56) | |
| Poor | 21 | 35.33 (26.05-145.77) | |
| Size | 0.527 | ||
| ≤ 5 cm | 101 | 129.59 (31.90-468.70) | |
| > 5 cm | 68 | 135.35 (35.87-524.93) | |
| Local invasion | < 0.001 | ||
| T1-T2 | 42 | 562.75 (299.65-999.73) | |
| T3-T4 | 127 | 88.17 (25.50-267.13) | |
| Lymph node metastasis | < 0.001 | ||
| No | 98 | 300.87 (72.88-740.23) | |
| Yes | 71 | 67.92 (21.22-200.36) | |
| TNM stage | < 0.001 | ||
| Stage I | 34 | 708.05 (362.41-1054.28) | |
| Stage II | 64 | 132.50 (34.53-421.89) | |
| Stage III | 71 | 67.92 (21.22-200.36) |
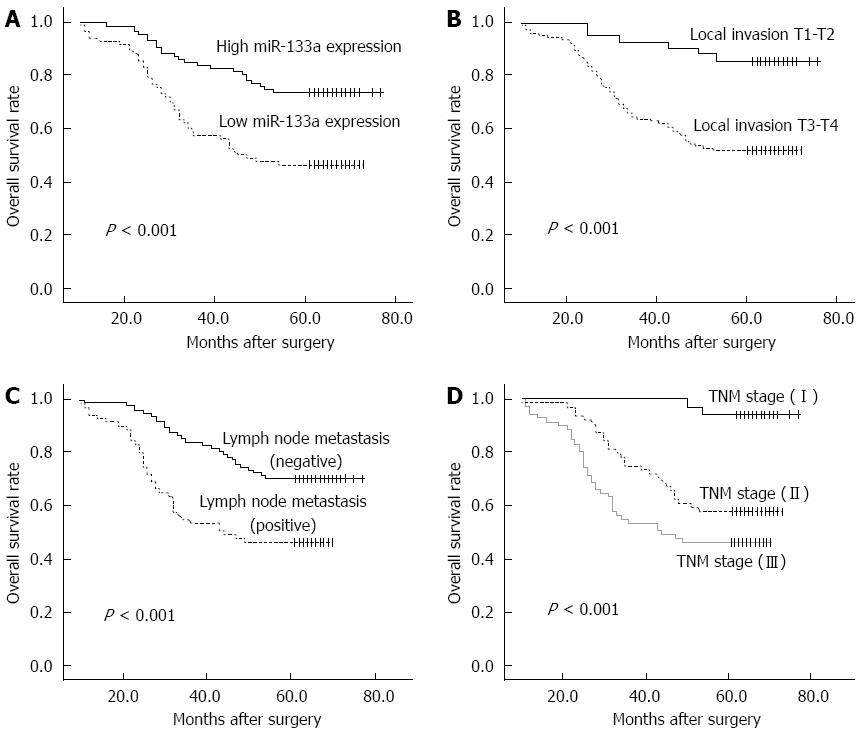
Of the 169 CRC patients, 67 died within the follow-up period, resulting in a cumulative 5-year OS rate of 60.4%. The prognostic value of miR-133a expression in CRC OS was evaluated between patients with low and high miR-133a expression levels. Patients with miR-133a expression levels below the median level (133.6) were assigned to the low expression group (median 33.73, n = 85), and those with values above the median were assigned to the high expression group (median 514.9, n = 84). Kaplan-Meier analysis showed that the cumulative 5-year OS rate was 46.3% in the low expression group, and 73.6% in the high expression group (Figure 2A). The log-rank test showed that the OS rate of patients with low miR-133a expression was significantly poorer than that of the remaining cases (P < 0.001). OS was also significantly associated with local invasion (Figure 2B; P < 0.001), regional lymph node metastasis (Figure 2C; P < 0.001) and TNM stage (Figure 2D; P < 0.001).
A Cox proportional hazards regression model analysis was performed to determine the independent prognostic indicators for patients with CRC. The results of univariate analyses revealed statistically significant correlations between OS and miR-133a level (HR = 0.385, 95%CI: 0.232-0.638, P < 0.001), tumor local invasion (HR = 4.328, 95%CI: 1.780-10.020, P < 0.05), lymph node metastasis (HR = 2.416, 95%CI: 1.488-3.923, P < 0.001) and TNM stage (HR = 2.336, 95%CI: 1.609-3.393, P < 0.001) (Table 2). A multivariate analysis of these factors showed that miR-133a level and TNM stage maintained their significance as independent prognostic factors for OS (HR = 0.590, 95%CI: 0.350-0.995, P < 0.05; and HR = 6.111, 95%CI: 1.029-36.278, P < 0.05, respectively).
| Variable | Univariate analysis | Multivariate analysis | |||||
| HR | 95%CI | P value | HR | 95%CI | P value | ||
| Age (yr) | 1.107 | 0.685-1.789 | 0.679 | ||||
| Sex (male female) | 1.154 | 0.714-1.863 | 0.559 | ||||
| Tumor location (rectum vs colon) | 1.006 | 0.623-1.625 | 0.981 | ||||
| Tumor size | 1.318 | 0.814-2.134 | 0.261 | ||||
| Differentiation | 0.788 | 0.490-1.268 | 0.326 | ||||
| Local invasion (T3-4 vs T1-2) | 4.328 | 1.780-10.020 | 0.001 | 1.196 | 0.424-3.370 | 0.735 | |
| Lymph node metastasis | 2.416 | 1.488-3.923 | < 0.001 | 0.238 | 0.033-1.717 | 0.154 | |
| TNM stage | 2.336 | 1.609-3.393 | < 0.001 | 6.111 | 1.029-36.278 | < 0.05 | |
| miR-133a expression (high vs low) | 0.385 | 0.232-0.638 | < 0.001 | 0.590 | 0.350-0.995 | < 0.05 | |
There is mounting evidence demonstrating the tissue-specificity and stability of miRNA expression patterns in various tumors, which has provided insights into the molecular mechanisms involved[14]. Although the precise mechanisms are unknown, many functional studies suggest that miRNA dysregulation is involved in the initiation and progression of cancer[15-17]. miR-21 and miR-92a, two highly investigated miRNAs that function as promoters for cell proliferation, invasion and metastasis, are significantly overexpressed in CRC[18]. Recently, the downregulation of other miRNAs, such as miR-126 and miR-218, has also been implicated in CRC[19,20]. Contradictory data concerning the role of miR-133a in the development and progression of CRC have emerged, with evidence that it can behave both as a promoter and a suppressor. To reconcile this discrepancy, the expression patterns of miR-133a in CRC tissues were examined, along with their association to CRC development.
The expression of miR-133 has been characterized in multiple species and found to play roles in the development of different types of malignant tumors. Results of this study indicated that miR-133a is significantly downregulated in CRC tissues compared with the adjacent normal tissues, which is consistent with previous studies showing a reduction in miR-133a in CRC patients compared to healthy controls. Furthermore, the levels of miR-133a were significantly lower in tumors with poor differentiation, greater depth of local invasion, positive lymph node metastasis and advanced disease. Hamara et al[21] reported that miR-133a is homologous to the 3’-UTRs of iron-related genes and consistently reduced in CRC patients in comparison to healthy colon mucosa. These findings suggest that the downregulation of miR-133a is related to the tumorigenesis and progression of CRC. The precise mechanism for this regulation is unclear, however, there is evidence to suggest that it involves the inhibition of the MAPK pathway[12].
To explore the potential prognostic value of miR-133a, the relationship between expression levels and OS in CRC patients was analyzed. Results indicate that patients with low expression had poorer survival compared to those with high expression of miR-133a. Furthermore, miR-133a and TNM stage could serve as independent prognostic indicators for the survival of CRC patients, indicating that the detection of miR-133a together with pathologic diagnosis in tumor tissues could be used to evaluate patient outcome and help design optimal individual treatment strategies.
A previous study reported that miR-133a in CRC patients may serve as a gene promoter for brain metastasis[13]. However, six of the CRC patients in the present study with liver metastasis had lower expression of miR-133a, and were not analyzed for the prognostic factor due to the limited number of cases. This discrepancy may indicate distinct organ-specific miR-133a functions, induced by different target genes. For example, miR-133a induces G2 arrest in renal cell carcinomas[7], and inhibits breast cancer cell growth and invasion by regulating fascin 1 expression, and thus its downregulation is associated with poor survival in breast cancer patients[6]. In esophageal squamous cell carcinoma, miR-133a was reported to regulate the expression of CD47 and function in the development of this cancer[22]. Other validated oncogenic targets of miR-133a include fascin homolog 1, glutathione-S-transferase pi 1 and transgelin 2 in bladder cancer[23-25], and actin-related protein 2/3 complex, subunit 5 in lung squamous cell carcinoma[5].
In conclusion, the results of this study confirm the clinical and prognostic significance of miR-133a in CRC. Although this study was limited by the retrospective design and the relatively small number of CRC patients, the data suggest that miR-133a plays a critical role in the development and progression of CRC and therefore may serve as a potential therapeutic target. Future studies evaluating the role of miR-133a in the metastasis of various cancers will help to further elucidate the mechanisms controlling miR-133a dysregulation and define the oncogenic targets.
Colorectal cancer (CRC) is one of the most common malignancies worldwide, with over 1.2 million new cases and an estimated 608700 deaths occurring in 2008 alone. Although recent advances have been achieved in comprehensive therapeutic strategies, CRC outcome remains poor. Biomarkers such as carcinoembryonic antigen have been recommended for CRC prognosis and postoperative surveillance, despite limited sensitivity. Therefore, the identification of novel and reliable biomarkers is urgently needed for CRC prognosis.
There is increasing evidence for microRNA (miRNA) dysregulation in tumor development, which can be utilized as valuable biomarkers. Previous data have indicated that the ectopic expression of miR-133a is associated with various human malignancies. However, one study showed that miR-133a repressed tumor growth and metastasis, while another study indicated that miR-133a serves as a gene promoter for brain metastasis. The present study confirms the repression of miR-133a in CRC tissues, and provides evidence that miR-133a expression can serve as a reliable prognostic indicator for the progression of CRC.
In the present study, miR-133a levels were measured in CRC tissues by quantitative real-time PCR, and the clinical significance of miR-133a expression was investigated. The data indicate that the expression of miR-133a was reduced in CRC tissues and was significantly associated with tumor differentiation, local invasion, regional lymph node metastasis and TNM stage. Moreover, CRC patients with low miR-133a expression had poorer overall survival than those with high miR-133a expression. Multivariate analysis suggested that miR-133a is an independent factor for CRC prognosis.
The confirmation of miR-133a dysregulation in CRC and its correlation with disease progression suggest that miR-133a is a potential novel target for therapeutic strategies. Moreover, results of this study may help to develop the use of miR-133a levels as a biomarker to independently predict clinical outcome.
MicroRNAs are a class of short non-coding ribonucleotides that have been shown to regulate gene expression. Multiple miRNAs have been implicated in the initiation and progression of tumors through dysregulation of oncogenes and tumor suppressor genes, including miR-133a, which may be involved in the development of different types of malignant tumors. The three known genes of miR-133 include miR-133a-1, miR-133a-2 and miR-133b, located on chromosomes 18, 20 and 6, respectively.
The authors present a study concerning the clinical implication of miR-133a expression in colorectal cancer. The results demonstrate that miR-133a is reduced in CRC tissues, and low expression is associated with poorer overall survival of CRC patients. These data indicate that miR-133a levels may serve as a useful prognostic biomarker for clinical assessment of patient outcome, and implicate miR-133a as a potential therapeutic target for CRC.
P- Reviewer: Haier J, Il Kim T, Seow-Choen F S- Editor: Ma YJ L- Editor: Wang TQ E- Editor: Wang CH
| 1. | Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D. Global cancer statistics. CA Cancer J Clin. 2010;61:69-90. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 23762] [Cited by in F6Publishing: 25182] [Article Influence: 1937.1] [Reference Citation Analysis (3)] |
| 2. | Jemal A, Siegel R, Xu J, Ward E. Cancer statistics, 2010. CA Cancer J Clin. 2011;60:277-300. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 10002] [Cited by in F6Publishing: 10353] [Article Influence: 739.5] [Reference Citation Analysis (0)] |
| 3. | Duffy MJ, Lamerz R, Haglund C, Nicolini A, Kalousová M, Holubec L, Sturgeon C. Tumor markers in colorectal cancer, gastric cancer and gastrointestinal stromal cancers: European group on tumor markers 2014 guidelines update. Int J Cancer. 2014;134:2513-2522. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 207] [Cited by in F6Publishing: 224] [Article Influence: 20.4] [Reference Citation Analysis (1)] |
| 4. | Ma Y, Zhang P, Yang J, Liu Z, Yang Z, Qin H. Candidate microRNA biomarkers in human colorectal cancer: systematic review profiling studies and experimental validation. Int J Cancer. 2012;130:2077-2087. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 96] [Cited by in F6Publishing: 107] [Article Influence: 8.2] [Reference Citation Analysis (0)] |
| 5. | Moriya Y, Nohata N, Kinoshita T, Mutallip M, Okamoto T, Yoshida S, Suzuki M, Yoshino I, Seki N. Tumor suppressive microRNA-133a regulates novel molecular networks in lung squamous cell carcinoma. J Hum Genet. 2012;57:38-45. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 92] [Cited by in F6Publishing: 96] [Article Influence: 7.4] [Reference Citation Analysis (0)] |
| 6. | Wu ZS, Wang CQ, Xiang R, Liu X, Ye S, Yang XQ, Zhang GH, Xu XC, Zhu T, Wu Q. Loss of miR-133a expression associated with poor survival of breast cancer and restoration of miR-133a expression inhibited breast cancer cell growth and invasion. BMC Cancer. 2012;12:51. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 75] [Cited by in F6Publishing: 77] [Article Influence: 6.4] [Reference Citation Analysis (0)] |
| 7. | Kawakami K, Enokida H, Chiyomaru T, Tatarano S, Yoshino H, Kagara I, Gotanda T, Tachiwada T, Nishiyama K, Nohata N. The functional significance of miR-1 and miR-133a in renal cell carcinoma. Eur J Cancer. 2012;48:827-836. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 101] [Cited by in F6Publishing: 115] [Article Influence: 9.6] [Reference Citation Analysis (0)] |
| 8. | Kojima S, Chiyomaru T, Kawakami K, Yoshino H, Enokida H, Nohata N, Fuse M, Ichikawa T, Naya Y, Nakagawa M. Tumour suppressors miR-1 and miR-133a target the oncogenic function of purine nucleoside phosphorylase (PNP) in prostate cancer. Br J Cancer. 2012;106:405-413. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 154] [Cited by in F6Publishing: 166] [Article Influence: 12.8] [Reference Citation Analysis (0)] |
| 9. | Song T, Xia W, Shao N, Zhang X, Wang C, Wu Y, Dong J, Cai W, Li H. Differential miRNA expression profiles in bladder urothelial carcinomas. Asian Pac J Cancer Prev. 2010;11:905-911. [PubMed] [Cited in This Article: ] |
| 10. | Chen WS, Leung CM, Pan HW, Hu LY, Li SC, Ho MR, Tsai KW. Silencing of miR-1-1 and miR-133a-2 cluster expression by DNA hypermethylation in colorectal cancer. Oncol Rep. 2012;28:1069-1076. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 57] [Cited by in F6Publishing: 59] [Article Influence: 4.9] [Reference Citation Analysis (0)] |
| 11. | Dong Y, Zhao J, Wu CW, Zhang L, Liu X, Kang W, Leung WW, Zhang N, Chan FK, Sung JJ. Tumor suppressor functions of miR-133a in colorectal cancer. Mol Cancer Res. 2013;11:1051-1060. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 73] [Cited by in F6Publishing: 89] [Article Influence: 8.1] [Reference Citation Analysis (0)] |
| 12. | Wang H, An H, Wang B, Liao Q, Li W, Jin X, Cui S, Zhang Y, Ding Y, Zhao L. miR-133a represses tumour growth and metastasis in colorectal cancer by targeting LIM and SH3 protein 1 and inhibiting the MAPK pathway. Eur J Cancer. 2013;49:3924-3935. [PubMed] [DOI] [Cited in This Article: ] [Cited by in F6Publishing: 1] [Reference Citation Analysis (0)] |
| 13. | Li Z, Gu X, Fang Y, Xiang J, Chen Z. microRNA expression profiles in human colorectal cancers with brain metastases. Oncol Lett. 2012;3:346-350. [PubMed] [Cited in This Article: ] |
| 14. | Nohata N, Hanazawa T, Enokida H, Seki N. microRNA-1/133a and microRNA-206/133b clusters: dysregulation and functional roles in human cancers. Oncotarget. 2012;3:9-21. [PubMed] [Cited in This Article: ] |
| 15. | Nelson KM, Weiss GJ. MicroRNAs and cancer: past, present, and potential future. Mol Cancer Ther. 2008;7:3655-3660. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 205] [Cited by in F6Publishing: 222] [Article Influence: 14.8] [Reference Citation Analysis (0)] |
| 16. | Krol J, Loedige I, Filipowicz W. The widespread regulation of microRNA biogenesis, function and decay. Nat Rev Genet. 2010;11:597-610. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 3165] [Cited by in F6Publishing: 3434] [Article Influence: 245.3] [Reference Citation Analysis (0)] |
| 17. | Davis-Dusenbery BN, Hata A. MicroRNA in Cancer: The Involvement of Aberrant MicroRNA Biogenesis Regulatory Pathways. Genes Cancer. 2010;1:1100-1114. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 120] [Cited by in F6Publishing: 132] [Article Influence: 11.0] [Reference Citation Analysis (0)] |
| 18. | Liu GH, Zhou ZG, Chen R, Wang MJ, Zhou B, Li Y, Sun XF. Serum miR-21 and miR-92a as biomarkers in the diagnosis and prognosis of colorectal cancer. Tumour Biol. 2013;34:2175-2181. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 143] [Cited by in F6Publishing: 158] [Article Influence: 14.4] [Reference Citation Analysis (0)] |
| 19. | Yu H, Gao G, Jiang L, Guo L, Lin M, Jiao X, Jia W, Huang J. Decreased expression of miR-218 is associated with poor prognosis in patients with colorectal cancer. Int J Clin Exp Pathol. 2013;6:2904-2911. [PubMed] [Cited in This Article: ] |
| 20. | Zhou Y, Feng X, Liu YL, Ye SC, Wang H, Tan WK, Tian T, Qiu YM, Luo HS. Down-regulation of miR-126 is associated with colorectal cancer cells proliferation, migration and invasion by targeting IRS-1 via the AKT and ERK1/2 signaling pathways. PLoS One. 2013;8:e81203. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 55] [Cited by in F6Publishing: 65] [Article Influence: 5.9] [Reference Citation Analysis (0)] |
| 21. | Hamara K, Bielecka-Kowalska A, Przybylowska-Sygut K, Sygut A, Dziki A, Szemraj J. Alterations in expression profile of iron-related genes in colorectal cancer. Mol Biol Rep. 2013;40:5573-5585. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 13] [Cited by in F6Publishing: 14] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 22. | Suzuki S, Yokobori T, Tanaka N, Sakai M, Sano A, Inose T, Sohda M, Nakajima M, Miyazaki T, Kato H. CD47 expression regulated by the miR-133a tumor suppressor is a novel prognostic marker in esophageal squamous cell carcinoma. Oncol Rep. 2012;28:465-472. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 71] [Cited by in F6Publishing: 76] [Article Influence: 6.3] [Reference Citation Analysis (0)] |
| 23. | Chiyomaru T, Enokida H, Tatarano S, Kawahara K, Uchida Y, Nishiyama K, Fujimura L, Kikkawa N, Seki N, Nakagawa M. miR-145 and miR-133a function as tumour suppressors and directly regulate FSCN1 expression in bladder cancer. Br J Cancer. 2010;102:883-891. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 256] [Cited by in F6Publishing: 279] [Article Influence: 19.9] [Reference Citation Analysis (0)] |
| 24. | Uchida Y, Chiyomaru T, Enokida H, Kawakami K, Tatarano S, Kawahara K, Nishiyama K, Seki N, Nakagawa M. MiR-133a induces apoptosis through direct regulation of GSTP1 in bladder cancer cell lines. Urol Oncol. 2013;31:115-123. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 56] [Cited by in F6Publishing: 73] [Article Influence: 5.6] [Reference Citation Analysis (0)] |
| 25. | Yoshino H, Chiyomaru T, Enokida H, Kawakami K, Tatarano S, Nishiyama K, Nohata N, Seki N, Nakagawa M. The tumour-suppressive function of miR-1 and miR-133a targeting TAGLN2 in bladder cancer. Br J Cancer. 2011;104:808-818. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 204] [Cited by in F6Publishing: 223] [Article Influence: 17.2] [Reference Citation Analysis (0)] |