Published online Aug 7, 2014. doi: 10.3748/wjg.v20.i29.9912
Revised: February 13, 2014
Accepted: April 27, 2014
Published online: August 7, 2014
The gram-negative bacterium Helicobacter pylori (H. pylori) causes chronic gastritis, gastric and duodenal ulcers, gastric cancer and mucosa-associated lymphoid tissue lymphoma. Treatment is recommended in all symptomatic patients. The current treatment options for H. pylori infection are outlined in this review in light of the recent challenges in eradication success, largely due to the rapid emergence of antibiotic resistant strains of H. pylori. Antibiotic resistance is a constantly evolving process and numerous studies have shown that the prevalence of H. pylori antibiotic resistance varies significantly from country to country, and even between regions within the same country. In addition, recent data has shown that previous antibiotic use is associated with harbouring antibiotic resistant H. pylori. Local surveillance of antibiotic resistance is warranted to guide clinicians in their choice of therapy. Antimicrobial resistance is assessed by H. pylori culture and antimicrobial susceptibility testing. Recently developed molecular tests offer an attractive alternative to culture and allow for the rapid molecular genetic identification of H. pylori and resistance-associated mutations directly from biopsy samples or bacterial culture material. Accumulating evidence indicates that surveillance of antimicrobial resistance by susceptibility testing is feasible and necessary to inform clinicians in their choice of therapy for management of H. pylori infection.
Core tip: There has been a significant decrease in the success rate of empirical triple therapy to treat Helicobacter pylori (H. pylori) infection, largely due to a rapid increase in the prevalence of antibiotic resistant strains. Antibiotic resistance is a constantly evolving process and there are significant regional variations in H. pylori antibiotic resistance rates. As such, local surveillance of antibiotic resistance is warranted to guide clinicians in their therapeutic choice. Standard culture-based antimicrobial susceptibility testing and molecular methods provide key opportunities to tailor H. pylori treatment based on the detection of antibiotic resistant strains, thereby enhancing eradication rates and decreasing H. pylori-associated disease.
-
Citation: Smith SM, O’Morain C, McNamara D. Antimicrobial susceptibility testing for
Helicobacter pylori in times of increasing antibiotic resistance. World J Gastroenterol 2014; 20(29): 9912-9921 - URL: https://www.wjgnet.com/1007-9327/full/v20/i29/9912.htm
- DOI: https://dx.doi.org/10.3748/wjg.v20.i29.9912
Helicobacter pylori (H. pylori) specifically colonizes the gastric epithelium and is the most common human bacterial infection worldwide, infecting approximately half of the world’s population[1-3]. Infection is usually acquired in early childhood and persists for decades if left untreated[1,2]. Prevalence of H. pylori infection varies globally but increases with older age and with lower socioeconomic status[4,5]. The higher prevalence in older age groups likely reflects poorer childhood living conditions in previous decades[2]. Although most infected patients will not develop any clinically significant complications, H. pylori infection confers a 1%-10% risk of developing gastric or duodenal ulcers, a 0.1%-3% risk of developing gastric adenocarcinoma, and < 0.01% of developing mucosa-associated lymphoid tissue (MALT) lymphoma[2]. H. pylori has been designated a Class I carcinogen by the World Health Organization[6]. Disease risk in infected individuals varies greatly in different populations and is associated with host genotype and strain-specific bacterial factors.
Patients with uncomplicated dyspepsia are managed using a ‘‘test-and-treat’’ strategy. H. pylori infection is diagnosed using non-invasive methods, including the urea breath test (UBT), serologic tests and the stool antigen test[2,3]. The UBT involves ingesting 13C-labelled urea. If present, the H. pylori enzyme urease converts the 13C-labelled urea to labelled carbon dioxide, which is detected in a breath sample. The UBT is easy to perform and highly accurate with a specificity and sensitivity of 95%[7,8]. The stool antigen test detects H. pylori antigens in stool samples, with laboratory-based stool antigen tests displaying specificity and sensitivity of at least 95%[9-11]. Serologic tests detect IgG antibodies to H. pylori. There is significant variability in the accuracy of serology test kits for H. pylori[12], but validated commercial kits with an accuracy of over 90% are available[13].
Endoscopy is warranted for dyspeptic patients with accompanying alarm symptoms such as weight loss, persistent vomiting, gastrointestinal bleeding, abdominal mass or iron-deficient anaemia[2,3]. In addition, endoscopy is recommended for patients with new onset dyspepsia above the age of 45 (European guidelines[3,14]) or 55 (United States guidelines[15]). H. pylori infection can be detected in endoscopic gastric biopsy samples by several methods. The rapid urease-test for Campylobacter-like organisms (CLO) involves placing the biopsy specimen in a solution of urea and pH-sensitive dye. The pH change resulting from the H. pylori urease-mediated conversion of urea to ammonia results in a colour change indicative of infection[2]. The CLO test has a sensitivity of greater than 90% and a specificity of more than 95%[7]. Histology on the biopsy specimen by means of staining with hematoxylin and eosin or Giemsa will also detect the presence of H. pylori infection and the degree of inflammation[2,3]. Immunohistochemistry-based tests with improved sensitivity have also been developed for the visualisation of H. pylori in biopsy samples[16-18], but they are not yet used routinely. Culturing of the bacteria from biopsy samples is also possible and has the added advantage of allowing for antimicrobial susceptibility testing. Acid suppressing proton pump inhibitors (PPIs) should be avoided 2 wk prior to diagnostic testing by the UBT, stool antigen test or endoscopy as they increase the gastric pH, leading to a decrease in bacterial load, and may result in a false negative H. pylori diagnosis[3,19]. In addition, antimicrobials should be avoided for 4 wk prior to testing (UBT, stool antigen test or endoscopy) as these agents also supress infection and reduce test sensitivity[2]. Serological tests that detect antibodies to H. pylori are the only methods that are not affected by decreased gastric bacterial load, which may lead to false negative results using the other diagnostic testing methods outlined above[3].
Treatment for H. pylori is recommended in all symptomatic individuals. Eradication of H. pylori infection provides a long-term cure for both duodenal and gastric ulcers in the majority of patients whose ulcers are not associated with non-steroidal anti-inflammatory drug use[20,21]. In addition, evidence suggests that H. pylori eradication reduces the development of atrophic gastritis and the risk of cancer progression in infected individuals without premalignant gastric lesions[22-29]. Furthermore, eradication of infection leads to regression of most localized gastric MALT lymphomas[30-33].
The standard empirical triple therapy for H. pylori proposed at the first Maastricht conference on the management of H. pylori infection[34] has become widely used throughout the world[15,35,36]. This first-line therapy consists of a PPI with the antibiotics clarithromycin and amoxicillin taken twice daily for 7-14 d[3]. Metronidazole is used instead of amoxicillin in patients with a penicillin allergy[2]. The success rate of first-line treatment has fallen below the recommended 80% in recent years[3,5]. Non-compliance[37] and the emergence of antibiotic resistant strains of H. pylori[38-42] are considered to be the major factors contributing to treatment failure. Standard empirical triple therapy is now only recommended in regions where clarithromycin resistance is known to be less than 15%-20%[3]. While no new treatment has been developed as an alternative to the standard triple therapy, recent studies have described the advantages of using different combinations of known antibiotics or extended treatment durations. In regions where clarithromycin resistance is greater than 15%-20%, bismuth quadruple therapy consisting of a PPI, a bismuth salt, tetracycline and metronidazole is recommended[3]. As bismuth salts are not available in every country, non-bismuth quadruple therapies, namely sequential therapy (5 d PPI and amoxicillin; 5 d PPI with clarithromycin and metronidazole) or concomitant therapy (PPI with amoxicillin, metronidazole and clarithromycin) may be prescribed as alternative therapies[3]. Following treatment, eradication of H. pylori should be confirmed by the UBT, stool antigen test or by endoscopy if required. As antibodies persist for months following infection, serology testing is not recommended for eradication confirmation[2]. Following failure of initial therapy, a PPI with amoxicillin and levofloxacin is recommended. As non-compliance may lead to treatment failure, adherence should be strongly emphasised for subsequent therapies. If third-line treatment is required, a number of studies support quinolone or rifabutin-based regimens[3,43,44] but treatment should only be prescribed following antimicrobial susceptibility testing where possible[3].
H. pylori culture and antimicrobial susceptibility testing is carried out in an effort to predict antibiotic treatment outcome and guide clinicians in their choice of therapy. Culture and antimicrobial susceptibility testing guidelines have been outlined by the European Helicobacter Study Group (EHSG)[42]. As use of PPIs or antimicrobials inhibits the growth of H. pylori and reduces the chances of successful culture, patients should avoid taking PPIs for at least 2 wk and antimicrobials for 4 wk prior to endoscopy[3,19]. Biopsy specimens should be transported and processed for culture as soon as possible, ideally within 6 h[45]. If processing is delayed refrigeration is recommended[45,46]. Biopsy specimens are used to inoculate Columbia blood agar plates containing 10% laked horse blood and incubated under microaerophilic conditions at 37 °C for 7-10 d[19], although colonies are usually visible at 3-5 d[45]. Antimicrobial supplements may be added to media to inhibit overgrowth with contaminating bacteria and fungi[19,47]. The presence of H. pylori should be confirmed by the Gram stain, and positive oxidase, urease and catalase tests. Fresh cultures (48-72 h growth) at an inoculum concentration of McFarland 3[19] should be used for H. pylori culture-based antimicrobial susceptibility testing[45]. Culture medium manufactured specifically for antimicrobial susceptibility testing (e.g., Mueller Hinton agar; Oxoid, Basingstoke, United Kingdom)[19] should be used and the depth of the agar should be kept consistent across tests.
Several assays are commercially available to test the antimicrobials commonly used to treat H. pylori. The disc diffusion method (Oxoid) involves placing an antibiotic-coated disc directly onto the agar plate inoculated with H. pylori and determining the zone of bacterial growth inhibition. This cost-effective method is widely used for antimicrobial susceptibility testing for a variety of microorganisms, but disc diffusion criteria for antimicrobial susceptibility testing for H. pylori have not to date been defined by the European Committee on Antimicrobial Susceptibility Testing (EUCAST)[48]. Both EUCAST and the British Society for Antimicrobial Chemotherapy (http://bsac.org.uk) recommend Etest strips for H. pylori culture-based antimicrobial susceptibility testing. Etests (Biomerieux; Basingstoke, United Kingdom) are plastic strips calibrated with a predefined concentration gradient of antibiotic and enable the quantitative determination of the minimum inhibitory concentration (MIC) of an antimicrobial agent required to inhibit bacterial growth. The MIC can be read directly from the scale printed on the strip at the point where the edge of the inhibition ellipse of bacterial culture intersects with the strip. The EUCAST breakpoints for antimicrobial resistance to H. pylori are listed in Table 1. Etest MIC results may be validated subsequently using agar dilution approaches. When performing antimicrobial susceptibility testing, quality control tests involving reference strains with known susceptibility or resistance should be included.
While H. pylori culture allows an evaluation of antibiotic resistance irrespective of the intrinsic mechanism involved, H. pylori is a fastidious bacterium and culture is time-consuming and often difficult with sensitivity values of culture from gastric biopsies as low as 55%-73%[49-52]. Molecular testing for H. pylori offers an attractive alternative to culture and allows for molecular genetic identification of H. pylori directly from biopsy samples in addition to culture material. As such, it provides the opportunity for rapid analysis, enabling same-day diagnosis. Molecular testing has been recommended to detect H. pylori and both clarithromycin and quinolone resistance when standard culture and susceptibility testing are not possible[3].
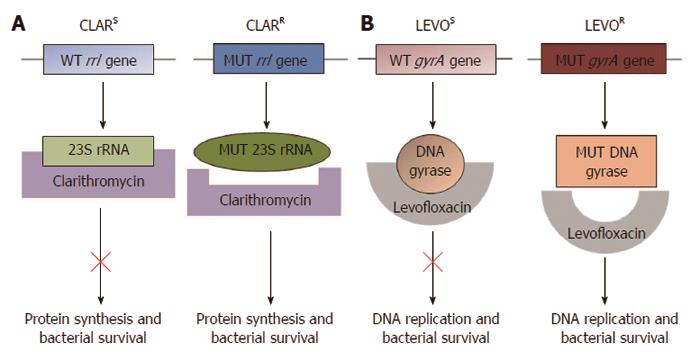
The genetic mutations conferring resistance to clarithromycin and quinolones have been well characterised[53]. Clarithromycin functions by binding and suppressing the activity of the bacterial ribosomal subunit, thus inhibiting protein synthesis[4] (Figure 1). Single point mutations within the H. pylori rrl gene encoding the 23S ribosomal RNA component of ribosomes result in clarithromycin resistance. The three most frequently occurring mutations, A2146C, A2146G and A2147G (Genbank Accession number NC_000915; formerly described as A2142C, A2142G and A2143G), are thought to account for 90% of primary clarithromycin resistance in Western countries[4,53-57]. These mutations can be detected by a number of polymerase chain reaction (PCR)-based molecular methods using both bacterial culture samples[54,58,59] and gastric biopsies[55,59-61]. In addition, these assays have been used to analyse stool samples[62-65], providing an opportunity for diagnosis of H. pylori infection and antimicrobial susceptibility testing through non-invasive procedures. Several molecular testing kits are commercially available for the detection of clarithromycin resistance, including the MutaREAL H. pylori kit[59] (Immunodiagnostik; Benshiem, Germany), the ClariRes real-time PCR assay[63] (Ingentix; Vienna Austria) and the Seeplex ClaR-H. pylori ACE detection system[61] (Seegene; Eschborn, Germany). Accumulating evidence has demonstrated that the presence of mutations detected by molecular tests correlates well with culture-based susceptibility testing[54,60,61].
The most significant mutations conferring quinolone resistance lie at positions 87 and 91 of the H. pylori gyrA gene, which encodes the A subunit of the DNA gyrase enzyme involved in regulating the topological state of bacterial DNA during replication (Figure 1)[53,57,66,67]. PCR-based molecular assays have successfully detected quinolone resistant strains of H. pylori[39,66,67]. The mechanism of resistance to metronidazole is less clear. Mutations in the rdxA and frxA genes have been implicated; however, a definitive panel of point mutations accounting for metronidazole resistance has not yet been described[4]. Although mutations in the DNA encoding the 16S rRNA involved in tetracycline resistance, the rpoB gene involved in rifabutin resistance and the pbp-1a gene involved in amoxicillin resistance have also been described[4,46,68-70], they are of less interest since the rates of H. pylori resistance to these antibiotics are low in most regions[19,42,70,71].
The recently developed GenoType HelicoDR assay (Hain Lifescience; Nehren, Germany) enables determination of resistance to clarithromycin and quinolones through a protocol involving DNA extraction from either bacterial cultures or biopsy material, multiplex PCR amplification of H. pylori DNA sequences and hybridization against probes that detect specific H. pylori wild-type or mutated elements. The GenoType HelicoDR assay is highly accurate with a sensitivity and specificity of 94%-100% and 86%-99% respectively for clarithromycin resistance and 83%-87% and 95%-98.5% respectively for quinolone resistance[57,72]. Studies defining the sensitivity and specificity of molecular tests for detecting H. pylori clarithromycin and levofloxacin resistance are summarized in Table 2.
| Molecular test | Antibiotic | Sensitivity | Specificity | Ref. |
| PCR | Clarithromycin | 82% | 100% | [64] |
| Genotype HelicoDR assay | Clarithromycin | 94% | 99% | [57] |
| Levofloxacin | 87% | 98.5% | ||
| PCR | Clarithromycin | 90.6% | 95.8% | [59] |
| PCR | Clarithromycin | 89.2% | 100% | [62] |
| Genotype HelicoDR assay | Clarithromycin | 100% | 86.2% | [72] |
| Levofloxacin | 82.6% | 95.1% | ||
| PCR | Clarithromycin | 83.3% | 100% | [65] |
Several issues are to be considered when choosing a molecular assay for H. pylori antimicrobial susceptibility testing, including cost, local expertise in molecular diagnosis, the equipment available and the sensitivity and specificity of the test[53]. Molecular-based techniques are highly accurate in detecting minimal traces of antibiotic resistant H. pylori strains, even in small tissue samples. Moreover these tools are accurate in detecting the co-existence of H. pylori strains susceptible and resistant to the same antibiotic within the same patient sample, known as hetero-resistance[4,55,73]. This is potentially important given that in one recent study molecular-based tests indicated that in relation to genetic clarithromycin resistance, H. pylori infection was cured less frequently in patients with pure resistant strains (46%) than those infected with hetero-resistant strains (78.5%) or susceptible strains (94.5%)[55].
It must be kept in mind that the majority of molecular tests available do not detect resistance based on uncommon genetic mechanisms. Additional rare mutations within the rrl and gyrA gene regions conferring clarithromycin and quinolone resistance respectively have been described[59,66]. In addition, resistances of H. pylori strains that lie outside the rrl and gyrA genes, as well as other possible mechanisms of clarithromycin and quinolone resistance must be considered. Further studies are required to determine if molecular tests predict H. pylori treatment failure more accurately than phenotypic culture and sensitivity tests.
Most patients are prescribed initial H. pylori eradication treatment without culture and antibiotic susceptibility testing as current guidance recommend a ‘‘test-and-treat’’ strategy based on non-invasive diagnostic methods[3,15,35]. As the most recent Maastricht IV consensus guidelines recommend that clarithromycin should not be used to treat H. pylori if resistance rates are above 15%-20%[3], surveillance of primary antibiotic resistance is warranted to guide clinicians in their choice of therapy. The EHSG have recently reported on the prevalence of primary H. pylori antibiotic resistance from 2008-2009 using a multicentre approach with standardised protocols[42]. The number of test centres involved in the study was proportional to population of each country, with 2204 patients included from 32 centres in 18 European countries. The overall primary resistance rates for clarithromycin, levofloxacin and metronidazole were 17.5%, 14.1% and 34.9% respectively, with a prevalence ≤ 1% for tetracycline, rifampicin and amoxicillin[42]. Combined resistance to metronidazole and clarithromycin was found in 7.8% of strains. The rate of clarithromycin resistance had almost doubled since the previous European survey[74] (Table 3), which has important implications considering that clarithromycin resistance decreases the efficacy of clarithromycin-amoxicillin-PPI triple therapy by up to 70%[75-77]. Prevalence of levofloxacin resistance was not tested in the previous European study[74] as levofloxacin-based treatment was introduced later, but several studies have shown an emergence in levofloxacin resistance in the last decade[42,67,78]. Metronidazole resistance was high at 34.9%[42] but the rate was similar to that of the previous Europe-wide study[74] (Table 3). The impact of metronidazole resistance on H. pylori eradication is less than that of clarithromycin resistance, and can be overcome by increasing the dose and duration of treatment or by prescription of bismuth-containing quadruple therapy that includes metronidazole[38,79,80]. The latest EHSG study on antibiotic resistance also indicated variations across European countries; the resistance rate for clarithromycin was < 10% in Northern European countries, while most countries in the rest of Europe (except Spain and Germany) had a resistance rate of > 15% (Table 4)[42]. Such variations in antibiotic resistance have also been reported on a regional basis. For example, a recent study within the United Kingdom indicated that the resistance rates to clarithromycin, metronidazole and quinolones were 18%, 43%, 13% respectively at a test centre in Wales but the rates were lower in an English test centre at 3%, 22%, 1%[19]. Recent findings from Irish patient cohorts indicate that the rates of clarithromycin, metronidazole and levofloxacin were 9.3%, 29.1%[81] and 11.7% respectively[82].
| Ref. | Region | Antibiotic resistance rates | ||
| Clar | Met | Levo | ||
| Tveit et al[94] | Alaska | 30%2 | 42%2 | 19%2 |
| Gao et al[71] | China; Beijing | 37.2%2 | 63.9%2 | 50.3%2 |
| Su et al[83] | China; South East coastal region | 21.5%2 | 95.4%2 | 20.6%2 |
| McNulty et al[19] | England | 3%2 | 22%2 | 1%2 |
| Megraud et al[42] | Europe; Northern countries | 7.7%1 | 28.6%1 | 7.7%1 |
| Megraud et al[42] | Europe; Southern countries | 21.5%1 | 29.7%1 | 13.1%1 |
| Megraud et al[42] | Europe; Western and central countries | 18.7%1 | 43.8%1 | 18.6%1 |
| Abadi et al[95] | Iran | 45.2%2 | 65.5%2 | 34.5%2 |
| O’Connor et al[81] | Ireland | 9.3%1 | 29.1%1 | ND |
| O’Connor et al[82] | 13.2%2 | 31.5%2 | 11.7%2 | |
| Yamade et al[84] | Japan | 38.8%1 | ND | 34%1 |
| 55.6%2 | ND | 38.6%2 | ||
| Lee et al[85] | Korea | 23.7%1 | ND | 28.1%1 |
| Seck et al[96] | Senegal | 1%1 | 85%1 | 15%1 |
| Vilachione et al[70] | Thailand | 3.7%1 | 36%1 | 7.2%1 |
| McNulty et al[19] | Wales | 18%2 | 43%2 | 13%2 |
Variations in resistance rates have also been reported outside Europe (Table 4)[4,5]. In Thailand for example, the resistance rates for clarithromycin, metronidazole and levofloxacin are 3.7%, 36% and 7.2% respectively, with significantly higher rates of metronidazole resistance in Southern Thailand than North Eastern Thailand (66.7% vs 33.3%)[70]. In the South East coastal region of China resistance rates for clarithromycin, metronidazole and levofloxacin are 21.5%, 95.4% and 20.6%[83] respectively, while in Beijing the rates are 37.2%, 63.9% and 50.3% for clarithromycin, metronidazole and levofloxacin respectively[71]. A recent study from Japan indicates that the prevalence of clarithromycin resistance is 55.6% and levofloxacin resistance is 38.6%[84], while in South Korea, an increase in the resistance rates for clarithromycin (17.2%-23.7%) and quinolones (4.7%-28.1%) was reported from 2003 to 2012[85].
Emergence of antibiotic resistance is thought to be associated with prior antibiotic use. Indeed, by analysing cumulative and yearly outpatient antibiotic consumption the pan-European study demonstrated a significant association between the use of long acting macrolides and resistance of H. pylori to clarithromycin, and between previous quinolone use and levofloxacin resistance[42]. Using an alternative approach to analyse data from GP and patient records, the United Kingdom study demonstrated that each previous course of clarithromycin, metronidazole or quinolone taken by an individual was associated with an increase in the risk harbouring an antibiotic resistant strain of H. pylori[19].
Consensus guidelines recommend abandoning clarithromycin in empirical triple therapy when the prevalence of clarithromycin resistance is higher than 15%-20%[3]. This level has now been reached in most countries in Western/Central and Southern Europe[42] as well as many countries in Asia[71,83-85]. Information on resistance rates is not widely available to clinicians, especially at a local level. As antibiotic resistance is a constantly evolving process and there is significant variation in resistance rates between countries and within different regions of the same country[19,70,71,83], it is important that local surveillance of primary antibiotic resistance is performed regularly and that anti-H. pylori treatment regimens should be chosen according to local resistance data. Increasing resistance rates for the second-line antibiotic levofloxacin have been reported in several countries[42,71,83,84]. Culture and antimicrobial susceptibility testing should be considered in all regions before second-line treatment is prescribed if endoscopy is performed[3]. Furthermore, antimicrobial susceptibility testing is recommended in all regions when a second-line treatment has failed[3]. If standard culture and susceptibility testing is not possible, molecular tests can be used to detect antibiotic resistance. The recent studies by McNulty et al[19] and the EHSG[42] indicate that on-going surveillance of antimicrobial resistance is indeed feasible and necessary to inform clinicians in the management of H. pylori infection. Alternatively, as previous antibiotic use is linked to an increased risk of antibiotic resistance[19,42], knowledge of previous antibiotic consumption may provide a tool to predict the susceptibility of H. pylori to antimicrobial agents and adapt treatment strategies where culture and susceptibility testing or molecular testing are not available.
Evidence from numerous studies provides a rationale for tailoring treatment based on antimicrobial susceptibility testing to improve eradication rates for primary and subsequent anti-H. pylori treatment regimens. A recent meta-analysis by Wenzhen et al[86] of five randomised control trials totalling 701 patients has shown that tailored treatments based on resistance data show better eradication rates than standard empirical triple therapy. Tailored triple therapy based on culture and sensitivity testing was also found to be more cost effective than standard triple therapy for first line treatment in a study by Romano et al[87]. However, others argue against the cost-effectiveness of culture-based treatment for first line therapy[88,89]. The economic benefits of tailoring first line therapy are likely to depend on the local antibiotic resistance levels as a recent study by Cosme et al[90] reported that performing culture and antimicrobial susceptibility testing lead to higher eradication rates and increased cost efficiency in an area where clarithromycin resistance was high (> 15%-20%).
With regard to tailoring rescue regimens for anti-H. pylori treatment, Fiorini et al[91] demonstrated that culture-based therapy eradicated H. pylori infection in 90% of patients who had not previously responded to treatment. Liou et al[92] showed increased efficacy of sequential therapy guided by PCR-based molecular tests in the third-line treatment of refractory H. pylori infection. Furthermore, newly developed methods allow the detection of H. pylori and antibiotic resistance in stool samples[62-65], providing a potential opportunity to assess the prevalence of antimicrobial resistance by non-invasive methods without the expense of endoscopy.
It is clear that the emerging rates of antimicrobial resistance represent a significant challenge in the successful management of H. pylori infection and that resistance surveillance is warranted. The recent evidence highlights the importance of antimicrobial susceptibility testing in both the on-going assessment of primary antibiotic resistance rates and in tailoring treatments to increase H. pylori eradication success. In addition to standard culture-based antimicrobial susceptibility testing, molecular methods provide key opportunities in detecting antibiotic resistant strains of H. pylori, enhancing eradication rates and decreasing H. pylori-associated disease.
P- Reviewer: Borgmann S, Kelesidis T, Mastroianni CM, Wang JT S- Editor: Gou SX L- Editor: A E- Editor: Zhang DN
| 1. | Polk DB, Peek RM. Helicobacter pylori: gastric cancer and beyond. Nat Rev Cancer. 2010;10:403-414. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 718] [Cited by in F6Publishing: 773] [Article Influence: 55.2] [Reference Citation Analysis (0)] |
| 2. | McColl KE. Clinical practice. Helicobacter pylori infection. N Engl J Med. 2010;362:1597-1604. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 529] [Cited by in F6Publishing: 517] [Article Influence: 36.9] [Reference Citation Analysis (0)] |
| 3. | Malfertheiner P, Megraud F, O’Morain CA, Atherton J, Axon AT, Bazzoli F, Gensini GF, Gisbert JP, Graham DY, Rokkas T. Management of Helicobacter pylori infection--the Maastricht IV/ Florence Consensus Report. Gut. 2012;61:646-664. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1541] [Cited by in F6Publishing: 1494] [Article Influence: 124.5] [Reference Citation Analysis (3)] |
| 4. | Ierardi E, Giorgio F, Losurdo G, Di Leo A, Principi M. How antibiotic resistances could change Helicobacter pylori treatment: A matter of geography? World J Gastroenterol. 2013;19:8168-8180. [PubMed] [DOI] [Cited in This Article: ] [Cited by in CrossRef: 70] [Cited by in F6Publishing: 78] [Article Influence: 7.1] [Reference Citation Analysis (0)] |
| 5. | Iwańczak F, Iwańczak B. Treatment of Helicobacter pylori infection in the aspect of increasing antibiotic resistance. Adv Clin Exp Med. 2012;21:671-680. [PubMed] [Cited in This Article: ] |
| 6. | IARC Working Group on the Evaluation of Carcinogenic Risks to Humans. Infection with Helicobacter pylori. IARC Monogr Eval Carcinog Risks Hum. 1994;61:177-240. [Cited in This Article: ] |
| 7. | Vaira D, Vakil N. Blood, urine, stool, breath, money, and Helicobacter pylori. Gut. 2001;48:287-289. [PubMed] [Cited in This Article: ] |
| 8. | Gisbert JP, Pajares JM. Review article: 13C-urea breath test in the diagnosis of Helicobacter pylori infection -- a critical review. Aliment Pharmacol Ther. 2004;20:1001-1017. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 237] [Cited by in F6Publishing: 242] [Article Influence: 12.1] [Reference Citation Analysis (0)] |
| 9. | Gisbert JP, Pajares JM. Stool antigen test for the diagnosis of Helicobacter pylori infection: a systematic review. Helicobacter. 2004;9:347-368. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 119] [Cited by in F6Publishing: 125] [Article Influence: 6.3] [Reference Citation Analysis (0)] |
| 10. | Gisbert JP, de la Morena F, Abraira V. Accuracy of monoclonal stool antigen test for the diagnosis of H. pylori infection: a systematic review and meta-analysis. Am J Gastroenterol. 2006;101:1921-1930. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 177] [Cited by in F6Publishing: 174] [Article Influence: 9.7] [Reference Citation Analysis (0)] |
| 11. | Deguchi R, Matsushima M, Suzuki T, Mine T, Fukuda R, Nishina M, Ozawa H, Takagi A. Comparison of a monoclonal with a polyclonal antibody-based enzyme immunoassay stool test in diagnosing Helicobacter pylori infection after eradication therapy. J Gastroenterol. 2009;44:713-716. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 22] [Cited by in F6Publishing: 23] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 12. | Loy CT, Irwig LM, Katelaris PH, Talley NJ. Do commercial serological kits for Helicobacter pylori infection differ in accuracy? A meta-analysis. Am J Gastroenterol. 1996;91:1138-1144. [PubMed] [Cited in This Article: ] |
| 13. | Feldman RA, Deeks JJ, Evans SJ. Multi-laboratory comparison of eight commercially available Helicobacter pylori serology kits. Helicobacter pylori Serology Study Group. Eur J Clin Microbiol Infect Dis. 1995;14:428-433. [PubMed] [Cited in This Article: ] |
| 14. | Malfertheiner P, Megraud F, O’Morain C, Bazzoli F, El-Omar E, Graham D, Hunt R, Rokkas T, Vakil N, Kuipers EJ. Current concepts in the management of Helicobacter pylori infection: the Maastricht III Consensus Report. Gut. 2007;56:772-781. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1348] [Cited by in F6Publishing: 1286] [Article Influence: 75.6] [Reference Citation Analysis (0)] |
| 15. | Chey WD, Wong BC. American College of Gastroenterology guideline on the management of Helicobacter pylori infection. Am J Gastroenterol. 2007;102:1808-1825. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 819] [Cited by in F6Publishing: 792] [Article Influence: 46.6] [Reference Citation Analysis (1)] |
| 16. | Jonkers D, Stobberingh E, de Bruine A, Arends JW, Stockbrügger R. Evaluation of immunohistochemistry for the detection of Helicobacter pylori in gastric mucosal biopsies. J Infect. 1997;35:149-154. [PubMed] [Cited in This Article: ] |
| 17. | Marzio L, Angelucci D, Grossi L, Diodoro MG, Di Campli E, Cellini L. Anti-Helicobacter pylori specific antibody immunohistochemistry improves the diagnostic accuracy of Helicobacter pylori in biopsy specimen from patients treated with triple therapy. Am J Gastroenterol. 1998;93:223-226. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 36] [Cited by in F6Publishing: 36] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 18. | Sepulveda AR, Patil M. Practical approach to the pathologic diagnosis of gastritis. Arch Pathol Lab Med. 2008;132:1586-1593. [PubMed] [Cited in This Article: ] |
| 19. | McNulty CA, Lasseter G, Shaw I, Nichols T, D’Arcy S, Lawson AJ, Glocker E. Is Helicobacter pylori antibiotic resistance surveillance needed and how can it be delivered? Aliment Pharmacol Ther. 2012;35:1221-1230. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 53] [Cited by in F6Publishing: 45] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 20. | Hentschel E, Brandstätter G, Dragosics B, Hirschl AM, Nemec H, Schütze K, Taufer M, Wurzer H. Effect of ranitidine and amoxicillin plus metronidazole on the eradication of Helicobacter pylori and the recurrence of duodenal ulcer. N Engl J Med. 1993;328:308-312. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 523] [Cited by in F6Publishing: 524] [Article Influence: 16.9] [Reference Citation Analysis (0)] |
| 21. | Axon AT, O’Moráin CA, Bardhan KD, Crowe JP, Beattie AD, Thompson RP, Smith PM, Hollanders FD, Baron JH, Lynch DA. Randomised double blind controlled study of recurrence of gastric ulcer after treatment for eradication of Helicobacter pylori infection. BMJ. 1997;314:565-568. [PubMed] [Cited in This Article: ] |
| 22. | Leung WK, Lin SR, Ching JY, To KF, Ng EK, Chan FK, Lau JY, Sung JJ. Factors predicting progression of gastric intestinal metaplasia: results of a randomised trial on Helicobacter pylori eradication. Gut. 2004;53:1244-1249. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 277] [Cited by in F6Publishing: 304] [Article Influence: 15.2] [Reference Citation Analysis (0)] |
| 23. | Wong BC, Lam SK, Wong WM, Chen JS, Zheng TT, Feng RE, Lai KC, Hu WH, Yuen ST, Leung SY. Helicobacter pylori eradication to prevent gastric cancer in a high-risk region of China: a randomized controlled trial. JAMA. 2004;291:187-194. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1015] [Cited by in F6Publishing: 1107] [Article Influence: 55.4] [Reference Citation Analysis (0)] |
| 24. | Malfertheiner P, Sipponen P, Naumann M, Moayyedi P, Mégraud F, Xiao SD, Sugano K, Nyrén O. Helicobacter pylori eradication has the potential to prevent gastric cancer: a state-of-the-art critique. Am J Gastroenterol. 2005;100:2100-2115. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 143] [Cited by in F6Publishing: 146] [Article Influence: 7.7] [Reference Citation Analysis (0)] |
| 25. | Correa P, Fontham ET, Bravo JC, Bravo LE, Ruiz B, Zarama G, Realpe JL, Malcom GT, Li D, Johnson WD. Chemoprevention of gastric dysplasia: randomized trial of antioxidant supplements and anti-helicobacter pylori therapy. J Natl Cancer Inst. 2000;92:1881-1888. [PubMed] [Cited in This Article: ] |
| 26. | Mera R, Fontham ET, Bravo LE, Bravo JC, Piazuelo MB, Camargo MC, Correa P. Long term follow up of patients treated for Helicobacter pylori infection. Gut. 2005;54:1536-1540. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 239] [Cited by in F6Publishing: 259] [Article Influence: 13.6] [Reference Citation Analysis (0)] |
| 27. | You WC, Brown LM, Zhang L, Li JY, Jin ML, Chang YS, Ma JL, Pan KF, Liu WD, Hu Y. Randomized double-blind factorial trial of three treatments to reduce the prevalence of precancerous gastric lesions. J Natl Cancer Inst. 2006;98:974-983. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 310] [Cited by in F6Publishing: 300] [Article Influence: 16.7] [Reference Citation Analysis (0)] |
| 28. | Take S, Mizuno M, Ishiki K, Nagahara Y, Yoshida T, Yokota K, Oguma K, Okada H, Shiratori Y. The effect of eradicating helicobacter pylori on the development of gastric cancer in patients with peptic ulcer disease. Am J Gastroenterol. 2005;100:1037-1042. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 149] [Cited by in F6Publishing: 159] [Article Influence: 8.4] [Reference Citation Analysis (0)] |
| 29. | Fuccio L, Zagari RM, Eusebi LH, Laterza L, Cennamo V, Ceroni L, Grilli D, Bazzoli F. Meta-analysis: can Helicobacter pylori eradication treatment reduce the risk for gastric cancer? Ann Intern Med. 2009;151:121-128. [PubMed] [Cited in This Article: ] |
| 30. | Fischbach W, Goebeler-Kolve ME, Dragosics B, Greiner A, Stolte M. Long term outcome of patients with gastric marginal zone B cell lymphoma of mucosa associated lymphoid tissue (MALT) following exclusive Helicobacter pylori eradication therapy: experience from a large prospective series. Gut. 2004;53:34-37. [PubMed] [Cited in This Article: ] |
| 31. | Wotherspoon AC, Doglioni C, Diss TC, Pan L, Moschini A, de Boni M, Isaacson PG. Regression of primary low-grade B-cell gastric lymphoma of mucosa-associated lymphoid tissue type after eradication of Helicobacter pylori. Lancet. 1993;342:575-577. [PubMed] [Cited in This Article: ] |
| 32. | Chen LT, Lin JT, Tai JJ, Chen GH, Yeh HZ, Yang SS, Wang HP, Kuo SH, Sheu BS, Jan CM. Long-term results of anti-Helicobacter pylori therapy in early-stage gastric high-grade transformed MALT lymphoma. J Natl Cancer Inst. 2005;97:1345-1353. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 140] [Cited by in F6Publishing: 154] [Article Influence: 8.1] [Reference Citation Analysis (0)] |
| 33. | Wündisch T, Thiede C, Morgner A, Dempfle A, Günther A, Liu H, Ye H, Du MQ, Kim TD, Bayerdörffer E. Long-term follow-up of gastric MALT lymphoma after Helicobacter pylori eradication. J Clin Oncol. 2005;23:8018-8024. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 217] [Cited by in F6Publishing: 245] [Article Influence: 12.9] [Reference Citation Analysis (0)] |
| 34. | Malfertheiner P, Mégraud F, O’Morain C, Bell D, Bianchi Porro G, Deltenre M, Forman D, Gasbarrini G, Jaup B, Misiewicz JJ. Current European concepts in the management of Helicobacter pylori infection--the Maastricht Consensus Report. The European Helicobacter Pylori Study Group (EHPSG). Eur J Gastroenterol Hepatol. 1997;9:1-2. [PubMed] [Cited in This Article: ] |
| 35. | Fock KM, Katelaris P, Sugano K, Ang TL, Hunt R, Talley NJ, Lam SK, Xiao SD, Tan HJ, Wu CY. Second Asia-Pacific Consensus Guidelines for Helicobacter pylori infection. J Gastroenterol Hepatol. 2009;24:1587-1600. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 384] [Cited by in F6Publishing: 405] [Article Influence: 27.0] [Reference Citation Analysis (1)] |
| 36. | O’Connor A, Molina-Infante J, Gisbert JP, O’Morain C. Treatment of Helicobacter pylori infection 2013. Helicobacter. 2013;18 Suppl 1:58-65. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 41] [Cited by in F6Publishing: 41] [Article Influence: 3.7] [Reference Citation Analysis (0)] |
| 37. | O’Connor JP, Taneike I, O’Morain C. Improving compliance with helicobacter pylori eradication therapy: when and how? Therap Adv Gastroenterol. 2009;2:273-279. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 43] [Cited by in F6Publishing: 47] [Article Influence: 3.6] [Reference Citation Analysis (0)] |
| 38. | Graham DY, Fischbach L. Helicobacter pylori treatment in the era of increasing antibiotic resistance. Gut. 2010;59:1143-1153. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 652] [Cited by in F6Publishing: 694] [Article Influence: 49.6] [Reference Citation Analysis (0)] |
| 39. | Wueppenhorst N, Stueger HP, Kist M, Glocker E. Identification and molecular characterization of triple- and quadruple-resistant Helicobacter pylori clinical isolates in Germany. J Antimicrob Chemother. 2009;63:648-653. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 51] [Cited by in F6Publishing: 59] [Article Influence: 3.9] [Reference Citation Analysis (1)] |
| 40. | Wüppenhorst N, Lenze F, Ross M, Kist M. Isolation and eradication of a clinical isolate of Helicobacter pylori resistant to five antimicrobials in Germany. J Antimicrob Chemother. 2011;66:222-223. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 9] [Cited by in F6Publishing: 9] [Article Influence: 0.6] [Reference Citation Analysis (1)] |
| 41. | Buzás GM. First-line eradication of Helicobacter pylori: are the standard triple therapies obsolete? A different perspective. World J Gastroenterol. 2010;16:3865-3870. [PubMed] [DOI] [Cited in This Article: ] [Cited by in CrossRef: 15] [Cited by in F6Publishing: 17] [Article Influence: 1.2] [Reference Citation Analysis (1)] |
| 42. | Megraud F, Coenen S, Versporten A, Kist M, Lopez-Brea M, Hirschl AM, Andersen LP, Goossens H, Glupczynski Y. Helicobacter pylori resistance to antibiotics in Europe and its relationship to antibiotic consumption. Gut. 2013;62:34-42. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 588] [Cited by in F6Publishing: 607] [Article Influence: 55.2] [Reference Citation Analysis (2)] |
| 43. | Toracchio S, Capodicasa S, Soraja DB, Cellini L, Marzio L. Rifabutin based triple therapy for eradication of H. pylori primary and secondary resistant to tinidazole and clarithromycin. Dig Liver Dis. 2005;37:33-38. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 41] [Cited by in F6Publishing: 40] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 44. | Gisbert JP, Bermejo F, Castro-Fernández M, Pérez-Aisa A, Fernández-Bermejo M, Tomas A, Barrio J, Bory F, Almela P, Sánchez-Pobre P. Second-line rescue therapy with levofloxacin after H. pylori treatment failure: a Spanish multicenter study of 300 patients. Am J Gastroenterol. 2008;103:71-76. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 66] [Cited by in F6Publishing: 69] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 45. | Health Protection Agency UK. Investigation of gastric biopsies for Helicobacter pylori. Health Protection Agency Standards Unit, Standards and Evaluation laboratory 2012. Available from: http://www.hpa.org.uk/webc/hpawebfile/hpaweb_c/1317132860197. [Cited in This Article: ] |
| 46. | Mégraud F, Lehours P. Helicobacter pylori detection and antimicrobial susceptibility testing. Clin Microbiol Rev. 2007;20:280-322. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 476] [Cited by in F6Publishing: 459] [Article Influence: 27.0] [Reference Citation Analysis (0)] |
| 47. | Dent JC, McNulty CA. Evaluation of a new selective medium for Campylobacter pylori. Eur J Clin Microbiol Infect Dis. 1988;7:555-558. [PubMed] [Cited in This Article: ] |
| 48. | EUCAST. Breakpoint tables for interpretation of MICs and zone diameters. 2013; Version 3.1. Available from: http://www.eucast.org/fileadmin/src/media/PDFs/EUCAST_files/Consultation/EUCAST_clinical_breakpoints_for_Helicobacter_pylori.pdf. [Cited in This Article: ] |
| 49. | Savarino V, Zentilin P, Pivari M, Bisso G, Raffaella Mele M, Bilardi C, Borro P, Dulbecco P, Tessieri L, Mansi C. The impact of antibiotic resistance on the efficacy of three 7-day regimens against Helicobacter pylori. Aliment Pharmacol Ther. 2000;14:893-900. [PubMed] [Cited in This Article: ] |
| 50. | Pilotto A, Rassu M, Leandro G, Franceschi M, Di Mario F. Prevalence of Helicobacter pylori resistance to antibiotics in Northeast Italy: a multicentre study. GISU. Interdisciplinary Group for the Study of Ulcer. Dig Liver Dis. 2000;32:763-768. [PubMed] [Cited in This Article: ] |
| 51. | Schwartz H, Krause R, Sahba B, Haber M, Weissfeld A, Rose P, Siepman N, Freston J. Triple versus dual therapy for eradicating Helicobacter pylori and preventing ulcer recurrence: a randomized, double-blind, multicenter study of lansoprazole, clarithromycin, and/or amoxicillin in different dosing regimens. Am J Gastroenterol. 1998;93:584-590. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 48] [Cited by in F6Publishing: 53] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 52. | Chisholm SA, Owen RJ. Application of polymerase chain reaction-based assays for rapid identification and antibiotic resistance screening of Helicobacter pylori in gastric biopsies. Diagn Microbiol Infect Dis. 2008;61:67-71. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 20] [Cited by in F6Publishing: 22] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 53. | Lehours P, Mégraud F. Helicobacter pylori molecular diagnosis. Expert Rev Mol Diagn. 2011;11:351-355. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 18] [Cited by in F6Publishing: 19] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 54. | Elviss NC, Lawson AJ, Owen RJ. Application of 3’-mismatched reverse primer PCR compared with real-time PCR and PCR-RFLP for the rapid detection of 23S rDNA mutations associated with clarithromycin resistance in Helicobacter pylori. Int J Antimicrob Agents. 2004;23:349-355. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 15] [Cited by in F6Publishing: 14] [Article Influence: 0.7] [Reference Citation Analysis (0)] |
| 55. | De Francesco V, Zullo A, Ierardi E, Giorgio F, Perna F, Hassan C, Morini S, Panella C, Vaira D. Phenotypic and genotypic Helicobacter pylori clarithromycin resistance and therapeutic outcome: benefits and limits. J Antimicrob Chemother. 2010;65:327-332. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 70] [Cited by in F6Publishing: 77] [Article Influence: 5.1] [Reference Citation Analysis (0)] |
| 56. | Sakinc T, Baars B, Wüppenhorst N, Kist M, Huebner J, Opferkuch W. Influence of a 23S ribosomal RNA mutation in Helicobacter pylori strains on the in vitro synergistic effect of clarithromycin and amoxicillin. BMC Res Notes. 2012;5:603. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 12] [Cited by in F6Publishing: 15] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 57. | Cambau E, Allerheiligen V, Coulon C, Corbel C, Lascols C, Deforges L, Soussy CJ, Delchier JC, Megraud F. Evaluation of a new test, genotype HelicoDR, for molecular detection of antibiotic resistance in Helicobacter pylori. J Clin Microbiol. 2009;47:3600-3607. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 105] [Cited by in F6Publishing: 119] [Article Influence: 7.9] [Reference Citation Analysis (0)] |
| 58. | Gibson JR, Saunders NA, Burke B, Owen RJ. Novel method for rapid determination of clarithromycin sensitivity in Helicobacter pylori. J Clin Microbiol. 1999;37:3746-3748. [PubMed] [Cited in This Article: ] |
| 59. | Agudo S, Alarcón T, Urruzuno P, Martínez MJ, López-Brea M. Detection of Helicobacter pylori and clarithromycin resistance in gastric biopsies of pediatric patients by using a commercially available real-time polymerase chain reaction after NucliSens semiautomated DNA extraction. Diagn Microbiol Infect Dis. 2010;67:213-219. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 23] [Cited by in F6Publishing: 24] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 60. | Chisholm SA, Owen RJ, Teare EL, Saverymuttu S. PCR-based diagnosis of Helicobacter pylori infection and real-time determination of clarithromycin resistance directly from human gastric biopsy samples. J Clin Microbiol. 2001;39:1217-1220. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 95] [Cited by in F6Publishing: 103] [Article Influence: 4.5] [Reference Citation Analysis (0)] |
| 61. | Lehours P, Siffré E, Mégraud F. DPO multiplex PCR as an alternative to culture and susceptibility testing to detect Helicobacter pylori and its resistance to clarithromycin. BMC Gastroenterol. 2011;11:112. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 51] [Cited by in F6Publishing: 57] [Article Influence: 4.4] [Reference Citation Analysis (0)] |
| 62. | Vécsei A, Innerhofer A, Binder C, Gizci H, Hammer K, Bruckdorfer A, Riedl S, Gadner H, Hirschl AM, Makristathis A. Stool polymerase chain reaction for Helicobacter pylori detection and clarithromycin susceptibility testing in children. Clin Gastroenterol Hepatol. 2010;8:309-312. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 40] [Cited by in F6Publishing: 39] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 63. | Lottspeich C, Schwarzer A, Panthel K, Koletzko S, Rüssmann H. Evaluation of the novel Helicobacter pylori ClariRes real-time PCR assay for detection and clarithromycin susceptibility testing of H. pylori in stool specimens from symptomatic children. J Clin Microbiol. 2007;45:1718-1722. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 55] [Cited by in F6Publishing: 55] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 64. | Schabereiter-Gurtner C, Hirschl AM, Dragosics B, Hufnagl P, Puz S, Kovách Z, Rotter M, Makristathis A. Novel real-time PCR assay for detection of Helicobacter pylori infection and simultaneous clarithromycin susceptibility testing of stool and biopsy specimens. J Clin Microbiol. 2004;42:4512-4518. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 137] [Cited by in F6Publishing: 146] [Article Influence: 7.3] [Reference Citation Analysis (0)] |
| 65. | Scaletsky IC, Aranda KR, Garcia GT, Gonçalves ME, Cardoso SR, Iriya K, Silva NP. Application of real-time PCR stool assay for Helicobacter pylori detection and clarithromycin susceptibility testing in Brazilian children. Helicobacter. 2011;16:311-315. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 38] [Cited by in F6Publishing: 35] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 66. | Wang LH, Cheng H, Hu FL, Li J. Distribution of gyrA mutations in fluoroquinolone-resistant Helicobacter pylori strains. World J Gastroenterol. 2010;16:2272-2277. [PubMed] [Cited in This Article: ] |
| 67. | Glocker E, Stueger HP, Kist M. Quinolone resistance in Helicobacter pylori isolates in Germany. Antimicrob Agents Chemother. 2007;51:346-349. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 65] [Cited by in F6Publishing: 69] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 68. | Glocker E, Berning M, Gerrits MM, Kusters JG, Kist M. Real-time PCR screening for 16S rRNA mutations associated with resistance to tetracycline in Helicobacter pylori. Antimicrob Agents Chemother. 2005;49:3166-3170. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 36] [Cited by in F6Publishing: 40] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 69. | Lawson AJ, Elviss NC, Owen RJ. Real-time PCR detection and frequency of 16S rDNA mutations associated with resistance and reduced susceptibility to tetracycline in Helicobacter pylori from England and Wales. J Antimicrob Chemother. 2005;56:282-286. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 36] [Cited by in F6Publishing: 39] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 70. | Vilaichone RK, Gumnarai P, Ratanachu-Ek T, Mahachai V. Nationwide survey of Helicobacter pylori antibiotic resistance in Thailand. Diagn Microbiol Infect Dis. 2013;77:346-349. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 30] [Cited by in F6Publishing: 28] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 71. | Gao W, Cheng H, Hu F, Li J, Wang L, Yang G, Xu L, Zheng X. The evolution of Helicobacter pylori antibiotics resistance over 10 years in Beijing, China. Helicobacter. 2010;15:460-466. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 101] [Cited by in F6Publishing: 100] [Article Influence: 7.1] [Reference Citation Analysis (0)] |
| 72. | Miendje Deyi VY, Burette A, Bentatou Z, Maaroufi Y, Bontems P, Lepage P, Reynders M. Practical use of GenoType® HelicoDR, a molecular test for Helicobacter pylori detection and susceptibility testing. Diagn Microbiol Infect Dis. 2011;70:557-560. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 29] [Cited by in F6Publishing: 35] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 73. | Zullo A, Hassan C, Cristofari F, Andriani A, De Francesco V, Ierardi E, Tomao S, Stolte M, Morini S, Vaira D. Effects of Helicobacter pylori eradication on early stage gastric mucosa-associated lymphoid tissue lymphoma. Clin Gastroenterol Hepatol. 2010;8:105-110. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 194] [Cited by in F6Publishing: 181] [Article Influence: 12.9] [Reference Citation Analysis (0)] |
| 74. | Glupczynski Y, Mégraud F, Lopez-Brea M, Andersen LP. European multicentre survey of in vitro antimicrobial resistance in Helicobacter pylori. Eur J Clin Microbiol Infect Dis. 2001;20:820-823. [PubMed] [Cited in This Article: ] |
| 75. | Fischbach LA, van Zanten S, Dickason J. Meta-analysis: the efficacy, adverse events, and adherence related to first-line anti-Helicobacter pylori quadruple therapies. Aliment Pharmacol Ther. 2004;20:1071-1082. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 140] [Cited by in F6Publishing: 141] [Article Influence: 7.1] [Reference Citation Analysis (0)] |
| 76. | Fischbach L, Evans EL. Meta-analysis: the effect of antibiotic resistance status on the efficacy of triple and quadruple first-line therapies for Helicobacter pylori. Aliment Pharmacol Ther. 2007;26:343-357. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 246] [Cited by in F6Publishing: 264] [Article Influence: 15.5] [Reference Citation Analysis (0)] |
| 77. | Dore MP, Leandro G, Realdi G, Sepulveda AR, Graham DY. Effect of pretreatment antibiotic resistance to metronidazole and clarithromycin on outcome of Helicobacter pylori therapy: a meta-analytical approach. Dig Dis Sci. 2000;45:68-76. [PubMed] [Cited in This Article: ] |
| 78. | Oleastro M, Cabral J, Ramalho PM, Lemos PS, Paixão E, Benoliel J, Santos A, Lopes AI. Primary antibiotic resistance of Helicobacter pylori strains isolated from Portuguese children: a prospective multicentre study over a 10 year period. J Antimicrob Chemother. 2011;66:2308-2311. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 38] [Cited by in F6Publishing: 44] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 79. | Malfertheiner P, Bazzoli F, Delchier JC, Celiñski K, Giguère M, Rivière M, Mégraud F. Helicobacter pylori eradication with a capsule containing bismuth subcitrate potassium, metronidazole, and tetracycline given with omeprazole versus clarithromycin-based triple therapy: a randomised, open-label, non-inferiority, phase 3 trial. Lancet. 2011;377:905-913. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 350] [Cited by in F6Publishing: 346] [Article Influence: 26.6] [Reference Citation Analysis (0)] |
| 80. | Bardhan K, Bayerdörffer E, Veldhuyzen Van Zanten SJ, Lind T, Mégraud F, Delchier JC, Hellblom M, Stubberöd A, Burman CF, Gromark P. The HOMER Study: the effect of increasing the dose of metronidazole when given with omeprazole and amoxicillin to cure Helicobacter pylori infection. Helicobacter. 2000;5:196-201. [PubMed] [Cited in This Article: ] |
| 81. | O’Connor A, Taneike I, Nami A, Fitzgerald N, Murphy P, Ryan B, O’connor H, Qasim A, Breslin N, O’moráin C. Helicobacter pylori resistance to metronidazole and clarithromycin in Ireland. Eur J Gastroenterol Hepatol. 2010;22:1123-1127. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 25] [Cited by in F6Publishing: 28] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 82. | O’Connor A, Taneike I, Nami A, Fitzgerald N, Ryan B, Breslin N, O’Connor H, McNamara D, Murphy P, O’Morain C. Helicobacter pylori resistance rates for levofloxacin, tetracycline and rifabutin among Irish isolates at a reference centre. Ir J Med Sci. 2013;182:693-695. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 17] [Cited by in F6Publishing: 24] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 83. | Su P, Li Y, Li H, Zhang J, Lin L, Wang Q, Guo F, Ji Z, Mao J, Tang W. Antibiotic resistance of Helicobacter pylori isolated in the Southeast Coastal Region of China. Helicobacter. 2013;18:274-279. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 106] [Cited by in F6Publishing: 108] [Article Influence: 9.8] [Reference Citation Analysis (1)] |
| 84. | Yamade M, Sugimoto M, Uotani T, Nishino M, Kodaira C, Furuta T. Resistance of Helicobacter pylori to quinolones and clarithromycin assessed by genetic testing in Japan. J Gastroenterol Hepatol. 2011;26:1457-1461. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 11] [Cited by in F6Publishing: 15] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 85. | Lee JW, Kim N, Kim JM, Nam RH, Chang H, Kim JY, Shin CM, Park YS, Lee DH, Jung HC. Prevalence of primary and secondary antimicrobial resistance of Helicobacter pylori in Korea from 2003 through 2012. Helicobacter. 2013;18:206-214. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 181] [Cited by in F6Publishing: 163] [Article Influence: 14.8] [Reference Citation Analysis (0)] |
| 86. | Wenzhen Y, Yumin L, Quanlin G, Kehu Y, Lei J, Donghai W, Lijuan Y. Is antimicrobial susceptibility testing necessary before first-line treatment for Helicobacter pylori infection? Meta-analysis of randomized controlled trials. Intern Med. 2010;49:1103-1109. [PubMed] [Cited in This Article: ] |
| 87. | Romano M, Marmo R, Cuomo A, De Simone T, Mucherino C, Iovene MR, Montella F, Tufano MA, Del Vecchio Blanco C, Nardone G. Pretreatment antimicrobial susceptibility testing is cost saving in the eradication of Helicobacter pylori. Clin Gastroenterol Hepatol. 2003;1:273-278. [PubMed] [Cited in This Article: ] |
| 88. | Qasim A, Sebastian S, Buckley M, O’Connor H, O’Morain C. Pretreatment antimicrobial susceptibility testing is not cost saving in the standard eradication of Helicobacter pylori. Clin Gastroenterol Hepatol. 2004;2:85; discussion 85. [PubMed] [Cited in This Article: ] |
| 89. | Faber J, Bar-Meir M, Rudensky B, Schlesinger Y, Rachman E, Benenson S, Sirota G, Stankiewic H, Halle D, Wilschanski M. Treatment regimens for Helicobacter pylori infection in children: is in vitro susceptibility testing helpful? J Pediatr Gastroenterol Nutr. 2005;40:571-574. [PubMed] [Cited in This Article: ] |
| 90. | Cosme A, Montes M, Martos M, Gil I, Mendarte U, Salicio Y, Piñeiro L, Recasens MT, Ibarra B, Sarasqueta C. Usefulness of antimicrobial susceptibility in the eradication of Helicobacter pylori. Clin Microbiol Infect. 2013;19:379-383. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 43] [Cited by in F6Publishing: 42] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 91. | Fiorini G, Vakil N, Zullo A, Saracino IM, Castelli V, Ricci C, Zaccaro C, Gatta L, Vaira D. Culture-based selection therapy for patients who did not respond to previous treatment for Helicobacter pylori infection. Clin Gastroenterol Hepatol. 2013;11:507-510. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 43] [Cited by in F6Publishing: 46] [Article Influence: 4.2] [Reference Citation Analysis (0)] |
| 92. | Liou JM, Chen CC, Chang CY, Chen MJ, Fang YJ, Lee JY, Chen CC, Hsu SJ, Hsu YC, Tseng CH. Efficacy of genotypic resistance-guided sequential therapy in the third-line treatment of refractory Helicobacter pylori infection: a multicentre clinical trial. J Antimicrob Chemother. 2013;68:450-456. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 56] [Cited by in F6Publishing: 65] [Article Influence: 5.4] [Reference Citation Analysis (0)] |
| 93. | Results of a multicentre European survey in 1991 of metronidazole resistance in Helicobacter pylori. European Study Group on Antibiotic Susceptibility of Helicobacter pylori. Eur J Clin Microbiol Infect Dis. 1992;11:777-781. [PubMed] [Cited in This Article: ] |
| 94. | Tveit AH, Bruce MG, Bruden DL, Morris J, Reasonover A, Hurlburt DA, Hennessy TW, McMahon B. Alaska sentinel surveillance study of Helicobacter pylori isolates from Alaska Native persons from 2000 to 2008. J Clin Microbiol. 2011;49:3638-3643. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 30] [Cited by in F6Publishing: 34] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 95. | Abadi AT, Taghvaei T, Mobarez AM, Carpenter BM, Merrell DS. Frequency of antibiotic resistance in Helicobacter pylori strains isolated from the northern population of Iran. J Microbiol. 2011;49:987-993. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 30] [Cited by in F6Publishing: 41] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 96. | Seck A, Burucoa C, Dia D, Mbengue M, Onambele M, Raymond J, Breurec S. Primary antibiotic resistance and associated mechanisms in Helicobacter pylori isolates from Senegalese patients. Ann Clin Microbiol Antimicrob. 2013;12:3. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 39] [Cited by in F6Publishing: 41] [Article Influence: 3.7] [Reference Citation Analysis (0)] |