Copyright
©2014 Baishideng Publishing Group Inc.
World J Gastroenterol. Jul 7, 2014; 20(25): 7993-8004
Published online Jul 7, 2014. doi: 10.3748/wjg.v20.i25.7993
Published online Jul 7, 2014. doi: 10.3748/wjg.v20.i25.7993
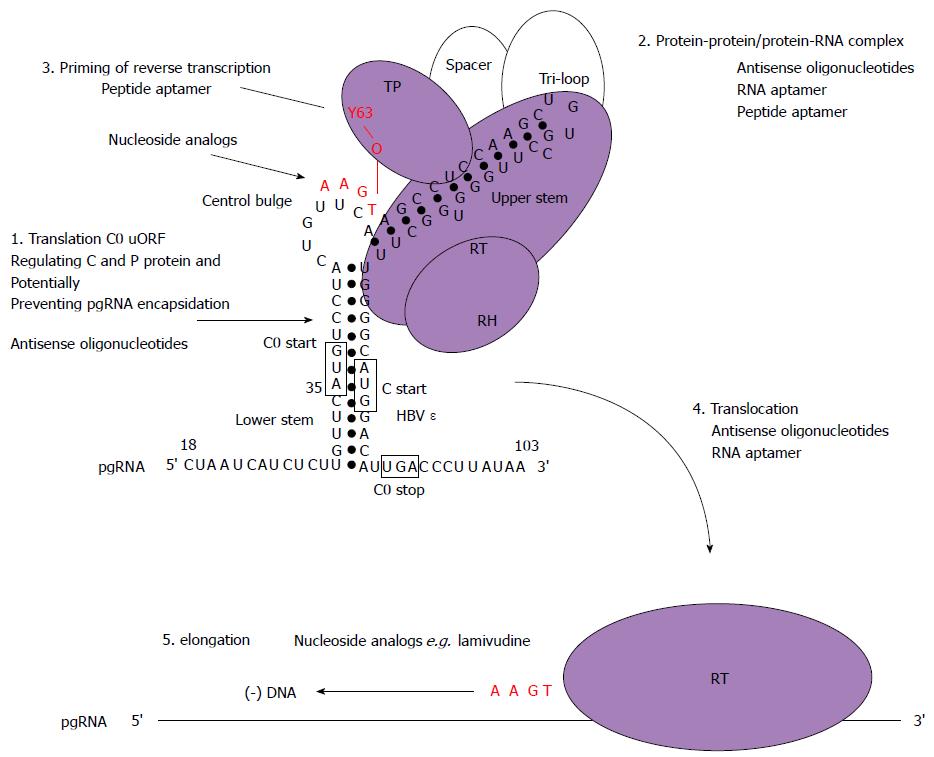
Figure 5 Schematic model for the initiation of reverse transcription in hepatitis B virus with stages targetable by existing drugs, as well as potential targets for antiviral compounds.
These may serve as a basis to design new strategies combining drugs with different mechanisms of action. Domains of the P protein are abbreviated as follows: terminal protein (TP), RNase H domain (RH), reverse transcriptase domain (RT). Open circles represent cellular chaperones. The initiation of reverse transcription to generate a dsDNA genome involves the pgRNA which is also the template for C and P protein translation. In this model, the C0 ORF which spans the ε structure is also translated (albeit weakly) and could potentially discriminate the selection of pgRNA required for encapsidation and translation (step 1). Secondly, the hepatitis B virus (HBV) P protein would then interact with cellular chaperones (heat shock proteins), the ε structure and gain enzymatic activity (step 2). Next, the reverse transcription step is primed by the formation of covalent bond between conserved tyrosine of the TP domain and the first nt of the minus-strand DNA followed by additions of three more nucleotides, GAA in a template dependent manner (step 3). Following an arrest in the viral DNA synthesis, a strand transfer occurs and the complex of P protein and short DNA primer is translocated to the 3′ end of the template DR1 (step 4). Then, elongation of the minus-strand DNA continues (step 5). Currently, the triphosphate forms of nucleoside analogs have shown inhibitory effects on the priming reaction (targeting step 3) or interfere with viral minus-strand DNA elongation (targeting step 5).
- Citation: Chen A, T-Thienprasert NP, Brown CM. Prospects for inhibiting the post-transcriptional regulation of gene expression in hepatitis B virus. World J Gastroenterol 2014; 20(25): 7993-8004
- URL: https://www.wjgnet.com/1007-9327/full/v20/i25/7993.htm
- DOI: https://dx.doi.org/10.3748/wjg.v20.i25.7993