Published online Mar 28, 2014. doi: 10.3748/wjg.v20.i12.3208
Revised: December 20, 2013
Accepted: February 20, 2014
Published online: March 28, 2014
Processing time: 180 Days and 16.2 Hours
Inflammatory bowel disease (IBD), which includes Crohn’s disease (CD) and ulcerative colitis (UC), represents a group of chronic inflammatory disorders caused by dysregulated immune responses in genetically predisposed individuals. Genetic markers are associated with disease phenotype and long-term evolution, but their value in everyday clinical practice is limited at the moment. IBD has a clear immunological background and interleukins play key role in the process. Almost 130 original papers were revised including meta-analysis. It is clear these data are very important for understanding the base of the disease, especially in terms of clinical utility and validity, but text often do not available for the doctors use these in the clinical practice nowadays. We conducted a systematic review of the current literature on interleukin and interleukin receptor gene polymorphisms associated with IBD, performing an electronic search of PubMed Database from publications of the last 10 years, and used the following medical subject heading terms and/or text words: IBD, CD, UC, interleukins and polymorphisms.
Core tip: Inflammatory bowel diseases (Crohn’s disease and ulcerative colitis) are chronic, progressive disorders of the gastrointestinal tract. Different genes, including interleukin genes play central role in mediating and modulating of inflammation in inflammatory bowel diseases. In this review we summarized the interleukin and the interleukin receptor genes associated with Crohn’s disease and/or ulcerative colitis performing an electronic search on the PubMed database focusing on the following terminology: inflammatory bowel disease, Crohn’s disease, ulcerative colitis, interleukin and interleukin receptor.
- Citation: Magyari L, Kovesdi E, Sarlos P, Javorhazy A, Sumegi K, Melegh B. Interleukin and interleukin receptor gene polymorphisms in inflammatory bowel diseases susceptibility. World J Gastroenterol 2014; 20(12): 3208-3222
- URL: https://www.wjgnet.com/1007-9327/full/v20/i12/3208.htm
- DOI: https://dx.doi.org/10.3748/wjg.v20.i12.3208
Inflammatory bowel disease (IBD) - clinically classified as Crohn’s disease (CD; OMIM 26600) and ulcerative colitis (UC; OMIM 191390) - is a common chronic, relapsing inflammatory disorder of the gastrointestinal tract[1]. In Europe the highest annual incidence of CD is 12.7/100000 and 24.3/100000 for UC. In Asia and in the Middle East both rates are much lower (CD: 5.0/100000 and UC: 6.3/100000). However in North America the incidence for UC is 19.2/100000 and they have the highest rate for CD in the world with 20.2/100000[2]. Although the precise etiology of IBD still remains obscure, the accepted hypothesis is that in genetically predisposed individuals the commensal luminal flora trigger an inappropriate, overactive mucosal immune response causing intestinal tissue damage that is further modified by specific environmental factors (e.g., smoking)[3].
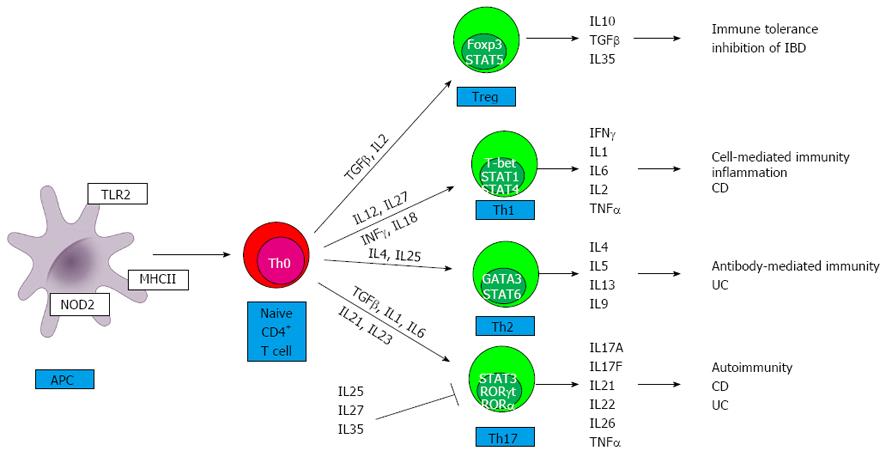
The location of inflammatory lesions and the types of cytokines involved in the pathogenesis mainly distinguish CD from UC. Whereas CD is a segmental, transmural disorder involving any part of the gastrointestinal tract, UC is characterized by superficial, continuous mucosal ulcers restricted to the colon. Imbalances between pro- and anti-inflammatory cytokines in the mucosa have been established for both CD and UC[4]. CD is associated with a T helper type 1 (Th1)[4] and T helper type 17 (Th17)[5] immune response, thus interferon gamma/interleukin 12 (IFNγ/IL12) and IL23/IL17 cytokines assign the downstream release of complex network of further pro-inflammatory cytokines (e.g., IL18, IL2, IL1, IL21, IL22) (Figure 1). Th17 and a modified Th2 cytokine profile (IL13 and IL5) are characteristic for UC. In addition, IL6 and tumor necrosis alpha (TNFα) are produced by both Th1 and Th2 cells as well as by macrophages in both IBD entities. A further group of T cells, regulatory T cells (Treg) cells are important for the control of immune responses to self-antigens preventing autoimmunity and maintaining self-tolerance[6]. The final result of this activated cytokine network is the recruitment of more effector cells and the beginning of mucosal inflammation, which will eventually become chronic due to defective regulation of the immune response[6].
First, genome-wide association studies (GWAS) resulted in the identification of many novel susceptibility loci CD and later for UC[7,8]. To date, the number of known risk loci has expanded to 163[9]. Some loci seem to be specific either to CD or to UC, whereas others confer common susceptibility to IBD; approximately 30% of IBD-related genetic loci are shared[10,11]. The IBD-associated loci encode genes involved in innate pattern recognition {nucleotide-binding oligomerization domain-containing protein 2 (NOD2), autophagy (autophagy-related protein 16-1 (ATG16L1), immunity-related GTPase family M protein (IRGM), differentiation of Th17-T lymphocytes (IL23R), maintenance of epithelial barrier integrity IBD5 locus], and coordination of adaptive immune responses [human leukocyte antigen (HLA)-region]}[12]. Polymorphisms in genes encoding cytokines and cytokine receptors may affect the course of the inflammatory cascade and thereby increase the risk of developing IBD.
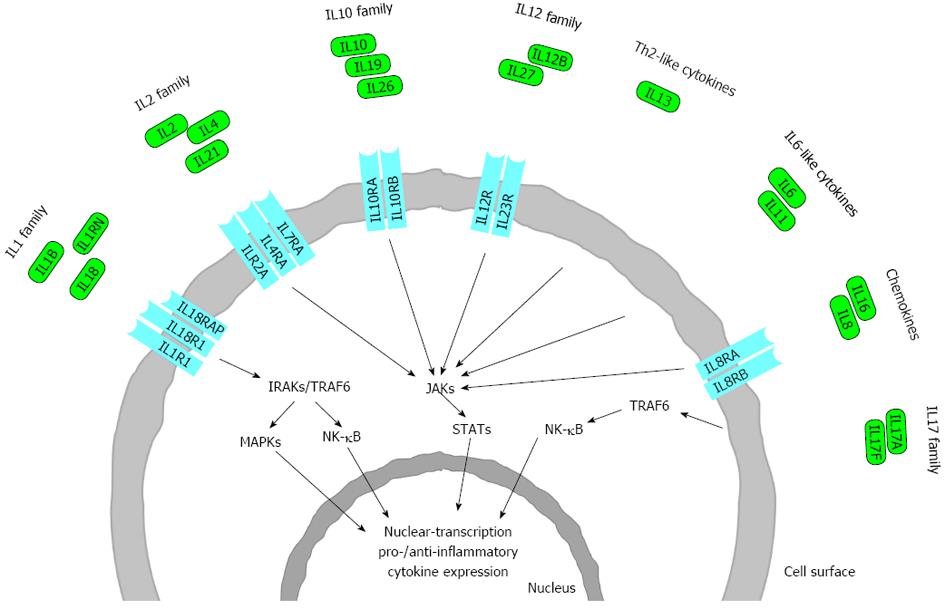
In this review we discuss in each of the IL families only those interleukins or interleukin receptors in detail, which have relevant polymorphisms in IBD, CD or UC (Figure 2).
We conducted a systematic review of the literature of the last 10 years on interleukin susceptibility genes to IBD. PubMed was searched for papers and abstracts published in English-language journals. We used the following medical subject heading terms and/or text words: “inflammatory bowel disease”, “ulcerative colitis”, “Crohn’s disease” and “cytokines”. The search was focused on interleukin susceptibility genes polymorphism resulting in IBD. No restrictions were placed on race, ethnicity, or geographic area. Extraction from each study was conducted independently by all authors, and consensus was achieved for all data.
ILs are the subset of a larger group of cellular messenger molecules called cytokines, which are humoral, small (4-15 kDa) inducible immune-regulatory proteins or glycoproteins which mediate communication between cells, regulate cell growth and differentiation, and play a central role in the development and homeostasis of the immune system[6]. They act on target cells by binding to specific IL receptors, initiating signal transduction and second messenger pathways within the target cell. This can result in gene activation, lead to mitotic division, growth and differentiation, migration, or apoptosis. Cytokines act in a highly complex coordinated network in which they induce or repress their own synthesis as well as that of other cytokines and cytokine receptors. The nomenclature of ILs is continuously evolving (http://www.genenames.org/genefamilies/il); they are assigned to each family based on sequence homology and receptor chain similarities or functional properties (Table 1)[13].
| Family | Cytokine | Receptor | Cytogenetic location | Molecular weight | Cell source | Disease association (IBD) |
| IL1 | IL1A | IL1R1 | 2q14 | 17 kD | Macrophages, monocytes, lymphocytes, keratinocytes, microglia, megakaryocytes, neutrophils, fibroblasts and synovial lining cells | CD, UC |
| (IL1F1) | IL1R2 | |||||
| IL1B | IL1R1 | 2q14 | 17 kD | |||
| (IL1F2) | IL1R2 | CD, UC | ||||
| IL1RN | IL1R1 | 2q14.2 | 16.1-20 kD | Monocytes, macrophages, fibroblasts, neutrophils, epithelial cells and keratinocytes | UC | |
| (IL1F3) | IL1R2 | |||||
| IL18 | IL18R1 | 11q22.2-q22.3 | 22.3 kD | Macrophages, Kupffer cells, keratinocytes, osteoblasts, astrocytes, and DCs | CD, UC | |
| (IL1F4) | IL18RAP | |||||
| IL2 | IL2 | IL2R | 4q26-q27 | 15.5 kD | CD4+, CD8+ activated T cells, DCs, NK and NKT cells | CD, UC |
| IL4 | IL4RI | 5q23-q31 | 15 kD | Th2 cells, basophils, eosinophils, mast cells, NKT and γ/δ T cells | CD | |
| IL4RII | ||||||
| IL21 | IL21R | 4q26-q27 | 15 kD | T and NKT cells | CD, UC | |
| IL10 | IL10 | IL10RA/IL10RB | 1q31-q32 | 18.6 kD | T and B cells, monocytes, macrophages and DCs | CD, UC |
| IL19 | IL20RA/IL20RB | 1q32.2 | 35-40 kD | Monocytes, keratinocytes, airway epithelial cells and B cells | UC | |
| IL26 | IL10R2/IL20R1 | 12q15 | 38 kD | Activated T cells | CD, UC | |
| IL12 | IL12 | IL12RB1/IL12RB2 | 5q33.3 | IL12A: 35 kD, IL12B: 40 kD | Monocytes, macrophages, neutrophils, microglia, DCs and B cells | CD, UC |
| IL23 | IL12RB1/IL23R | 12q13.13 | 19 kD | Macrophages and activated DCs | CD, UC | |
| IL27 | IL27RA/ IL6ST | 16p11 | IL27A: 2 8 kDIL27B: 25.4 kD | Activated DCs, macrophages, and epithelial cells | CD, UC | |
| IL6-like cytokines | IL6 | IL6R/IL6ST | 7p21-p15 | 19-26 kD | Endothelial cells, fibroblasts, monocytes/macrophages | CD |
| IL17 | IL17A | IL17RA/ IL17RC | 6p12 | 35 kD | Th17, CD8+ T cells, NK cells, NKT cells, γ/δ T cells and neutrophils | UC |
| IL17F | IL17RA/IL17RC | 6p12 | 44 kD | Th17, CD8+ T cells, NK cells, NKT cells, γ/δ T cells and neutrophils | CD, UC | |
| Chemokines | IL8 | IL8RA/IL8RB | 4q13-q21 | 16 kD | Monocytes, macrophages, neutrophils, lymphocytes, endothelial cells, epithelial cells, fibroblasts, keratinocytes, chondrocytes, synovial cells, and hepatocytes | CD, UC |
| IL16 | CD4 | 15q26.3 | 56 kD | T cells, eosinophils, mast cells, eosinophils, monocytes, DCs, fibroblasts and epithelial cells | CD | |
| Th2-like cytokines | IL13 | IL13RA1/IL13RA2 | 5q31 | 10 kD | T, NKT, mast cells, basophils and eosinophils | CD, UC |
The IL1 family is a group of 11 cytokines (IL1A, IL1B, IL1RN, IL18, IL33, IL36A, IL36B, IL36G, IL36RN, IL37 and IL38), which have similar gene structure and induce a complex network of proinflammatory cytokines. The interleukin 1 receptor (IL1R) family also expands to 9 distinct genes and includes coreceptors, decoy receptors, binding proteins, and inhibitory receptors[14].
IL1 is a potent proinflammatory cytokine, which affects cell proliferation, differentiation, and the function of many innate and specific immunocompetent cells, and acts as an endogenous pyrogen. It also mediates many inflammatory diseases by initiating and potentiating immune and inflammatory responses[13].
IL1 is made up of two major proteins: IL1A (OMIM 147760) and IL1B (OMIM 147720)[15]. These proteins exert similar effects, first, by binding to the first extracellular chain of the IL1 type I receptor (IL1RI) (OMIM 147810) that recruits the IL1 receptor accessory protein (IL1RAP) (OMIM 602626), which serves as a coreceptor and is necessary for signal transduction. IL1A and IL1B are also able bind to the IL1 type II receptor (OMIM 147811), which acts as a decoy receptor and is not involved in signal transduction[13].
IL1B has an important role in initiating and amplifying the inflammatory response[16]. Normal colonic mucosa cells produce very little mature IL1B, however in the mucosa of affected IBD patients, a large amount of mature IL1B is produced[17,18]. The inability of normal intestinal macrophages to produce mature IL1B could result from regulation at one or more steps from gene activation to post-translational processing of the propeptide by IL1B converting enzyme and release of the mature peptide[19,20].
The IL1 receptor antagonist (IL1RN) (OMIM 147679) is an anti-inflammatory cytokine, which is synthesized and released in response to the same stimuli that lead to IL1 production[21]. IL1RN lacks the IL1RAP interacting domain, so that binding of the IL1RN to IL1RI inhibits IL1 signaling[15]. In IBD and several other inflammatory diseases, an imbalance the IL1RN/IL1 ratio contributes to the chronic inflammatory response[22-24]. Polymorphisms of the IL1RN gene, which can lead to changes in the IL1RN and IL1 balance, are associated with susceptibility to UC[25]. Moreover, it is well accepted that IBD patients have a decreased ratio of IL1RN/IL1B in their colonic mucosal tissue[26].
The variant alleles of two IL1B promoter polymorphisms, IL1B T-31C and IL1B C-511T, have been found to be in almost complete linkage disequilibrium, and the haplotypes encompassing the IL1B T-31C variant conferred higher transcription of IL1B compared to the wild type haplotype[27].
Four polymorphisms (rs315951, rs315952, rs419598 and rs16944) in the IL1B and IL1RN genes were analyzed in Mexican Mestizo UC patients. The first 3 single nucleotide polymorphisms (SNPs) are located in the IL1RN and the fourth one in the IL1B gene. The first two (rs315951 and rs315952) are associated with the risk of developing UC. They found significant increased frequencies of IL1RN6/1TC (rs315952) and RN6/2CC (rs315951), and decreased frequency of IL1B-511 TC (rs16944) genotypes in UC patients. UC patients showed increased frequencies of IL1RN CTC and TCG haplotypes, whereas TTG and CTG haplotypes showed decreased frequency in UC patients. They also found decreased gene expression of IL1RN level in the mucosa from UC patients carrying the rs315951 GG genotype when compared with UC patients with the rs315951 CC genotype[28].
One of the main function of IL18 (OMIM 600953) is to promote the production of IFNγ from T and natural killer (NK) cells, particularly in the presence of IL12p70. First it binds to its ligand binding chain the interleukin 18 receptor 1 (OMIM 604494), recruits its coreceptor the IL18 receptor accessory protein (IL18RAP) (OMIM 604509), and the activation of nuclear factor kappa-light-chain-enhancer of activated B cells/mitogen activated protein 8 is initiated. IL18 expression correlates with the activities of CD[29].
IL18 binding protein (IL18BP, OMIM 604113) is able to prevent the binding of IL18 to its receptor, and thereby blocks its downstream functional effects. IL18BP has neutralizing isoforms, which have increased levels in the intestinal tissue of active CD patients[30].
Several polymorphisms were studied in the IL18 gene: the A105C, the T113G and the C127T in the coding region, and the G-137C, the C-607A and the G-656T in the promoter region. In the Japanese population significant difference was found in the allele frequency of A105C between CD patients and healthy controls. However, there was no association between A105C and UC[31]. In another Japanese study the G allele at 113 and the T allele at 127 were significantly higher in patients with IBD compared to the control[32]. In the third Japanese study allele and genotype frequency of G-137C were significantly higher in the proctitis-type UC patients than in controls[33]. The frequency of haplotype 2 (-607A, -137C), which have lower promoter activity and IFNγ- mRNA level was significantly increased in the proctitis-type patients than in the control group[33]. Any significant differences in allele or genotype frequencies were observed in the CD group[33]. The C-607A and the G-137C SNPs in the promoter region were associated with the development of UC but not with CD in Tunisian patients. The -137GG genotype frequency was significantly higher in UC than in controls. Statistically significant association was found between -607AA genotype in UC patients and the distal localization of the lesions[34]. However the polymorphism G-137C was not found a susceptibility factor for IBD in a German population[35].
Recent GWAS study[36] and meta-analysis confirmed the IL18RAP region as CD locus[8]. The rs6708413 G allele is a shared risk locus for CD and Celiac disease[37]. In individuals homozygous for the risk allele, the genotypes strongly correlate with lower IL18RAP expression which may lead to differential IL18-mediated innate immune responses to infection[38]. Strong association of rs917997 SNP was demonstrated for both CD and UC[39]. In a new GWAS study association of CD and IBD with coding variant V527L was found. This rare missense increased high the risk for CD[40].
The IL2 family consists of IL2, IL4, IL7, IL9, IL15 and IL21. This family of cytokines encompasses a group of interleukins which share a common receptor subunit, the “common γ chain”, which acts in unison with a subtype specific α-chain to initiate the signaling cascade. This ILs act mainly as growth and proliferation factors for progenitors and mature cells and also have roles in lineage-specific cell differentiation[13].
IL2 acts as a T cell growth factor and promotes proliferation and differentiation of NK cells to increase their cytolytic functions. IL2 is essential for the development of Th1, Th2, Treg, and Th17 differentiation[41].
The IL2 receptor (IL2R) consists of three non-covalently associated proteins: IL2RA (OMIM 147730), IL2RB (OMIM 146710) and IL2RG (OMIM 308380). The α-chain is produced when the T cell is activated by antigen and constitutes the high affinity receptor together with the other two subunits[42]. The β- and γ-chains form the intermediate affinity IL2 receptor[43].
Several SNPs (rs6822844, rs13151961, rs13119723 and rs6840978) in the IL2/IL21 block were analyzed in different populations. In a Dutch cohort the minor alleles of these SNPs were associated with IBD. In UC patients the effect was even stronger. However in the CD subgroup, the rs13119723 SNP was only borderline significant, while only a trend towards association was found for the other SNPs. Testing of all four SNPs in the Italian cohort, the same strong association of the minor alleles in UC was found as in the Dutch cohort. The CD subgroup of the Italian cohort showed only a trend towards association with the same alleles. However in the Jewish population there was any significant association between any of the SNPs and CD. Similarly a North American study showed that these alleles have an influence on IBD. The effect was strongest in the UC subgroup likewise. In the CD subgroup of the North American cohort moderate association with the same alleles was also observed[44].
IL4 (OMIM 147780), a pleiotropic cytokine, is the major stimulus of Th2-cell development, which regulates allergic conditions and the protective immune response against helminthes and other extracellular parasites[45]. There are 2 types of IL4Rs. IL4R Type I binds only IL4 and consists of 2 receptor chains: IL4RA (OMIM 147781) and the common γc, IL4R Type II binds IL4 and IL13 and consists of the IL4RA and the IL13RA1 chains[46].
Functional polymorphism in the IL4 gene promoter C-34T was associated with CD in a British population[47]. The same polymorphism was tested in a New Zealand population, where no significant difference was observed in the genotype frequencies of controls vs CD patients[48].
IL21 (OMIM 605384) is a cytokine with potent regulatory effects on cells of the immune system, including NK cells and cytotoxic T cells, which can destroy virus infected and cancerous cells[49]. In contrast with its anticancer effects, IL21 also contributes to inflammation in several disorders, as can be expected for a Th17-related cytokine. The functional receptor of IL21 consists of γc and the IL21RA (OMIM 605383)[13].
GWAS provides evidence for 4q27 region IL2/IL21 association with UC[44] and CD[50]. This region contains four genes in strong linkage disequilibrium: KIAA1109-TENR-IL2-IL21.
The members of the IL10 cytokine family (IL10, IL19, IL20, IL22, IL24, IL26, IL28, and IL29) are mainly linked through their similar intron-exon structure. This family can be divided into viral and cellular homologs, where this last named group contains the above mentioned ILs[51].
IL10 (OMIM 124092) is an anti-inflammatory cytokine, which is produced by monocytes, T cells, B cells, NK cells, macrophages, and dendritic cells. It inhibits both antigen presentation and subsequent release of pro-inflammatory cytokines. Thereby attenuates the activated immune system. The IL10 gene maps to a cytokine cluster that includes IL19, IL20, IL24, and IL6 genes. Two IL10RA (OMIM 146933) and two IL10RB (OMIM 123889) chains forms the heterotetrameric IL10 receptor complex. The IL10RB chain is shared with other cytokine receptors[52].
In a GWAS the rs3024505 showed the most significant association in the combined verification UC samples. Association between rs3024505 and CD was weak. These results suggest that defective IL10 function is central to the pathogenesis of the UC subtype of IBD[53]. In a latest study from 29 SNPs conferring high genetic susceptibility to CD, the rs3024505 of the IL10 gene was associated with susceptibility to UC in Australian population[54]. Similarly to this, the rs3024505 was associated with the risk of UC and CD in a Danish study[55].
The IL10 promoter polymorphisms G-1082A, C-819T, and C-592A have been most extensively studied. They are in tight linkage disequilibrium[56]. Studies on the IL10 promoter polymorphisms and IBD susceptibility have been controversial[57-60]. In Spanish population the G-1082A and the C-819T polymorphisms in the IL10 gene contribute to susceptibility to CD[61].
In IBD patiens in Tunisia the polymorphisms A-627C and G-1117A were analyzed and found as potential factors influencing IBD susceptibility and phenotype. However, no significant variations in genotypes frequencies were found comparing the CD and UC patients[62].
The A-1082G variant was analyzed in Caucasian population in many studies. It was suggested that G carriers were more susceptible to UC[63], whereas in another study G carriers were associated with lower UC incidence[64]. Only two datasets concerning the relationship between A-1082G polymorphism and UC susceptibility in Asian subjects[65,66] were identified. A meta-analysis showed no association between -A1082G polymorphism and UC susceptibility under any genetic models in overall analysis or in subgroup analysis[67]. In an another study this variant was significantly associated with the colonic localization of the disease in Caucasian CD patients (children) with French-Canadian origin[68].
IL19 (OMIM 605687) might promote Th2-cell responses, because it induces expression of IL4, IL5, IL10, and IL13 by activated T-cells[69]. IL19 functions as a monomer, binds to a heterodimeric receptor made up of IL20RA (OMIM 605620) and IL20RB (OMIM 605621). This complex also binds to IL20 and IL24[70].
In GWAS, 14 previously identified UC susceptibility loci were analyzed. Association including the polymorphism rs3024505 in the IL19 was confirmed[71].
Expression of IL26 (OMIM 605679) seems to be restricted to memory T cells, NK cells, and Th17 cells. Thereby it could have proinflammatory effects in CD[72]. The receptor for IL26 consists of IL10RB and IL20RA chains. Dambacher reported expression of both IL26 receptor subunits IL20RA and IL10RB by several intestinal epithelial cells (IEC) lines in CD[73].
First time, the rs2870946-G and the rs1558744-A association were described with UC[74]. Further meta-analysis study confirmed the association of rs1558744-A with UC[71].
The IL12 family of cytokines includes IL12, IL23, IL27, IL30 and IL35, which are important mediators of inflammatory diseases. Each member is heterodimeric complex composed of two subunits whose expression is regulated independently and have very different biological activities[75].
IL12 (also known as IL12p70) was first described as a NK stimulating factor. It mediates development and maintenance of Th1 cells by inducing production of IFNγ by Th1 and NK cells. IL12 indirectly activates the antimicrobial, antiparasitic, and antitumor activity of macrophages and promotes cytolytic activity of NK cells and lymphokine-activated killer cells[76]. Reduced production of IL12 impairs Th1 responses and increases susceptibility to infection with intracellular pathogens. IL12 consist of p35 (IL12A, OMIM 161560) and p40 (IL12B, OMIM 161561) subunits, which is shared by IL12 and IL23 cytokines. The IL12 receptor is composed of two subunits, IL12RB1 (OMIM 601604) and IL12RB2 (OMIM 601642), which is homologous to the gp130 subunit[77,78].
In a German population four IL12B SNPs (rs3212227, rs17860508, rs10045431 and rs6887695) were analyzed. The rs6887695 showed association with increased IBD susceptibility, and there was also trend for association with CD and UC. However just a trend was found for association of rs10045431 with UC. A haplotype of all four investigated SNPs showed a trend for association with CD[79].
From these SNPs the rs6887695 was investigated in Spanish and Japanese population, but with different results. An association was found with CD (rs6887695) and UC susceptibility (rs6887695) in the Japanese cohort, but not to CD susceptibility in the Spanish cohort[80,81]. Examining rs1363670 and rs6887695 SNPs in New Zealand population differential effect was found. Carrying the rs1363670 C variant increases risk for CD, while carrying the rs6887695 C variant decreases risk for CD[82].
In a British cohort an association was found at rs6556416, which encodes a subunit shared by IL12 and IL23. Thus, the Th17 pathway seems as relevant to UC and CD[83].
IL23 is a disulfide-linked heterodimer of the p40 (IL12B) and p19 (IL23A, OMIM 605580). IL23 interacts with a receptor composed of IL12RB1 and IL23R chain (IL23R, OMIM 607562)[78]. IL23 functions in innate and adaptive immunity to regulate Th17 function and expansion[84]. In addition, this cytokine induces CD8+ memory T cells to proliferate and produce IL17. Dysregulation of the IL23/IL17 immune axis has been linked to immunopathology and autoimmune inflammation, like IBD. IL23R polymorphisms play role in many autoimmune diseases[85-87] especially in IBD[88]. Polymorphisms in the IL23R represent one of the strongest associations in CD, and they have also been linked to the pathogenesis of UC[89].
The IL23R gene was identified as a CD susceptibility gene in a North American population. Several independent functional SNPs of the gene and its neighboring region were determined, several were found susceptible to (rs10889677, rs11209032, rs11465804, rs11805303, rs1495965, rs2201841, rs1004819) and the others were protective (rs10489629, rs11209026, rs1343151, rs7517847) against IBD in non-Jewish subjects[89]. After the primary publication, numerous replication studies have been published these IL23R genetic polymorphisms in IBD.
From the susceptibility variants the rs1004819 and rs1495965 were found as important risk factors for CD in Koreans[90]. Similarly to these results the rs1004819 had the most significant association with CD in Germans, and the another rs10889677, rs2201841, rs11209032 showed increased genotype and allele frequencies comparing the CD cohort to the controls[91]. Positive association was described of IL23R rs10889677 and rs1004819 SNPs with CD in Brazilian population, where the allele frequencies of the patients’ group differ significantly from the controls[92]. Another susceptibility factors were studied in Chinese cohort and the findings showed that rs7530511 and rs11805303 of IL23R gene showed positive association with UC susceptibility[93]. In Jiangsu Han population the rs11805303 was found as a susceptibility polymorphism with UC too[94].
The protective variants of the IL23R gene were analyzed in different populations. Association with rs11209026 and rs7517847 SNPs were confirmed in English subject, where the most significant SNP was the rs7517847[95]. Similarly in Spanish population the rs7517847 and the rs11209026 showed association with IBD too, the rs7517847 showed the most protective effect against CD and UC[80,96]. In another study the rs11209026 coding variant was found as a strong protection against CD in German pediatric CD patients[97].
Five polymorphisms were analyzed in Hungarian IBD population (rs1884444, rs11805303, rs7517847, rs2201841, rs10889677 and rs11209032)[98]. The rs2201841 and rs10889677 homozygous variants confer risk for the disease, while rs7517847 GG genotype has a protective effect against the development of CD. In Hungarian CD population two IL23R gene risk variants the rs2201841 and rs1004819 were found to be a susceptibility factor for CD[99]. In another study with Hungarian CD patients increased prevalence of the homozygous rs10889677 AA and homozygous rs2201841 CC genotypes were found. The rs10889677 AA genotype was significantly increased in CD patients. The logistic regression analysis showed the AA genotype represents an independent risk factor for the development of CD[100].
IL27 is a heterodimeric cytokine consisting of Epstein-Barr virus-induced gene 3 (EBI3, OMIM 605816) and p28 (also known as IL30, OMIM 608273). It binds a unique receptor subunit IL27RA (OMIM 605350), which is associated with gp130 (IL6ST, OMIM 600694), a common chain utilized by IL6 family cytokines. IL27 suppresses Th2 and Th17 differentiation and proliferation[101,102].
In a Korean population the -A965G SNP was described as a susceptibility factor for IBD[103]. In a GWAS five new regions were identified near the IL27 gene associated with early-onset IBD susceptibility (rs8049439, rs2412973, rs1250550, rs4676410 and rs10500264)[104].
Cytokines in this family (IL6, IL11, IL27 and IL31) signal through receptors containing gp130 which are commonly referred to as the IL6-like or gp130 utilizing cytokines family[105]. They show pleiotropic biological activities with immune, hematopoietic, and neural systems[105].
IL6 (OMIM 147620) is a multifunctional, pleiotropic cytokine involved in regulation of immune responses, acute-phase responses, hematopoiesis, and inflammation. IL6 signals through a cell-surface type I cytokine receptor complex consisting of the ligand-binding IL6R chain (OMIM 147880), and the shared signal-transducing component IL6ST (also called gp130; OMIM 600694)[106].
English and Swedish children with CD and IL6-174 GG genotype were more growth retarded at diagnosis and had higher levels of the IL6-induced inflammatory marker C-reactive protein (CRP) than children with GC or CC genotypes, concluded that IL6-174 genotype mediates growth failure in CD[107]. In an Irish cohort from Dublin, significant difference was found in the frequency of IL6-174 genotypes in the UC group compared with the CD group[57]. In a Caucasian population from Canada the same polymorphism was analyzed in CD and UC patients. There were significant difference IBD, UC and CD susceptibility, but it has influence on the clinical phenotype of CD[108]. In a Spanish population[109] homozygous for the IL6 G-174C polymorphism showed six-fold higher risk for CD. The GG genotype is associated with a greater production of IL6 compared with GC or CC genotypes[110].
This cytokine family is a recently discovered group of cytokines with six members (IL17A, IL17B, IL17C, IL17D, IL17E and IL17F). IL17A was the original member of this family. The others were discovered primarily from the genome sequences within a short time-period (2000-2002), and were sequentially named in the order of discovery[111-114]. They share the highest amino acid sequence homology and perform distinct biological functions[115].
IL17A (OMIM 603149) acts on a variety of cells, which respond by upregulating expression of proinflammatory cytokines, chemokines, and metalloproteases. It is involved in the development of autoimmunity, inflammation, and tumors. IL17E (IL25, OMIM 605658) is an amplifier of Th2 immune responses. IL17F (OMIM 606496) is mainly involved in mucosal host defense mechanisms. The functions of IL17B (OMIM 604627), IL17C (OMIM 604628), and IL17D (OMIM 607587) are still largely elusive. Increased levels of IL17A[116] and IL17F[117] have been found in patients with IBD. It inhibits the proliferation of IECs, suggesting it might interfere with the repair mechanism important for the maintenance of the tissue integrity[118].
The IL17 receptor (IL17R) family includes five members: IL17RA (OMIM 605461), IL17RB (OMIM 605458), IL17RC (OMIM 610925), IL17RD (OMIM 606807), and IL17RE (OMIM 614995)[119].
In a Japanese UC patients the rs2275913 polymorphism in the IL17A gene and the rs763780 in the IL17F gene were analyzed. The frequencies of -197A/A and 7488T/T genotypes were found significantly higher in UC patients than in controls[120]. In a Caucasian (German) population, despite the increased colonic IL17F expression in CD, any significant differences could be found in the frequency of rs763780 on IBD susceptibility[117].
This group contains only two ILs, the IL8 and IL16[13].
IL8 was identified first as a neutrophil-specific chemotactic factor and later classified as a member of the CXC chemokine family[121]. Its receptors are: CXCR1 (IL8RA) and CXCR2 (IL8RB)[122]. IL8 plays crucial role in the chemotaxis and migration of neutrophils, monocytes, lymphocytes, and fibroblasts[123].
In a Polish population significant association was found between the genotype frequencies for the heterozygote of the IL8 T-251A and IBD. When IBD patients were subdivided in UC and CD subgroups this association was also observed. Significant difference was found between the A allele and the UC and CD cases, but not in the summarized IBD group[124]. In a Chinese population any association was found between the T-251A polymorphism and UC[125]. They also investigated the role of other polymorphisms of the IL8 gene and its impact on the level of IL8 in serum. The frequency of the -353A/-251A/+678T haplotype was significantly higher in UC patients than in healthy controls. This haplotype tends to be more common in severe UC patients than in mild to- moderate cases[125].
IL16 (OMIM 603035) is a proinflammatory cytokine, which inhibits T-cell proliferation, promotes Th1-mediated responses, and reduces Th2-mediated inflammation[126]. IL16 mediates its biological activity via CD4 molecule, which is present on T cells, monocytes, macrophages, and dendritic cells. Patients with CD have elevated levels of IL16[127].
Regardless of the disease phenotype or the site of intestinal involvement, the T allele and the TT genotype in the IL16 promoter region T-295C were found significantly increased in German CD cohort, but not in UC patients[128].
Cytokines produced during the induction and function of Th2 response include IL4, IL5, IL9, IL13, IL25 (IL17E), IL31, and IL33[13].
IL13’s (OMIM 147683) receptors are IL13RA1 (OMIM 300119) and IL13RA2 (OMIM 300130), signaling occurs via the IL4R complex type II, which consists of IL4RA and IL13RA1. IL13RA2 acts as a decoy receptor of IL13. IL13 activates the same signal transduction pathways as IL4 and induces IgE production, influences eosinophils and cause their prolonged survival, activation, and migration to inflammatory lesions[129]. IL13 plays an opposite role to IL8. In monocytes and macrophages, it inhibits the secretion of pro-inflammatory mediators such as prostaglandins, reactive oxygen species (ROS) and nitrogen species, TNFα, IL1, -6, -8, and -12[130].
Presence of the IL13 -1112 CT (rs1800925) genotypes in a Polish population showed higher risk of IBD as well as UC occurrence. The statistically significant differences in the T-allele distribution were observed in all the investigated groups[124].
In this review we discuss IL and IL gene polymorphisms which contribute to IBD, CD or UC in different ethnical population. The cytokine network is highly complex with interactive cascades of gene activation and suppression. Not only the IL and ILR gene polymorphisms are in relation with IBD pathogenesis but also the downstream signaling components of several ILs (i.e., JAKs, STATs) which could be potential targets of novel treatment strategies. Many IBD loci are also implicated in other immune-mediated disorders, most notably with ankylosing spondylitis and psoriasis[9]. Since the individual associations may be non-informative, specific combinations of cytokine genotypes might predispose to disease susceptibility or outcome. Therefore, polymorphisms in cytokine genes and receptors should not in all cases be studied strictly in isolation. More complete understanding of the immunopathogenic role of the various ILs in intestinal inflammation will help in the development of more effective novel therapeutic strategies in IBD. Albeit genotyping these interleukin variants are often offer on the palette of several direct-to-costumer companies, their diagnostic or therapeutic clinical use is very limited due to the limited clinical utility and validity of them. Meanwhile, the next generation techniques in combination with the data analysis by systems-biology approach hopefully will contribute to the personalized therapy of the patients in the near future.
P- Reviewers: D'Elios MM, Rocha R S- Editor: Gou SX L- Editor: A E- Editor: Ma S
| 1. | Podolsky DK. Inflammatory bowel disease. N Engl J Med. 2002;347:417-429. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2693] [Cited by in RCA: 2744] [Article Influence: 119.3] [Reference Citation Analysis (2)] |
| 2. | Molodecky NA, Soon IS, Rabi DM, Ghali WA, Ferris M, Chernoff G, Benchimol EI, Panaccione R, Ghosh S, Barkema HW. Increasing incidence and prevalence of the inflammatory bowel diseases with time, based on systematic review. Gastroenterology. 2012;142:46-54.e42; quiz e30. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3134] [Cited by in RCA: 3507] [Article Influence: 269.8] [Reference Citation Analysis (5)] |
| 3. | Van Limbergen J, Russell RK, Nimmo ER, Ho GT, Arnott ID, Wilson DC, Satsangi J. Genetics of the innate immune response in inflammatory bowel disease. Inflamm Bowel Dis. 2007;13:338-355. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 54] [Cited by in RCA: 50] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 4. | Fuss IJ, Neurath M, Boirivant M, Klein JS, de la Motte C, Strong SA, Fiocchi C, Strober W. Disparate CD4+ lamina propria (LP) lymphokine secretion profiles in inflammatory bowel disease. Crohn’s disease LP cells manifest increased secretion of IFN-gamma, whereas ulcerative colitis LP cells manifest increased secretion of IL-5. J Immunol. 1996;157:1261-1270. [PubMed] |
| 5. | Fujino S, Andoh A, Bamba S, Ogawa A, Hata K, Araki Y, Bamba T, Fujiyama Y. Increased expression of interleukin 17 in inflammatory bowel disease. Gut. 2003;52:65-70. [PubMed] |
| 6. | Fina D, Pallone F. What is the role of cytokines and chemokines in IBD? Inflamm Bowel Dis. 2008;14 Suppl 2:S117-S118. [PubMed] |
| 7. | Anderson CA, Boucher G, Lees CW, Franke A, D’Amato M, Taylor KD, Lee JC, Goyette P, Imielinski M, Latiano A. Meta-analysis identifies 29 additional ulcerative colitis risk loci, increasing the number of confirmed associations to 47. Nat Genet. 2011;43:246-252. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1095] [Cited by in RCA: 1041] [Article Influence: 74.4] [Reference Citation Analysis (1)] |
| 8. | Franke A, McGovern DP, Barrett JC, Wang K, Radford-Smith GL, Ahmad T, Lees CW, Balschun T, Lee J, Roberts R. Genome-wide meta-analysis increases to 71 the number of confirmed Crohn’s disease susceptibility loci. Nat Genet. 2010;42:1118-1125. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2110] [Cited by in RCA: 1993] [Article Influence: 132.9] [Reference Citation Analysis (0)] |
| 9. | Jostins L, Ripke S, Weersma RK, Duerr RH, McGovern DP, Hui KY, Lee JC, Schumm LP, Sharma Y, Anderson CA. Host-microbe interactions have shaped the genetic architecture of inflammatory bowel disease. Nature. 2012;491:119-124. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 3979] [Cited by in RCA: 3562] [Article Influence: 274.0] [Reference Citation Analysis (0)] |
| 10. | Khor B, Gardet A, Xavier RJ. Genetics and pathogenesis of inflammatory bowel disease. Nature. 2011;474:307-317. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2019] [Cited by in RCA: 1855] [Article Influence: 132.5] [Reference Citation Analysis (2)] |
| 11. | Thompson AI, Lees CW. Genetics of ulcerative colitis. Inflamm Bowel Dis. 2011;17:831-848. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 96] [Cited by in RCA: 109] [Article Influence: 7.8] [Reference Citation Analysis (0)] |
| 12. | Van Limbergen J, Wilson DC, Satsangi J. The genetics of Crohn’s disease. Annu Rev Genomics Hum Genet. 2009;10:89-116. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 200] [Cited by in RCA: 191] [Article Influence: 11.9] [Reference Citation Analysis (0)] |
| 13. | Akdis M, Burgler S, Crameri R, Eiwegger T, Fujita H, Gomez E, Klunker S, Meyer N, O’Mahony L, Palomares O. Interleukins, from 1 to 37, and interferon-γ: receptors, functions, and roles in diseases. J Allergy Clin Immunol. 2011;127:701-21.e1-70. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 499] [Cited by in RCA: 574] [Article Influence: 41.0] [Reference Citation Analysis (0)] |
| 14. | Dinarello CA. Interleukin-1 in the pathogenesis and treatment of inflammatory diseases. Blood. 2011;117:3720-3732. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1309] [Cited by in RCA: 1565] [Article Influence: 111.8] [Reference Citation Analysis (0)] |
| 15. | Dinarello CA. Immunological and inflammatory functions of the interleukin-1 family. Annu Rev Immunol. 2009;27:519-550. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2289] [Cited by in RCA: 2602] [Article Influence: 162.6] [Reference Citation Analysis (0)] |
| 16. | Dinarello CA. Interleukin-1, interleukin-1 receptors and interleukin-1 receptor antagonist. Int Rev Immunol. 1998;16:457-499. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 565] [Cited by in RCA: 548] [Article Influence: 20.3] [Reference Citation Analysis (0)] |
| 17. | Mahida YR, Lamming CE, Gallagher A, Hawthorne AB, Hawkey CJ. 5-Aminosalicylic acid is a potent inhibitor of interleukin 1 beta production in organ culture of colonic biopsy specimens from patients with inflammatory bowel disease. Gut. 1991;32:50-54. [PubMed] |
| 18. | Ligumsky M, Simon PL, Karmeli F, Rachmilewitz D. Role of interleukin 1 in inflammatory bowel disease--enhanced production during active disease. Gut. 1990;31:686-689. [PubMed] |
| 19. | Cerretti DP, Kozlosky CJ, Mosley B, Nelson N, Van Ness K, Greenstreet TA, March CJ, Kronheim SR, Druck T, Cannizzaro LA. Molecular cloning of the interleukin-1 beta converting enzyme. Science. 1992;256:97-100. [PubMed] |
| 20. | Thornberry NA, Bull HG, Calaycay JR, Chapman KT, Howard AD, Kostura MJ, Miller DK, Molineaux SM, Weidner JR, Aunins J. A novel heterodimeric cysteine protease is required for interleukin-1 beta processing in monocytes. Nature. 1992;356:768-774. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1857] [Cited by in RCA: 1976] [Article Influence: 59.9] [Reference Citation Analysis (0)] |
| 21. | Eisenberg SP, Evans RJ, Arend WP, Verderber E, Brewer MT, Hannum CH, Thompson RC. Primary structure and functional expression from complementary DNA of a human interleukin-1 receptor antagonist. Nature. 1990;343:341-346. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 801] [Cited by in RCA: 783] [Article Influence: 22.4] [Reference Citation Analysis (0)] |
| 22. | Casini-Raggi V, Kam L, Chong YJ, Fiocchi C, Pizarro TT, Cominelli F. Mucosal imbalance of IL-1 and IL-1 receptor antagonist in inflammatory bowel disease. A novel mechanism of chronic intestinal inflammation. J Immunol. 1995;154:2434-2440. [PubMed] |
| 23. | Tountas NA, Casini-Raggi V, Yang H, Di Giovine FS, Vecchi M, Kam L, Melani L, Pizarro TT, Rotter JI, Cominelli F. Functional and ethnic association of allele 2 of the interleukin-1 receptor antagonist gene in ulcerative colitis. Gastroenterology. 1999;117:806-813. [PubMed] |
| 24. | Arend WP, Guthridge CJ. Biological role of interleukin 1 receptor antagonist isoforms. Ann Rheum Dis. 2000;59 Suppl 1:i60-i64. [PubMed] |
| 25. | Witkin SS, Gerber S, Ledger WJ. Influence of interleukin-1 receptor antagonist gene polymorphism on disease. Clin Infect Dis. 2002;34:204-209. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 199] [Cited by in RCA: 211] [Article Influence: 9.2] [Reference Citation Analysis (0)] |
| 26. | Andus T, Daig R, Vogl D, Aschenbrenner E, Lock G, Hollerbach S, Köllinger M, Schölmerich J, Gross V. Imbalance of the interleukin 1 system in colonic mucosa--association with intestinal inflammation and interleukin 1 receptor antagonist [corrected] genotype 2. Gut. 1997;41:651-657. [PubMed] |
| 27. | Chen H, Wilkins LM, Aziz N, Cannings C, Wyllie DH, Bingle C, Rogus J, Beck JD, Offenbacher S, Cork MJ. Single nucleotide polymorphisms in the human interleukin-1B gene affect transcription according to haplotype context. Hum Mol Genet. 2006;15:519-529. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 219] [Cited by in RCA: 242] [Article Influence: 12.7] [Reference Citation Analysis (0)] |
| 28. | Yamamoto-Furusho JK, Santiago-Hernández JJ, Pérez-Hernández N, Ramírez-Fuentes S, Fragoso JM, Vargas-Alarcón G. Interleukin 1 β (IL-1B) and IL-1 antagonist receptor (IL-1RN) gene polymorphisms are associated with the genetic susceptibility and steroid dependence in patients with ulcerative colitis. J Clin Gastroenterol. 2011;45:531-535. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 37] [Cited by in RCA: 49] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 29. | Arend WP, Palmer G, Gabay C. IL-1, IL-18, and IL-33 families of cytokines. Immunol Rev. 2008;223:20-38. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 608] [Cited by in RCA: 676] [Article Influence: 42.3] [Reference Citation Analysis (0)] |
| 30. | Corbaz A, ten Hove T, Herren S, Graber P, Schwartsburd B, Belzer I, Harrison J, Plitz T, Kosco-Vilbois MH, Kim SH. IL-18-binding protein expression by endothelial cells and macrophages is up-regulated during active Crohn’s disease. J Immunol. 2002;168:3608-3616. [PubMed] |
| 31. | Tamura K, Fukuda Y, Sashio H, Takeda N, Bamba H, Kosaka T, Fukui S, Sawada K, Tamura K, Satomi M. IL18 polymorphism is associated with an increased risk of Crohn’s disease. J Gastroenterol. 2002;37 Suppl 14:111-116. [PubMed] |
| 32. | Aizawa Y, Sutoh S, Matsuoka M, Negishi M, Torii A, Miyakawa Y, Sugisaka H, Nakamura M, Toda G. Association of interleukin-18 gene single-nucleotide polymorphisms with susceptibility to inflammatory bowel disease. Tissue Antigens. 2005;65:88-92. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 22] [Cited by in RCA: 26] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 33. | Takagawa T, Tamura K, Takeda N, Tomita T, Ohda Y, Fukunaga K, Hida N, Ohnishi K, Hori K, Kosaka T. Association between IL-18 gene promoter polymorphisms and inflammatory bowel disease in a Japanese population. Inflamm Bowel Dis. 2005;11:1038-1043. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 34] [Cited by in RCA: 35] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 34. | Ben Aleya W, Sfar I, Habibi I, Mouelhi L, Aouadi H, Makhlouf M, Ayed-Jendoubi S, Najjar T, Ben Abdallah T, Ayed K. Interleukin-18 gene polymorphisms in tunisian patients with inflammatory bowel disease. Digestion. 2011;83:269-274. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 12] [Cited by in RCA: 14] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 35. | Haas SL, Andreas Koch W, Schreiber S, Reinhard I, Koyama N, Singer MV, Böcker U. -137 (G/C) IL-18 promoter polymorphism in patients with inflammatory bowel disease. Scand J Gastroenterol. 2005;40:1438-1443. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 17] [Cited by in RCA: 17] [Article Influence: 0.9] [Reference Citation Analysis (0)] |
| 36. | Wang K, Baldassano R, Zhang H, Qu HQ, Imielinski M, Kugathasan S, Annese V, Dubinsky M, Rotter JI, Russell RK. Comparative genetic analysis of inflammatory bowel disease and type 1 diabetes implicates multiple loci with opposite effects. Hum Mol Genet. 2010;19:2059-2067. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 147] [Cited by in RCA: 151] [Article Influence: 10.1] [Reference Citation Analysis (0)] |
| 37. | Festen EA, Goyette P, Green T, Boucher G, Beauchamp C, Trynka G, Dubois PC, Lagacé C, Stokkers PC, Hommes DW. A meta-analysis of genome-wide association scans identifies IL18RAP, PTPN2, TAGAP, and PUS10 as shared risk loci for Crohn’s disease and celiac disease. PLoS Genet. 2011;7:e1001283. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 187] [Cited by in RCA: 178] [Article Influence: 12.7] [Reference Citation Analysis (1)] |
| 38. | Zhernakova A, Elbers CC, Ferwerda B, Romanos J, Trynka G, Dubois PC, de Kovel CG, Franke L, Oosting M, Barisani D. Evolutionary and functional analysis of celiac risk loci reveals SH2B3 as a protective factor against bacterial infection. Am J Hum Genet. 2010;86:970-977. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 157] [Cited by in RCA: 141] [Article Influence: 9.4] [Reference Citation Analysis (0)] |
| 39. | Zhernakova A, Festen EM, Franke L, Trynka G, van Diemen CC, Monsuur AJ, Bevova M, Nijmeijer RM, van ‘t Slot R, Heijmans R. Genetic analysis of innate immunity in Crohn’s disease and ulcerative colitis identifies two susceptibility loci harboring CARD9 and IL18RAP. Am J Hum Genet. 2008;82:1202-1210. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 177] [Cited by in RCA: 199] [Article Influence: 11.7] [Reference Citation Analysis (0)] |
| 40. | Rivas MA, Beaudoin M, Gardet A, Stevens C, Sharma Y, Zhang CK, Boucher G, Ripke S, Ellinghaus D, Burtt N. Deep resequencing of GWAS loci identifies independent rare variants associated with inflammatory bowel disease. Nat Genet. 2011;43:1066-1073. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 629] [Cited by in RCA: 591] [Article Influence: 42.2] [Reference Citation Analysis (0)] |
| 41. | Malek TR. The biology of interleukin-2. Annu Rev Immunol. 2008;26:453-479. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 733] [Cited by in RCA: 822] [Article Influence: 48.4] [Reference Citation Analysis (0)] |
| 42. | Gaffen SL. Signaling domains of the interleukin 2 receptor. Cytokine. 2001;14:63-77. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 134] [Cited by in RCA: 142] [Article Influence: 5.9] [Reference Citation Analysis (0)] |
| 43. | Kim HP, Imbert J, Leonard WJ. Both integrated and differential regulation of components of the IL-2/IL-2 receptor system. Cytokine Growth Factor Rev. 2006;17:349-366. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 230] [Cited by in RCA: 236] [Article Influence: 12.4] [Reference Citation Analysis (0)] |
| 44. | Festen EA, Goyette P, Scott R, Annese V, Zhernakova A, Lian J, Lefèbvre C, Brant SR, Cho JH, Silverberg MS. Genetic variants in the region harbouring IL2/IL21 associated with ulcerative colitis. Gut. 2009;58:799-804. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 107] [Cited by in RCA: 108] [Article Influence: 6.8] [Reference Citation Analysis (0)] |
| 45. | Howard M, Farrar J, Hilfiker M, Johnson B, Takatsu K, Hamaoka T, Paul WE. Identification of a T cell-derived b cell growth factor distinct from interleukin 2. J Exp Med. 1982;155:914-923. [PubMed] |
| 46. | Wang X, Lupardus P, Laporte SL, Garcia KC. Structural biology of shared cytokine receptors. Annu Rev Immunol. 2009;27:29-60. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 339] [Cited by in RCA: 306] [Article Influence: 19.1] [Reference Citation Analysis (0)] |
| 47. | Aithal GP, Craggs A, Day CP, Welfare M, Daly AK, Mansfield JC, Hudson M. Role of polymorphisms in the interleukin-10 gene in determining disease susceptibility and phenotype in inflamatory bowel disease. Dig Dis Sci. 2001;46:1520-1525. [PubMed] |
| 48. | Hong J, Leung E, Fraser AG, Merriman TR, Vishnu P, Krissansen GW. IL4, IL10, IL16, and TNF polymorphisms in New Zealand Caucasian Crohn's disease patients. Int J Colorectal Dis. 2008;23:335-337. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 10] [Cited by in RCA: 11] [Article Influence: 0.6] [Reference Citation Analysis (0)] |
| 49. | Parrish-Novak J, Foster DC, Holly RD, Clegg CH. Interleukin-21 and the IL-21 receptor: novel effectors of NK and T cell responses. J Leukoc Biol. 2002;72:856-863. [PubMed] |
| 50. | Márquez A, Orozco G, Martínez A, Palomino-Morales R, Fernández-Arquero M, Mendoza JL, Taxonera C, Díaz-Rubio M, Gómez-García M, Nieto A. Novel association of the interleukin 2-interleukin 21 region with inflammatory bowel disease. Am J Gastroenterol. 2009;104:1968-1975. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 35] [Cited by in RCA: 38] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 51. | Zdanov A. Structural features of the interleukin-10 family of cytokines. Curr Pharm Des. 2004;10:3873-3884. [PubMed] |
| 52. | Deniz G, Erten G, Kücüksezer UC, Kocacik D, Karagiannidis C, Aktas E, Akdis CA, Akdis M. Regulatory NK cells suppress antigen-specific T cell responses. J Immunol. 2008;180:850-857. [PubMed] |
| 53. | Franke A, Balschun T, Karlsen TH, Sventoraityte J, Nikolaus S, Mayr G, Domingues FS, Albrecht M, Nothnagel M, Ellinghaus D. Sequence variants in IL10, ARPC2 and multiple other loci contribute to ulcerative colitis susceptibility. Nat Genet. 2008;40:1319-1323. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 486] [Cited by in RCA: 469] [Article Influence: 27.6] [Reference Citation Analysis (0)] |
| 54. | Doecke JD, Simms LA, Zhao ZZ, Huang N, Hanigan K, Krishnaprasad K, Roberts RL, Andrews JM, Mahy G, Bampton P. Genetic susceptibility in IBD: overlap between ulcerative colitis and Crohn’s disease. Inflamm Bowel Dis. 2013;19:240-245. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 30] [Cited by in RCA: 32] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 55. | Andersen V, Ernst A, Christensen J, Østergaard M, Jacobsen BA, Tjønneland A, Krarup HB, Vogel U. The polymorphism rs3024505 proximal to IL-10 is associated with risk of ulcerative colitis and Crohns disease in a Danish case-control study. BMC Med Genet. 2010;11:82. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 39] [Cited by in RCA: 46] [Article Influence: 3.1] [Reference Citation Analysis (0)] |
| 56. | Turner DM, Williams DM, Sankaran D, Lazarus M, Sinnott PJ, Hutchinson IV. An investigation of polymorphism in the interleukin-10 gene promoter. Eur J Immunogenet. 1997;24:1-8. [PubMed] |
| 57. | Balding J, Livingstone WJ, Conroy J, Mynett-Johnson L, Weir DG, Mahmud N, Smith OP. Inflammatory bowel disease: the role of inflammatory cytokine gene polymorphisms. Mediators Inflamm. 2004;13:181-187. [PubMed] |
| 58. | Amre DK, Mack DR, Morgan K, Israel D, Lambrette P, Costea I, Krupoves A, Fegury H, Dong J, Grimard G. Interleukin 10 (IL-10) gene variants and susceptibility for paediatric onset Crohn’s disease. Aliment Pharmacol Ther. 2009;29:1025-1031. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 35] [Cited by in RCA: 38] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 59. | Koss K, Satsangi J, Fanning GC, Welsh KI, Jewell DP. Cytokine (TNF alpha, LT alpha and IL-10) polymorphisms in inflammatory bowel diseases and normal controls: differential effects on production and allele frequencies. Genes Immun. 2000;1:185-190. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 150] [Cited by in RCA: 164] [Article Influence: 6.6] [Reference Citation Analysis (0)] |
| 60. | Klein W, Tromm A, Griga T, Fricke H, Folwaczny C, Hocke M, Eitner K, Marx M, Runte M, Epplen JT. The IL-10 gene is not involved in the predisposition to inflammatory bowel disease. Electrophoresis. 2000;21:3578-3582. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 3] [Reference Citation Analysis (0)] |
| 61. | Fernandez L, Martinez A, Mendoza JL, Urcelay E, Fernandez-Arquero M, Garcia-Paredes J, Diaz-Rubio M, de la Concha EG. Interleukin-10 polymorphisms in Spanish patients with IBD. Inflamm Bowel Dis. 2005;11:739-743. [PubMed] |
| 62. | Marrakchi R, Moussa A, Ouerhani S, Bougatef K, Bouhaha R, Messai Y, Rouissi K, Khadimallah I, Khodjet-el-Khil H, Najar T. Interleukin 10 promoter region polymorphisms in inflammatory bowel disease in Tunisian population. Inflamm Res. 2009;58:155-160. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 6] [Cited by in RCA: 9] [Article Influence: 0.6] [Reference Citation Analysis (0)] |
| 63. | Garza-González E, Pérez-Pérez GI, Mendoza-Ibarra SI, Flores-Gutiérrez JP, Bosques-Padilla FJ. Genetic risk factors for inflammatory bowel disease in a North-eastern Mexican population. Int J Immunogenet. 2010;37:355-359. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 16] [Cited by in RCA: 21] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 64. | Tedde A, Laura Putignano A, Bagnoli S, Congregati C, Milla M, Sorbi S, Genuardi M, Papi L. Interleukin-10 promoter polymorphisms influence susceptibility to ulcerative colitis in a gender-specific manner. Scand J Gastroenterol. 2008;43:712-718. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 35] [Cited by in RCA: 39] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 65. | Ahirwar DK, Kesarwani P, Singh R, Ghoshal UC, Mittal RD. Role of tumor necrosis factor-alpha (C-863A) polymorphism in pathogenesis of inflammatory bowel disease in Northern India. J Gastrointest Cancer. 2012;43:196-204. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 12] [Cited by in RCA: 16] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 66. | Kim EJ, Chung WC, Lee KM, Paik CN, Jung SH, Lee BI, Chae HS, Choi KY. Association between toll-like receptors/CD14 gene polymorphisms and inflammatory bowel disease in Korean population. J Korean Med Sci. 2012;27:72-77. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 19] [Cited by in RCA: 23] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 67. | Zhu H, Lei X, Liu Q, Wang Y. Interleukin-10-1082A/G polymorphism and inflammatory bowel disease susceptibility: a meta-analysis based on 17,585 subjects. Cytokine. 2013;61:146-153. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 28] [Cited by in RCA: 34] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 68. | Sanchez R, Levy E, Costea F, Sinnett D. IL-10 and TNF-alpha promoter haplotypes are associated with childhood Crohn’s disease location. World J Gastroenterol. 2009;15:3776-3782. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 33] [Cited by in RCA: 46] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 69. | Oral HB, Kotenko SV, Yilmaz M, Mani O, Zumkehr J, Blaser K, Akdis CA, Akdis M. Regulation of T cells and cytokines by the interleukin-10 (IL-10)-family cytokines IL-19, IL-20, IL-22, IL-24 and IL-26. Eur J Immunol. 2006;36:380-388. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 99] [Cited by in RCA: 110] [Article Influence: 5.8] [Reference Citation Analysis (0)] |
| 70. | Commins S, Steinke JW, Borish L. The extended IL-10 superfamily: IL-10, IL-19, IL-20, IL-22, IL-24, IL-26, IL-28, and IL-29. J Allergy Clin Immunol. 2008;121:1108-1111. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 247] [Cited by in RCA: 241] [Article Influence: 14.2] [Reference Citation Analysis (0)] |
| 71. | McGovern DP, Gardet A, Törkvist L, Goyette P, Essers J, Taylor KD, Neale BM, Ong RT, Lagacé C, Li C. Genome-wide association identifies multiple ulcerative colitis susceptibility loci. Nat Genet. 2010;42:332-337. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 525] [Cited by in RCA: 518] [Article Influence: 34.5] [Reference Citation Analysis (0)] |
| 72. | Pène J, Chevalier S, Preisser L, Vénéreau E, Guilleux MH, Ghannam S, Molès JP, Danger Y, Ravon E, Lesaux S. Chronically inflamed human tissues are infiltrated by highly differentiated Th17 lymphocytes. J Immunol. 2008;180:7423-7430. [PubMed] |
| 73. | Dambacher J, Beigel F, Zitzmann K, De Toni EN, Göke B, Diepolder HM, Auernhammer CJ, Brand S. The role of the novel Th17 cytokine IL-26 in intestinal inflammation. Gut. 2009;58:1207-1217. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 167] [Cited by in RCA: 187] [Article Influence: 11.7] [Reference Citation Analysis (0)] |
| 74. | Silverberg MS, Cho JH, Rioux JD, McGovern DP, Wu J, Annese V, Achkar JP, Goyette P, Scott R, Xu W. Ulcerative colitis-risk loci on chromosomes 1p36 and 12q15 found by genome-wide association study. Nat Genet. 2009;41:216-220. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 317] [Cited by in RCA: 320] [Article Influence: 20.0] [Reference Citation Analysis (0)] |
| 75. | Vignali DA, Kuchroo VK. IL-12 family cytokines: immunological playmakers. Nat Immunol. 2012;13:722-728. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 773] [Cited by in RCA: 977] [Article Influence: 75.2] [Reference Citation Analysis (0)] |
| 76. | Hsieh CS, Macatonia SE, Tripp CS, Wolf SF, O’Garra A, Murphy KM. Development of TH1 CD4+ T cells through IL-12 produced by Listeria-induced macrophages. Science. 1993;260:547-549. [PubMed] |
| 77. | Wolf SF, Sieburth D, Sypek J. Interleukin 12: a key modulator of immune function. Stem Cells. 1994;12:154-168. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 131] [Cited by in RCA: 128] [Article Influence: 4.1] [Reference Citation Analysis (0)] |
| 78. | Parham C, Chirica M, Timans J, Vaisberg E, Travis M, Cheung J, Pflanz S, Zhang R, Singh KP, Vega F. A receptor for the heterodimeric cytokine IL-23 is composed of IL-12Rbeta1 and a novel cytokine receptor subunit, IL-23R. J Immunol. 2002;168:5699-5708. [PubMed] |
| 79. | Glas J, Seiderer J, Wagner J, Olszak T, Fries C, Tillack C, Friedrich M, Beigel F, Stallhofer J, Steib C. Analysis of IL12B gene variants in inflammatory bowel disease. PLoS One. 2012;7:e34349. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 44] [Cited by in RCA: 47] [Article Influence: 3.6] [Reference Citation Analysis (0)] |
| 80. | Márquez A, Mendoza JL, Taxonera C, Díaz-Rubio M, De La Concha EG, Urcelay E, Martínez A. IL23R and IL12B polymorphisms in Spanish IBD patients: no evidence of interaction. Inflamm Bowel Dis. 2008;14:1192-1196. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 33] [Cited by in RCA: 33] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 81. | Yamazaki K, Takahashi A, Takazoe M, Kubo M, Onouchi Y, Fujino A, Kamatani N, Nakamura Y, Hata A. Positive association of genetic variants in the upstream region of NKX2-3 with Crohn’s disease in Japanese patients. Gut. 2009;58:228-232. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 49] [Cited by in RCA: 57] [Article Influence: 3.6] [Reference Citation Analysis (0)] |
| 82. | Ferguson LR, Han DY, Fraser AG, Huebner C, Lam WJ, Morgan AR. IL23R and IL12B SNPs and Haplotypes Strongly Associate with Crohn's Disease Risk in a New Zealand Population. Gastroenterol Res Pract. 2010;2010:539461. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 24] [Cited by in RCA: 29] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 83. | Fisher SA, Tremelling M, Anderson CA, Gwilliam R, Bumpstead S, Prescott NJ, Nimmo ER, Massey D, Berzuini C, Johnson C. Genetic determinants of ulcerative colitis include the ECM1 locus and five loci implicated in Crohn’s disease. Nat Genet. 2008;40:710-712. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 333] [Cited by in RCA: 338] [Article Influence: 19.9] [Reference Citation Analysis (0)] |
| 84. | Bin C, Zhirong Z, Xiaoqin W, Minhu C, Mei L, Xiang G, Baili C, Pinjin H. Contribution of rs11465788 in IL23R gene to Crohn’s disease susceptibility and phenotype in Chinese population. J Genet. 2009;88:191-196. [PubMed] |
| 85. | Cargill M, Schrodi SJ, Chang M, Garcia VE, Brandon R, Callis KP, Matsunami N, Ardlie KG, Civello D, Catanese JJ. A large-scale genetic association study confirms IL12B and leads to the identification of IL23R as psoriasis-risk genes. Am J Hum Genet. 2007;80:273-290. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 858] [Cited by in RCA: 855] [Article Influence: 47.5] [Reference Citation Analysis (0)] |
| 86. | Sáfrány E, Pazár B, Csöngei V, Járomi L, Polgár N, Sipeky C, Horváth IF, Zeher M, Poór G, Melegh B. Variants of the IL23R gene are associated with ankylosing spondylitis but not with Sjögren syndrome in Hungarian population samples. Scand J Immunol. 2009;70:68-74. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 52] [Cited by in RCA: 56] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 87. | Safrany E, Szell M, Csongei V, Jaromi L, Sipeky C, Szabo T, Kemeny L, Nagy J, Melegh B. Polymorphisms of the IL23R gene are associated with psoriasis but not with immunoglobulin A nephropathy in a Hungarian population. Inflammation. 2011;34:603-608. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 14] [Cited by in RCA: 18] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 88. | Safrany E, Melegh B. Functional variants of the interleukin-23 receptor gene in non-gastrointestinal autoimmune diseases. Curr Med Chem. 2009;16:3766-3774. [PubMed] |
| 89. | Duerr RH, Taylor KD, Brant SR, Rioux JD, Silverberg MS, Daly MJ, Steinhart AH, Abraham C, Regueiro M, Griffiths A. A genome-wide association study identifies IL23R as an inflammatory bowel disease gene. Science. 2006;314:1461-1463. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2428] [Cited by in RCA: 2300] [Article Influence: 121.1] [Reference Citation Analysis (0)] |
| 90. | Yang SK, Park M, Lim J, Park SH, Ye BD, Lee I, Song K. Contribution of IL23R but not ATG16L1 to Crohn’s disease susceptibility in Koreans. Inflamm Bowel Dis. 2009;15:1385-1390. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 60] [Cited by in RCA: 45] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 91. | Glas J, Seiderer J, Wetzke M, Konrad A, Török HP, Schmechel S, Tonenchi L, Grassl C, Dambacher J, Pfennig S. rs1004819 is the main disease-associated IL23R variant in German Crohn’s disease patients: combined analysis of IL23R, CARD15, and OCTN1/2 variants. PLoS One. 2007;2:e819. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 99] [Cited by in RCA: 113] [Article Influence: 6.3] [Reference Citation Analysis (0)] |
| 92. | Baptista ML, Amarante H, Picheth G, Sdepanian VL, Peterson N, Babasukumar U, Lima HC, Kugathasan S. CARD15 and IL23R influences Crohn’s disease susceptibility but not disease phenotype in a Brazilian population. Inflamm Bowel Dis. 2008;14:674-679. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 36] [Cited by in RCA: 39] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 93. | Yu P, Shen F, Zhang X, Cao R, Zhao X, Liu P, Tu H, Yang X, Shi R, Zhang H. Association of single nucleotide polymorphisms of IL23R and IL17 with ulcerative colitis risk in a Chinese Han population. PLoS One. 2012;7:e44380. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 18] [Cited by in RCA: 23] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 94. | Zhao XD, Shen FC, Zhang HJ, Shen XY, Wang YM, Yang XZ, Tu HM, Tai YH, Shi RH. [Association of interleukin-23 receptor gene polymorphisms with susceptibility and phenotypes of inflammatory bowel diseases in Jiangsu Han population]. Zhonghua Nei Ke Zazhi. 2011;50:935-941. [PubMed] |
| 95. | Cummings JR, Ahmad T, Geremia A, Beckly J, Cooney R, Hancock L, Pathan S, Guo C, Cardon LR, Jewell DP. Contribution of the novel inflammatory bowel disease gene IL23R to disease susceptibility and phenotype. Inflamm Bowel Dis. 2007;13:1063-1068. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 70] [Cited by in RCA: 71] [Article Influence: 3.9] [Reference Citation Analysis (0)] |
| 96. | Oliver J, Rueda B, López-Nevot MA, Gómez-García M, Martín J. Replication of an association between IL23R gene polymorphism with inflammatory bowel disease. Clin Gastroenterol Hepatol. 2007;5:977-81, 981.e1-2. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 45] [Cited by in RCA: 49] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 97. | Lacher M, Schroepf S, Helmbrecht J, von Schweinitz D, Ballauff A, Koch I, Lohse P, Osterrieder S, Kappler R, Koletzko S. Association of the interleukin-23 receptor gene variant rs11209026 with Crohn’s disease in German children. Acta Paediatr. 2010;99:727-733. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 10] [Cited by in RCA: 12] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 98. | Safrany E, Szabo M, Szell M, Kemeny L, Sumegi K, Melegh BI, Magyari L, Matyas P, Figler M, Weber A. Difference of interleukin-23 receptor gene haplotype variants in ulcerative colitis compared to Crohn’s disease and psoriasis. Inflamm Res. 2013;62:195-200. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 22] [Cited by in RCA: 26] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 99. | Csöngei V, Járomi L, Sáfrány E, Sipeky C, Magyari L, Faragó B, Bene J, Polgár N, Lakner L, Sarlós P. Interaction of the major inflammatory bowel disease susceptibility alleles in Crohn’s disease patients. World J Gastroenterol. 2010;16:176-183. [PubMed] |
| 100. | Faragó B, Magyari L, Sáfrány E, Csöngei V, Járomi L, Horvatovich K, Sipeky C, Maász A, Radics J, Gyetvai A. Functional variants of interleukin-23 receptor gene confer risk for rheumatoid arthritis but not for systemic sclerosis. Ann Rheum Dis. 2008;67:248-250. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 98] [Cited by in RCA: 103] [Article Influence: 6.1] [Reference Citation Analysis (0)] |
| 101. | Gong F, Pan YH, Huang X, Chen J, Xiao JH, Zhu HY. Interleukin-27 as a potential therapeutic target for rheumatoid arthritis: has the time come? Clin Rheumatol. 2013;32:1425-1428. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 7] [Cited by in RCA: 6] [Article Influence: 0.5] [Reference Citation Analysis (0)] |
| 102. | Zeitvogel J, Werfel T, Wittmann M. IL-27 acts as a priming signal for IL-23 but not IL-12 production on human antigen-presenting cells. Exp Dermatol. 2012;21:426-430. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 8] [Cited by in RCA: 10] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 103. | Li CS, Zhang Q, Lee KJ, Cho SW, Lee KM, Hahm KB, Choi SC, Yun KJ, Chung HT, Chae SC. Interleukin-27 polymorphisms are associated with inflammatory bowel diseases in a Korean population. J Gastroenterol Hepatol. 2009;24:1692-1696. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 61] [Cited by in RCA: 61] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 104. | Imielinski M, Baldassano RN, Griffiths A, Russell RK, Annese V, Dubinsky M, Kugathasan S, Bradfield JP, Walters TD, Sleiman P. Common variants at five new loci associated with early-onset inflammatory bowel disease. Nat Genet. 2009;41:1335-1340. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 405] [Cited by in RCA: 388] [Article Influence: 24.3] [Reference Citation Analysis (0)] |
| 105. | Heinrich PC, Behrmann I, Haan S, Hermanns HM, Müller-Newen G, Schaper F. Principles of interleukin (IL)-6-type cytokine signalling and its regulation. Biochem J. 2003;374:1-20. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2296] [Cited by in RCA: 2394] [Article Influence: 108.8] [Reference Citation Analysis (0)] |
| 106. | Boulanger MJ, Chow DC, Brevnova EE, Garcia KC. Hexameric structure and assembly of the interleukin-6/IL-6 alpha-receptor/gp130 complex. Science. 2003;300:2101-2104. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 455] [Cited by in RCA: 504] [Article Influence: 22.9] [Reference Citation Analysis (0)] |
| 107. | Sawczenko A, Azooz O, Paraszczuk J, Idestrom M, Croft NM, Savage MO, Ballinger AB, Sanderson IR. Intestinal inflammation-induced growth retardation acts through IL-6 in rats and depends on the -174 IL-6 G/C polymorphism in children. Proc Natl Acad Sci USA. 2005;102:13260-13265. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 94] [Cited by in RCA: 88] [Article Influence: 4.4] [Reference Citation Analysis (0)] |
| 108. | Cantor MJ, Nickerson P, Bernstein CN. The role of cytokine gene polymorphisms in determining disease susceptibility and phenotype in inflammatory bowel disease. Am J Gastroenterol. 2005;100:1134-1142. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 65] [Cited by in RCA: 73] [Article Influence: 3.7] [Reference Citation Analysis (0)] |
| 109. | Guerreiro CS, Ferreira P, Tavares L, Santos PM, Neves M, Brito M, Cravo M. Fatty acids, IL6, and TNFalpha polymorphisms: an example of nutrigenetics in Crohn’s disease. Am J Gastroenterol. 2009;104:2241-2249. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 46] [Cited by in RCA: 48] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 110. | Klein W, Tromm A, Griga T, Fricke H, Folwaczny C, Hocke M, Eitner K, Marx M, Epplen JT. The polymorphism at position -174 of the IL-6 gene is not associated with inflammatory bowel disease. Eur J Gastroenterol Hepatol. 2001;13:45-47. [PubMed] |
| 111. | Li H, Chen J, Huang A, Stinson J, Heldens S, Foster J, Dowd P, Gurney AL, Wood WI. Cloning and characterization of IL-17B and IL-17C, two new members of the IL-17 cytokine family. Proc Natl Acad Sci USA. 2000;97:773-778. [PubMed] |
| 112. | Shi Y, Ullrich SJ, Zhang J, Connolly K, Grzegorzewski KJ, Barber MC, Wang W, Wathen K, Hodge V, Fisher CL. A novel cytokine receptor-ligand pair. Identification, molecular characterization, and in vivo immunomodulatory activity. J Biol Chem. 2000;275:19167-19176. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 163] [Cited by in RCA: 171] [Article Influence: 6.8] [Reference Citation Analysis (0)] |
| 113. | Lee J, Ho WH, Maruoka M, Corpuz RT, Baldwin DT, Foster JS, Goddard AD, Yansura DG, Vandlen RL, Wood WI. IL-17E, a novel proinflammatory ligand for the IL-17 receptor homolog IL-17Rh1. J Biol Chem. 2001;276:1660-1664. [PubMed] |
| 114. | Starnes T, Broxmeyer HE, Robertson MJ, Hromas R. Cutting edge: IL-17D, a novel member of the IL-17 family, stimulates cytokine production and inhibits hemopoiesis. J Immunol. 2002;169:642-646. [PubMed] |
| 115. | Kawaguchi M, Adachi M, Oda N, Kokubu F, Huang SK. IL-17 cytokine family. J Allergy Clin Immunol. 2004;114:1265-173; quiz 1274. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 248] [Cited by in RCA: 245] [Article Influence: 12.3] [Reference Citation Analysis (0)] |
| 116. | Kobori A, Yagi Y, Imaeda H, Ban H, Bamba S, Tsujikawa T, Saito Y, Fujiyama Y, Andoh A. Interleukin-33 expression is specifically enhanced in inflamed mucosa of ulcerative colitis. J Gastroenterol. 2010;45:999-1007. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 183] [Cited by in RCA: 204] [Article Influence: 13.6] [Reference Citation Analysis (0)] |
| 117. | Seiderer J, Elben I, Diegelmann J, Glas J, Stallhofer J, Tillack C, Pfennig S, Jürgens M, Schmechel S, Konrad A. Role of the novel Th17 cytokine IL-17F in inflammatory bowel disease (IBD): upregulated colonic IL-17F expression in active Crohn’s disease and analysis of the IL17F p.His161Arg polymorphism in IBD. Inflamm Bowel Dis. 2008;14:437-445. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 246] [Cited by in RCA: 271] [Article Influence: 15.9] [Reference Citation Analysis (0)] |
| 118. | Schwartz S, Beaulieu JF, Ruemmele FM. Interleukin-17 is a potent immuno-modulator and regulator of normal human intestinal epithelial cell growth. Biochem Biophys Res Commun. 2005;337:505-509. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 34] [Cited by in RCA: 39] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 119. | Gaffen SL. Structure and signalling in the IL-17 receptor family. Nat Rev Immunol. 2009;9:556-567. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1174] [Cited by in RCA: 1130] [Article Influence: 70.6] [Reference Citation Analysis (0)] |
| 120. | Arisawa T, Tahara T, Shibata T, Nagasaka M, Nakamura M, Kamiya Y, Fujita H, Nakamura M, Yoshioka D, Arima Y. The influence of polymorphisms of interleukin-17A and interleukin-17F genes on the susceptibility to ulcerative colitis. J Clin Immunol. 2008;28:44-49. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 138] [Cited by in RCA: 146] [Article Influence: 8.1] [Reference Citation Analysis (0)] |
| 121. | Yoshimura T, Matsushima K, Tanaka S, Robinson EA, Appella E, Oppenheim JJ, Leonard EJ. Purification of a human monocyte-derived neutrophil chemotactic factor that has peptide sequence similarity to other host defense cytokines. Proc Natl Acad Sci USA. 1987;84:9233-9237. [PubMed] |
| 122. | Holmes WE, Lee J, Kuang WJ, Rice GC, Wood WI. Structure and functional expression of a human interleukin-8 receptor. Science. 1991;253:1278-1280. [PubMed] |
| 123. | Miller MD, Krangel MS. Biology and biochemistry of the chemokines: a family of chemotactic and inflammatory cytokines. Crit Rev Immunol. 1992;12:17-46. [PubMed] |
| 124. | Walczak A, Przybylowska K, Dziki L, Sygut A, Chojnacki C, Chojnacki J, Dziki A, Majsterek I. The lL-8 and IL-13 gene polymorphisms in inflammatory bowel disease and colorectal cancer. DNA Cell Biol. 2012;31:1431-1438. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 21] [Cited by in RCA: 24] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 125. | Li K, Yao S, Liu S, Wang B, Mao D. Genetic polymorphisms of interleukin 8 and risk of ulcerative colitis in the Chinese population. Clin Chim Acta. 2009;405:30-34. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 14] [Cited by in RCA: 18] [Article Influence: 1.1] [Reference Citation Analysis (0)] |
| 126. | Theodore AC, Center DM, Nicoll J, Fine G, Kornfeld H, Cruikshank WW. CD4 ligand IL-16 inhibits the mixed lymphocyte reaction. J Immunol. 1996;157:1958-1964. [PubMed] |
| 127. | Keates AC, Castagliuolo I, Cruickshank WW, Qiu B, Arseneau KO, Brazer W, Kelly CP. Interleukin 16 is up-regulated in Crohn’s disease and participates in TNBS colitis in mice. Gastroenterology. 2000;119:972-982. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 65] [Cited by in RCA: 67] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 128. | Glas J, Török HP, Unterhuber H, Radlmayr M, Folwaczny C. The -295T-to-C promoter polymorphism of the IL-16 gene is associated with Crohn’s disease. Clin Immunol. 2003;106:197-200. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 40] [Cited by in RCA: 41] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 129. | Rothenberg ME. Biology and treatment of eosinophilic esophagitis. Gastroenterology. 2009;137:1238-1249. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 215] [Cited by in RCA: 191] [Article Influence: 11.9] [Reference Citation Analysis (0)] |
| 130. | Minty A, Ferrara P, Caput D. Interleukin-13 effects on activated monocytes lead to novel cytokine secretion profiles intermediate between those induced by interleukin-10 and by interferon-gamma. Eur Cytokine Netw. 1997;8:189-201. [PubMed] |