Published online Dec 21, 2013. doi: 10.3748/wjg.v19.i47.8949
Revised: November 10, 2013
Accepted: November 28, 2013
Published online: December 21, 2013
In this review, we discuss recent advances in nucleic acid-based therapeutic technologies that target hepatitis C virus (HCV) infection. Because the HCV genome is present exclusively in RNA form during replication, various nucleic acid-based therapeutic approaches targeting the HCV genome, such as ribozymes, aptamers, siRNAs, and antisense oligonucleotides, have been suggested as potential tools against HCV. Nucleic acids are potentially immunogenic and typically require a delivery tool to be utilized as therapeutics. These limitations have hampered the clinical development of nucleic acid-based therapeutics. However, despite these limitations, nucleic acid-based therapeutics has clinical value due to their great specificity, easy and large-scale synthesis with chemical methods, and pharmaceutical flexibility. Moreover, nucleic acid therapeutics are expected to broaden the range of targetable molecules essential for the HCV replication cycle, and therefore they may prove to be more effective than existing therapeutics, such as interferon-α and ribavirin combination therapy. This review focuses on the current status and future prospects of ribozymes, aptamers, siRNAs, and antisense oligonucleotides as therapeutic reagents against HCV.
Core tip: Nucleic acids have emerged as new anti-hepatitis C virus (HCV) agents due to their great specificity, chemical synthesizability, pharmaceutical amenability, and broad targeting ability. Clinical applications of nucleic acids have been delayed due to their potential immunogenicity and toxicity, low efficacy, possible off-target effects, and lack of efficient delivery vehicles. However, recent advances in delivery carriers and chemical modification methods have improved the efficacy and bioavailability of nucleic acid-based agents. Hence, nucleic acids may be attractive anti-HCV options. In this report, the current status and future prospects of ribozymes, aptamers, siRNAs, and antisense oligonucleotides as anti-HCV regimens will be discussed.
- Citation: Lee CH, Kim JH, Lee SW. Prospects for nucleic acid-based therapeutics against hepatitis C virus. World J Gastroenterol 2013; 19(47): 8949-8962
- URL: https://www.wjgnet.com/1007-9327/full/v19/i47/8949.htm
- DOI: https://dx.doi.org/10.3748/wjg.v19.i47.8949
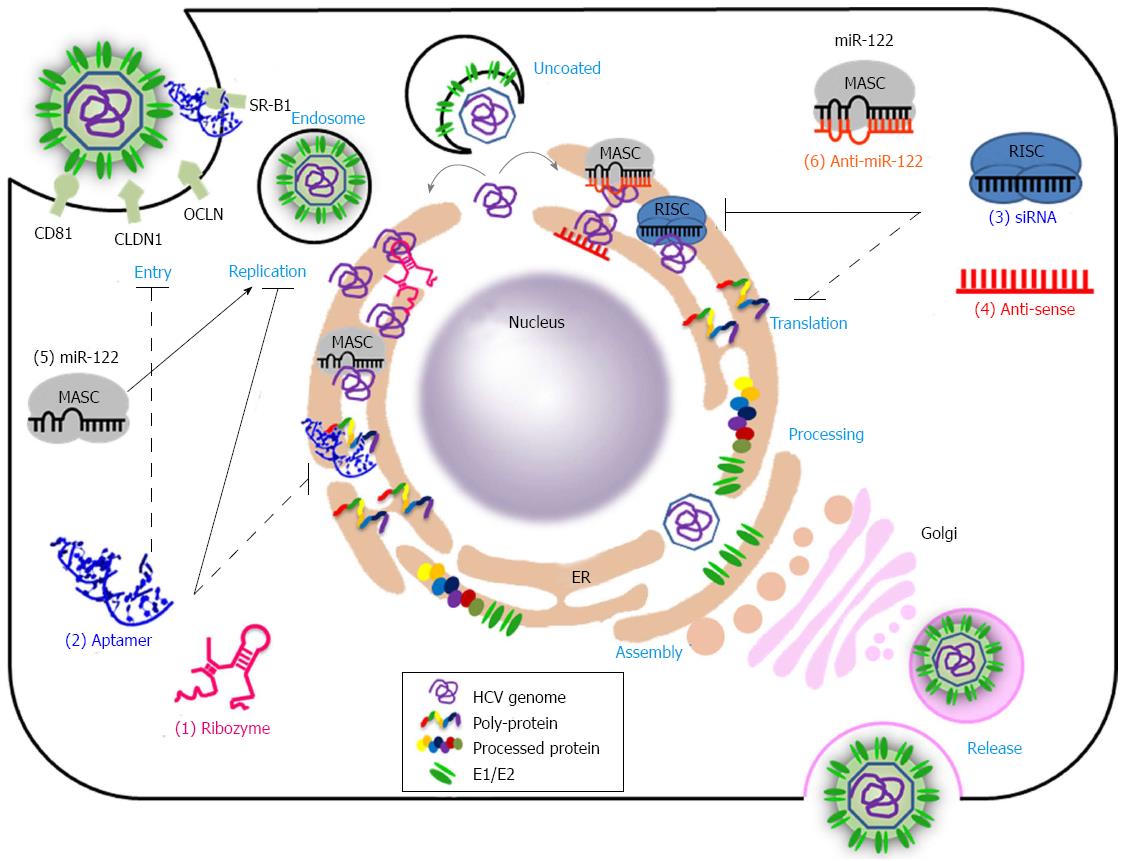
Hepatitis C Virus (HCV) infection is the main cause of chronic hepatitis, liver cirrhosis, and hepatocellular carcinoma[1,2]. Nearly 170 million people are chronically infected worldwide by HCV, and approximately 27% of all cases of liver cirrhosis and approximately 25% of hepatocellular carcinoma cases may be related to HCV infection[3]. Given this obvious therapeutic need, international efforts to develop new antiviral drugs and vaccines that are effective against all HCV genotypes have been prompted. However, HCV has seven major genotypes with numerous subtypes[4] and exists as a variable quasispecies because HCV NS5B displays an error-prone RNA-dependent RNA polymerase activity that lacks proofreading functions[5]. Unfortunately, this high variability in HCV genomic RNA hampers the development of prophylactic and therapeutic vaccines and antiviral drugs[5,6]. Until recently, the usual treatment option for HCV infection has been a combination of pegylated interferon-α (PEG-IFNα) and ribavirin. This treatment clears infections by genotypes 2 and 3 in up to approximately 85% of cases. However, in infections with genotype 1, approximately 45% of cases are able to support a sustained viral response after the combination treatment[7]. Moreover, this treatment is associated with many side effects including flu-like symptoms, severe depression and hemolytic anemia[8]. Recent approval of two direct-acting antivirals (DAA) targeting the HCV NS3 protease, telaprevir (VX-950) and boceprevir, gives hope for the treatment of HCV infection. However, these drugs, given in combination with PEG-IFNα and ribavirin, are prone to selecting for drug-resistant viruses[9,10]. Therefore, DAAs that are more specific, effective, and safer are required. Over the last three decades, nucleic acids have been developed as potential antiviral therapeutic agents. Nucleic acid-based drugs are theoretically capable of targeting many types of molecules such as DNA, RNA, protein, lipid and even small molecules[11]. This property could overcome the limitations of the current therapeutics, which target only a limited number of proteins. Nucleic acid-based agents bind to target molecules through sequence complementarity (antisense oligonucleotide, siRNA, ribozyme, and antimiR) or on the basis of three dimensional structure (aptamer) (Figure 1 and Table 1)[12]. For example, aptamers bind to target molecules and function as decoys and/or inhibitors, whereas siRNAs and miRNAs make use of the RNA-induced gene silencing complex (RISC), which induces target RNA cleavage or translation inhibition[13]. AntimiRs block miRNA activity and thus induce expression of miRNA target genes[14]. Antisense oligonucleotides bind to complementary RNAs and suppress access to the cellular machinery, thereby inhibiting expression or function of the targeted RNAs[12]. Ribozymes are catalytic RNAs that cleave target RNAs (for example, hairpin ribozyme and hammerhead ribozyme) or selectively replace target RNAs with desirable RNAs (trans-splicing ribozyme)[15]. These variable modes of action provide many opportunities and options for the treatment of intractable diseases including genetic disorders, cancers, and infectious diseases. Despite their great potential, only a few nucleic acid-based therapeutics have been approved; these include fomivirsen (an antisense oligonucleotide drug for the treatment of cytomegalovirus retinitis in patients with AIDS), pegaptanib (an aptamer for combating wet age-related macular degeneration), and mipomersen (an antisense oligonucleotide drug for the treatment of homozygous familial hypercholesterolemia)[16-18]. The problems involved in the application of RNA therapeutic agents include potential immunogenicity, inherent unstable nature, and the requirement for a delivery tool[11]. However, recent technological advances, such as the improvement of synthetic delivery carriers and the chemical modifications of nucleic acids, may help to overcome these obstacles. Recently, a phase II clinical trial with SPC3649 (formerly Miravirsen), an LNA-modified antimiR-122, was completed for the treatment of HCV[19]. Many other nucleic acid-based anti-HCV therapeutics are in the pre-clinical and clinical stage. In this review, we summarize the current status of nucleic acid-based therapeutics that target the HCV RNA genome or HCV-encoded proteins. Moreover, we summarize their mechanisms of action and discuss the prospects for their future application to the treatment of HCV infections.
| Class | Mode of action | Target | Status |
| Hairpin ribozyme | Cleave target RNAs | 5'-UTR[24,28], 3'-UTR[28], and core region[24] | Tested in in vitro[24] and in cell culture model[28] |
| HDV ribozyme | 5'-UTR[30] | Tested in in vitro | |
| Hammerhead ribozyme (Hepatozyme) | 5'-UTR[33-35] | Completion of phase II | |
| DNAzyme | 5'-UTR[41], core and NS5B region[39,40] | Tested in in vitro and in cell culture model | |
| RNase P | 5'-UTR[45-47] | ||
| Splicing riboyme | Selectively replace target RNAs with desirable RNAs | 5'-UTR[51] | |
| Allosteric ribozyme | Inhibit HCV replication[54] or cleave HCV RNA[31] through recognizing ligands | miR-122[54] and 5'-UTR[31] | |
| Aptamer | Bind to target molecule and function as decoys and/or inhibitors | NS3[68-70] | |
| NS5B[75-78] | |||
| E1E2[79] | |||
| Viral RNA[80,81] | |||
| RNAi | Target RNA cleavage or translation inhibition | 5'-UTR and 3'-UTR[93-96,100-102] | |
| Protein coding regions[103-109] | |||
| Antisense oligonucleotide | Bind to complementary RNAs and suppress the access to cellular machinery, thereby inhibiting expression or function of the targeted RNAs | 5'-UTR[134-137] | Completion of Phase II |
| AntimiR | Block miRNA activity | miR-122[19,138] | Completion of Phase II |
A ribozyme is a catalytic RNA that cleaves or reprograms a target RNA sequence specifically, thus inhibiting the target RNA’s expression or inducing new therapeutic gene expression only when the target RNA exists[20]. Since HCV has an RNA genome that replicates and exists exclusively in the cytoplasm, ribozymes are an attractive therapeutic option for HCV RNA clearance in infected cells. As HCV NS5B is an error-prone RNA-dependent RNA polymerase that lacks proofreading functions, viral replication is accompanied by the occurrence of mutations[5]. Therefore, sequence-specific therapeutics may induce escape mutant viruses. To overcome this obstacle, most ribozymes against the HCV genome have been designed to specifically target the HCV 5’- or 3’-untranslated regions (UTRs), which are highly conserved among all HCV genotypes and are essential for viral replication[21]. Promising in vitro results were obtained in the 1990s, using ribozymes directed against the HCV 5’- and 3’-UTRs[22-25]. Typically, naturally occurring ribozymes can be categorized into two groups depending on their mechanism of action: cleaving ribozymes (RNase P, hairpin ribozyme, and hammerhead ribozyme) and splicing ribozymes (group I and II introns)[20]. For therapeutic tools against HCV infection, researchers have modified and optimized these naturally occurring ribozymes or have engineered synthetic ribozymes to target the HCV 5’- or 3’-UTRs.
Cleaving ribozymes are divided into two subgroups according to their natural traits: self-cleaving or trans-cleaving[20]. Hairpin, hepatitis delta virus and hammerhead ribozymes are all naturally self-cleaving ribozymes required for the replication process of RNA genomes. RNase P is an essential enzyme in the biosynthesis of tRNAs that specifically cleaves the pre-tRNAs, releasing 5’-sequences and mature tRNAs. Except for the plant chloroplast and Trypanosomatid enzymes, all known RNase P enzymes are ribonucleoproteins that contain an RNA subunit essential for the catalysis[26].
Hairpin ribozymes consist of four helical domains and five loops. The cleavage site is flanked by the two substrate-binding sequences formed between the target RNA and the ribozyme, allowing the design of trans-acting ribozymes for target RNA sequence-specific cleavage[27]. The first effort to utilize engineered trans-cleaving hairpin ribozymes occurred in the 1990s[24]. Trans-cleaving hairpin ribozymes targeting HCV RNA 5’-UTR and capsid gene regions were generated and shown to inhibit the expression of a cotransfected reporter gene containing the HCV RNA target sequences. However, these ribozymes were not tested using an HCV cell culture replication system, such as the subgenomic replicon or the virus particle-producing JFH-1 system[6], due to the unavailability of those systems at that time. Therefore, the effects of these ribozymes on HCV replication are unknown. Recently, other hairpin ribozymes targeting the HCV 5’- or 3’-UTRs were reported[28]. In Huh-7 cells that stably express subgenomic HCV construct, I389/hyg-ubi/NS3-3’ 5.1, the inhibitory efficacy of these ribozymes on HCV replication was minimal (approximately 30%-40%). However, the inhibitory effects of these ribozymes were increased when combined with an HCV 3’-UTR targeting ribozyme (rather than HCV 5’-UTR targeting) and an HCV 5’-UTR targeting siRNA. This approach reduced HCV RNA and NS5B protein levels by 80%-90%. This result offers a promising combinatorial strategy for silencing HCV replication.
Among the different ribozymes, the hepatitis delta virus (HDV) ribozyme is the only catalytic cleaving RNA enzyme that has been discovered in humans[29]. The HDV ribozyme appears to be well adapted to the human cell environment, and hence, is a potential candidate for the development of anti-HCV therapeutics. The HDV ribozyme has been modified and designed to be able to cleave any specific RNA targets in trans possessing a complementary sequence to the recognition sequence of the ribozyme[20]. Unfortunately, the HDV ribozyme has not been developed extensively as an anti-HCV reagent; thus, its catalytic activity has been tested only in an in vitro trans-cleavage assay using the HCV 5’-UTR as a substrate[30]. Recently, an HDV ribozyme possessing a specific on/off adapter (SOFA), named SOFA-HDV ribozyme, was reported[31]. This SOFA-HDV ribozyme is discussed further in the allosteric ribozyme section.
The hammerhead ribozyme is one of the smallest ribozymes and likely the most widely studied ribozyme[26]. Due to its small size, specificity, and catalytic efficacy, the hammerhead ribozyme is the most commonly used ribozyme as a therapeutic agent for human diseases. Many attempts have been made to develop hammerhead ribozymes that can efficiently cleave the HCV RNA genome and inhibit HCV translation and replication[22,23,32]. For example, a nuclease-resistant synthetic ribozyme with modified nucleotides and phosphorothioate linkages that target the HCV 5’-UTR was developed by Ribozyme Pharmaceuticals (RPI) in collaboration with Eli Lilly. This hammerhead ribozyme, named Hepatozyme™, successfully inhibited viral replication in cell culture with a chimeric HCV-poliovirus in a dose-dependent manner, and this effect was potentiated by interferon[33,34]. In a phase I trial, Hepatozyme, administered either by subcutaneous injection or intravenously, was found to be safe[35]. A subsequent phase II trial assessing the safety and efficacy of this ribozyme in combination with IFNα has been initiated. While a reduction of HCV RNA in serum was observed in some patients, RPI opted to discontinue the development of this drug because of toxicology findings in primates[36].
Unfortunately, ribozymes have the disadvantages of being both short lived and prone to losing their biological activity when they encounter alternative base substitutions[37]. Deoxyribozyme (DNAzymes) can be an effective alternative because of their small size (30-40 bases), ease of synthesis, and increased resistance to chemical or nuclease degradation[38]. DNAzymes have been shown to efficiently cleave target RNAs at purine-pyrimidine junctions in vitro. Similar to the RNA-based ribozymes, DNAzymes were usually engineered to target highly conserved sequences in the HCV core and/or the NS5B protein coding region[39,40] and 5’-UTR[41]. Lee et al[42] constructed a pool of 10-23 DNAzymes that possessed randomized annealing arm sequences and then selected the most available site for DNAzyme cleavage. All reported DNAzymes targeting the HCV genome cleaved the target RNA and thus inhibited translation and replication of HCV in the cell culture system. However, the effects were minimal or not superior compared with those produced with RNA-based ribozymes. DNAzymes are yet in their infancy as therapeutics, and further improvements are needed.
Ribonuclease P (RNase P) is a ubiquitous endoribonuclease and is one of the most abundant and efficient enzymes in the cell. RNase P is a ribonucleoprotein complex that specifically cleaves pre-tRNAs, releasing 5’-sequences and mature tRNAs[43]. RNase P requires a short complementary oligonucleotide called an external guide sequence for its activity to recognize and cleave target RNA[44]. As an anti-HCV therapeutic, RNase P has been shown to display cleavage activity against the HCV 5’-UTR in vitro, but has not yet been extensively studied in a cell culture system[45-47]. Therefore, further evidences of its efficacy and safety in cell culture systems are needed to develop RNase P as an anti-HCV drug.
The self-splicing group I intron from Tetrahymena thermophila has been previously demonstrated to trans-splice an exon attached to its 3’ end onto a separate 5’ exon RNA not only in vitro[48] but also in Escherichia coli[49] and mammalian cells[50]. A promising advantage of trans-splicing group I intron is the cleavage of target RNA and the simultaneous induction of new therapeutic gene expression only when target RNA exists. Thus, trans-splicing ribozymes could potentially be used for the selective induction of new antiviral gene activities only in HCV-infected cells while simultaneously destroying the viral RNAs. Our group developed a trans-splicing group I ribozyme targeting the HCV 5’-UTR with the diphtheria toxin A (DTA) gene as a 3’ exon[51]. This trans-splicing ribozyme specifically cleaved the HCV 5’-UTR and ligated DTA RNA to the cleaved HCV 5’-UTR, thus inducing HCV RNA-specific cell death. To further improve the anti-HCV activities and the safety of the trans-splicing ribozyme, a more careful selection of an antiviral gene as 3’ exon, such as interferon instead of DTA, may be required as DTA can cause extensive death of HCV-infected hepatocytes.
An allosteric ribozyme is a ribozyme whose activity can be specifically regulated by ligands. Commonly, ribozymes recognize only a 7-15 nucleotide long target RNA, and thus, the possibility of nonspecific off-target side effects is significant. To overcome this limitation, specific sensing ligands, such as RNAs, proteins or small molecules, can be tagged to the ribozyme to specifically regulate its activity. The SOFA (a specific on/off adapter)-HDV ribozyme and aptazyme have been suggested as representative allosteric ribozymes for HCV therapeutics. The SOFA-HDV ribozyme can switch its cleavage activity from off to on solely in the presence of the desired RNA ligand. The SOFA module is composed of three domains: a blocker, a biosensor, and a stabilizer[52]. The blocker sequence inhibits the cleavage activity of the ribozyme by binding in cis. The biosensor must bind its complementary sequence on the substrate to unlock the SOFA module. This binding induces the folding of the catalytic core of the HDV ribozyme into the on conformation. Both the blocker and the biosensor increase the substrate specificity of the ribozyme’s cleavage by several orders of magnitude, compared with the wild-type HDV ribozyme[53]. Lévesque et al[31] attempted to develop a SOFA-HDV ribozyme to target HCV. They screened and identified the most active SOFA-HDV ribozyme against HCV RNA strands of both polarities. Unfortunately, the inhibition of HCV replication through targeting of the HCV replicon system with the SOFA-HDV ribozymes was not very effective, even though the SOFA-HDV ribozymes were active in an in vitro cleavage assay. Further elucidation of the reasons why SOFA-HDV ribozymes were not active in the cell culture HCV model is needed to optimize their activities against HCV. An aptazyme is composed of three independent modules: aptamer, communication module, and ribozyme. An aptamer binding to its ligand results in conformational change in the communication module, which can induce the on or off status of ribozyme activity. Recently, our group developed a specific aptazyme that can silence miR-122 activity only in HCV-infected cells[54]. MiR-122 is a positive regulator of HCV translation and replication. Functional sequestration of miR-122 effectively reduces the abundance of viral RNA, implicating miR-122 as a potential target for anti-HCV therapeutics[19]. However, miR-122 can also regulate the expression of a large number of genes involved in cellular physiological functions such as lipid metabolism and tumor suppression[55-60]. To overcome any possible nonspecific side effects due to miR-122 silencing in the normal liver, we created a hammerhead ribozyme-based aptazyme that can release antimiR-122 through self-cleavage activity, depending on the presence of the HCV NS5B protein[54]. This HCV NS5B-dependent antimiR-122 releasing aptazyme specifically inhibited miR-122 function only in the HCV-infected cells. Moreover, this aptazyme more efficaciously hampered HCV replication than the miRNA silencing approach did, as it contains an aptamer domain that can specifically bind and sequester the HCV NS5B protein. Through the combination of selective miR-122 silencing and specific sequestering of HCV NS5B, this aptazyme approach could be a promising anti-HCV therapeutic treatment.
Aptamers are small structured single-stranded nucleic acid sequences that have emerged as attractive and feasible alternatives to small molecule and antibody-based therapy, due to their great specificity, high affinity, easy and large-scale synthesis with a chemical method, pharmaceutically flexibility, and poor immunogenicity[61,62]. Aptamers can be evolved using systematic evolution of ligands by exponential enrichment, an iterative selection method, and can bind target proteins with high affinity and specificity[63,64] through formation of well-defined complementary three-dimensional structures[65]. The first aptamer drug, known as pegaptanib (Macugen), was approved for the treatment of wet age-related macular degeneration by the United States FDA[18]. Other aptamer drug candidates now in the clinical development phase include transcription factor decoys and aptamers against thrombin, factor IXa, and nucleolin[62]. Establishment of a robust HCV cell culture system[66,67] has allowed the identification and biochemical characterization of two viral enzymes, NS3-4A and NS5B, that are major targets for antiviral therapeutics. NS3-4A and NS5B are essential proteins for the HCV replication cycle, and therefore, most of the aptamers have been developed against these two viral proteins to clear HCV infection.
The HCV NS3 is a multifunctional protein with three known enzymatic activities. The serine protease activity (in conjunction with cofactor NS4A) is present within the first 180 N-terminal amino acids, while the nucleoside triphosphatase (NTPase) and helicase activities are in the carboxy-terminal region[6]. These three activities are important to HCV replication. Most DAAs, including the FDA-approved VX-950 and boceprevir, target NS3 protease activity, as DAAs targeting the NS3 helicase domain have met with limited success[6]. In contrast, aptamers have been developed to target not only the protease domain[68] but also the helicase[69,70] and NTPase domains. Moreover, simultaneous targeting of protease and helicase activities through conjugation of protease and helicase aptamers is possible[71]. So far, among the NS3 aptamers, only the helicase-specific aptamer developed by our group has been tested for its ability to inhibit HCV replication in an HCV cell culture system[70]. As the NS3 region of the HCV genome may be a hot spot for mutations that are not deleterious to HCV replication, and due to the similarity of the NS3 helicase to cellular RNA helicases[72,73], a more careful examination is required to develop NS3 protease and helicase targeting aptamers as anti-HCV drugs.
The RNA dependent RNA polymerase (RdRp) NS5B is the key enzyme in HCV RNA replication. Due to its essential role in the HCV life cycle, the NS5B protein is an attractive target for the development of specific anti-HCV drugs. Many nucleoside analogues and nonnucleoside inhibitors have been shown to inhibit RdRp activity in vitro, as well as in the replicon cell culture system[74]. Jones et al[75] developed a DNA aptamer against the HCV genotype 3a NS5B protein. They confirmed that selected DNA aptamers specifically inhibited the NS5B polymerase activity of genotype 3a, but not of genotypes 1a and 1b. The therapeutic effectiveness of such aptamers should be carefully considered, as the most prevalent HCV genotype throughout the world is genotype 1. Moreover, their inhibitory effects against HCV should be carefully tested using the genotype 3a cell culture system that has been recently developed[76]. Bellecave et al[77] also reported DNA aptamers against HCV NS5B, and their aptamers inhibited HCV JFH-1 replication and viral particle formation in the cell culture system. However, those aptamers were not examined with regard to cell toxicity profiles, distribution in animals, or side effects during long-term treatment. Therefore, concerns about safety and the possibility of escape mutant virus appearance during repeated treatment cannot be excluded with these DNA aptamers. Recently, our group reported two types of RNA aptamers against the HCV NS5B protein composed of 2’-hydroxyl ribonucleotides (2’-OH) or 2’-fluoro pyrimidine ribonucleotides (2’-F)[78]. Both aptamers avidly bound to the HCV NS5B replicases of genotypes 1b and 2a and efficiently inhibited HCV replication of both genotypes in cells without inducing the generation of escape mutant viruses, innate immunity, or cellular toxicity. In addition, therapeutically amenable quantities of 2’-F aptamer conjugated with galactose-PEG moiety were efficiently distributed in the mouse liver tissue. These results suggest that RNA aptamers against HCV NS5B have a potential as a new therapeutic tool and are a potentially feasible alternative or additive to the current HCV therapeutics.
In addition to aptamers against the HCV regulatory proteins, a DNA aptamer targeting the HCV E1E2 structural protein was recently reported[79]. The DNA aptamer exerted its antiviral effects through inhibition of virus binding to the host cell receptors and thus inhibited the viral life cycle. Other aptamers have been reported that target viral RNA to inhibit either HCV translation[80] or replication[81]. Efficacy of the aptamers was confirmed in the HCV cell culture system. However, issues about cell toxicity profiles, distribution in animals, escape mutant appearance or side effects during the long-term treatment were not addressed.
RNA interference (RNAi) is a sequence-specific cellular post-transcriptional gene silencing (PTGS) pathway that regulates gene expression and is considered as a defense mechanism against invading viral pathogens and transposable elements in multiple organisms from worms to plants to mammals[82,83]. RNAi is initiated by double-stranded RNA (dsRNA) that is processed in the cytoplasm by the RNase III enzyme Dicer to form 21-22 nucleotide (nt)-long small interfering RNA (siRNA) with 5’ phosphate groups and two nt 3’ overhangs[84,85]. siRNA is then incorporated into an Argonaut-containing RISC (RNA-induced silencing complex), which unwinds the siRNA into the sense (passenger) strand and the antisense (guide) strand. The passenger strand is then cleaved and removed, while the guide strand brings RISC to the mRNA, which has a sequence that is complementary to the guide strand[86,87]. The degree of complementarity between the target mRNA and the guide strand determines the extent to which RISC silences the expression of the target mRNA. If there is perfect complementarity of the guide strand with the target mRNA, RISC mediates site-specific cleavage that degrades the target mRNA. In contrast, partial complementarity represses translation of the target mRNA[88,89]. RNAi in mammalian cells was first described in 2001[90], and triggering of the RNAi pathway with synthetic (exogenous) siRNA has become the most powerful and essential tool for drug development against various human diseases such as viral infections, tumors, and metabolic disorders, due to its high knockdown efficacy and sequence specificity[91,92]. Because HCV has a single positive-stranded RNA genome that replicates in the cytoplasm, RNAi is an attractive therapeutic option for the treatment of HCV infection. Many attempts have been made to target HCV RNA with siRNA or with short hairpin RNA (shRNA) as an RNAi trigger.
Because siRNAs display high sequence specificity (up to a single nucleotide resolution), any mismatches between the siRNA and target RNA affect the activity of siRNA[91,92]. The 5’- and 3’-UTRs are the most highly conserved regions of HCV RNA and are also essential for HCV translation and replication. Therefore, both 5’- and 3’-UTRs are ideal regions for targeting with siRNAs[93,94]. Several groups have reported potent siRNA activity against HCV 5’-UTR in the subgenomic replicon system[93-95]. These reports demonstrated that siRNA targeting the HCV 5’-UTR resulted in 80%-90% inhibition of HCV. Prabhu et al[96] showed that siRNA that targets the highly conserved stem loop II region of the HCV IRES efficiently inhibited translation and replication of infectious full-length clones of HCV 1a and 1b strains. Moreover, this siRNA effectively mediated degradation of the HCV IRES RNA and inhibited GFP expression that was controlled by the IRES sequences of six different HCV genotypes. Compared with synthetic 21-22 nt siRNAs, expressed shRNAs can induce long term stable knockdown of their target RNAs as long as transcription of the shRNAs occurs[97,98]. Moreover, shRNAs can act as substrates for Dicer, which increases the incorporation rates of siRNAs into RISC. This process enhances RNAi potency and efficacy[99]. For these reasons, two groups have utilized HCV 5’-UTR-targeting vector-derived shRNAs instead of 21-22 nt siRNA[100,101]. In both cases, the shRNAs inhibited replication and decreased titers of HCV genotypes 1a and 2a. Ray et al[102] also reported that shRNA targeting the 5’-UTR suppressed the replication of different HCV genotypes in the replicon cell culture systems.
Because HCV RNA replicates in the cytoplasm, and its genome acts like mRNA, any region of the HCV genome is theoretically targetable with RNAi. A number of groups have demonstrated siRNAs or shRNAs that target the protein coding regions of HCV. Three different groups have shown that siRNA against the HCV core region reduced HCV RNA and protein expression[103-106]. Ansar et al[106] showed that siRNAs against the HCV core region showed a 70% reduction in viral titers, while siRNAs against E1 and E2 caused viral titers to drop by as much as 93% in HCV-infected liver cells. Moreover, Kim et al[105] demonstrated that siRNAs against the NS3, NS4A, and NS4B regions of HCV effectively inhibited HCV replication and translation. Ali Ashfaq et al[107] showed an 88% reduction in HCV replication with siRNA directed against HCV NS3 and a greater than 90% inhibition with siRNAs directed against the NS4B and NS5B regions. Two other studies also demonstrated that siRNAs against HCV coding regions significantly inhibited HCV RNA replication[108,109]. For example, Wilson et al[109] showed that siRNAs against the NS5A and NS5B regions dramatically reduced HCV replicon RNA levels by up to 99% and 94%, respectively.
Host genes that modulate HCV infection and replication have been identified[110-113], and, unlike HCV itself, these genes are not prone to mutations. Therefore, these genes could be important targets for anti-HCV therapeutics. Several studies have shown that siRNAs against HCV entry receptors, such as CD81, SRBI, Claudin I, or occludin, markedly decreased the susceptibility of human hepatoma cells to HCV infection[114-116]. In addition, cellular proteins with enzymatic functions have also been targeted by siRNA as an anti-HCV therapy[117-120]. Importantly, a combination of siRNAs directed against cellular HCV cofactors and the HCV genome had more pronounced effects on suppressing HCV replication than either treatment alone[116,118,121]. The instances of siRNAs targeting cellular factors for antiviral therapy against HCV has been more extensively reviewed in the literature[122,123].
Because HCV has an error prone RNA-dependent RNA polymerase, the occurrence of drug-resistant escape mutant viruses is one of the major concerns for the development of antiviral therapies against HCV. Because RNAi has a high sequence specificity, prolonged treatment with siRNA could result in the appearance of escape mutant viruses that cannot be targetable by the siRNA. Wilson et al[124] reported that continuous treatment with one siRNA to an HCV replicon could induce the emergence of multiple point mutations within the target sequence region. One strategy to prevent the formation of escape mutant viruses is to use multiple siRNAs targeting multiple regions of the HCV genome combined with siRNAs against cellular HCV cofactors[116,118,121]. Long shRNA can be processed by the host cell machinery into two or more siRNAs. A vector that directs expression of three shRNAs targeting the 5’-UTR and two NS5B regions of the HCV genome showed sequence-specific antiviral activity in the HCV replicon and in infectious virus systems[125,126]. When using a mutant virus with a genome containing an escape mutation against one siRNA, the remaining two siRNAs that could target the mutant virus displayed fully active and effective anti-HCV effects. Yang et al[127] designed a vector-derived shRNA that could be processed into multiple siRNAs, using the endogenous miRNA-17-92 cluster as scaffolds. These authors did this because a previous study had demonstrated that overexpression of exogenously introduced shRNA competed with endogenous miRNA and thus led to saturation of the endogenous miRNA pathway, resulting in serious toxicity in mouse liver, and in some instances, death[128]. This vector-derived shRNA consisted of five siRNAs targeted against HCV RNA; three target sequences in the 5’-UTR and two others in the core and NS5B regions of HCV. This vector-derived shRNA inhibited HCV RNA replication and translation up to between 93%-98% in the infectious virus systems without inducing toxicity.
RNAi-based antiviral therapeutics has a number of advantages. However, some limitations exist, such as the inherently unstable nature of RNA, the requirement of a delivery vehicle, off-target effects, potential immunogenicity, and toxicity resulting from interference with the endogenous miRNA machinery[11,119,128]. These barriers may be overcome with improved chemical modification of siRNAs and synthetic and viral delivery tools. Recent advances in chemical modification methods have increased the stability and efficiency and reduced the off-target effects, immunogenicity, and toxicity of siRNAs. The properties of chemically modified siRNAs have been extensively described in recent reviews[11,12]. Delivery methods are also important to consider when contemplating the use of an siRNA as an antiviral therapy. As vector-derived shRNAs are difficult to modify chemically, many researchers have manipulated the shRNA structure[127] and expression strategies by using tissue specific or inducible promoter to improve their usefulness as antivirals[129]. To test siRNA as an anti-HCV therapeutic in animal models, viral delivery systems have been employed[125-127,129]. Sakamoto et al[130] used adenovirus to deliver an shRNA expression vector into the livers of transgenic mice that could be induced to express HCV structural proteins by the Cre/loxP switching system. These authors showed that intravenous injection of the adenovirus expressing shRNA resulted in the specific suppression of virus protein synthesis in the liver. In other studies, adeno-associated virus (AAV) was used as a delivery vehicle[126,127]. Suhy et al[126] described an AAV serotype 8-based viral vector that expresses three shRNAs simultaneously. A single intravenous injection of AAV8 expressing the shRNAs showed comprehensive transduction into hepatocytes in a nonhuman primate model. In addition to viral delivery systems, Chandra et al[131] have demonstrated the efficacy of a nanosome (lipid nanoparticles)-based siRNA delivery system. Multiple siRNAs directed against the 5’-UTR of HCV and encapsulated into nanosomes efficiently inhibited HCV replication in a liver tumor-xenotransplanted mouse model. Recent advances in nanobiotechnology will increase the available repertoire of synthetic delivery carriers for siRNAs directed against HCV RNA.
Antisense oligonucleotide (ASO) refers to a short DNA or RNA molecule that is designed to base pair with a specific target gene sequence in a sequence-specific manner. Most ASOs are synthetic single-stranded DNA or modified derivatives. Therefore, sequence-specific hybridization of ASOs to the target mRNA induces inhibition of target gene expression through RNase H-dependent degradation of the hybridized mRNA or through steric hindrance that blocks the access of the cellular machinery necessary for mRNA processing or translation[12,132]. Various modifications have improved the efficacy of ASOs through enhancement of nuclease resistance, increase in tissue half-life, affinity, and potency, and reduction of non-sequence-specific toxicity[133]. To improve resistance against nuclease degradation, a phosphorothioate-modified backbone was used (first-generation ASO). In addition, to further enhance nuclease resistance and increase binding affinity, 2’-O-Methyl (2’-OME) and 2’-O-Methoxyethyl (2’-MOE) modifications were developed (second-generation ASO). Peptide nucleic acid (PNA), locked nucleic acid (LNA), and phosphoramidate morpholino oligomer (PMO) have been recently developed as third-generation ASOs to further improve target affinity, nuclease resistance, biostability, and pharmacokinetics[132,133]. The United States FDA has approved ASOs Fomivirsen (ISIS 2922: Isis Pharmaceuticals) and, more recently, Mipomersen (ISIS301012: Isis Pharmaceuticals) for the treatment of cytomegalovirus retinitis in patients with AIDS and in patients with homozygous familial hypercholesterolemia, respectively[16,17]. In addition, a number of other ASOs are also undergoing clinical trials[133]. Several ASOs have been reported to inhibit HCV replication and translation. A phase II clinical trial with ISIS 14803 (Isis Pharmaceuticals), a phosphorothioate oligodeoxynucleotide against the HCV 5’-UTR IRES, was completed in 2007, although results were not announced[134,135]. McCaffrey et al[136] demonstrated that morpholino phosphoramidate antisense oligonucleotides (morpholinos) complementary to the HCV 5’-UTR specifically inhibited HCV IRES-dependent luciferase translation by up to 95% for at least 6 d in mouse liver. Moreover, an adenoviral vector-expressing an RNA ASO has been reported to block HCV replication in the HCV replicon and in the infectious HCV JFH-1 cell culture system by up to 40% and 76%, respectively[137]. Recently, a very promising ASO against HCV was reported with LNA-modified Miravirsen (SPC3649; Santaris Pharmaceuticals), which is directed against microRNA 122 (miR-122)[57]. MiR-122 has been reported to promote HCV replication through an increase in either stability or translation of HCV RNA by interacting with the 5’-UTR of the viral genome[138-140]. Therefore, silencing of miR-122 is a new plausible approach for anti-HCV therapeutics. LNA-modified ASO (SPC3649) caused a long-lasting suppression of HCV viremia in chronically HCV infected chimpanzees[141]. Moreover, a phase II clinical trial showed that SPC3649 treatment resulted in a dose-dependent prolonged reduction of up to 2-3 logs of HCV RNA in patients chronically infected with HCV genotype 1[19]. More studies are needed regarding the long-term suppression of miR-122, as miR-122 functions as a tumor suppressor miRNA[58-60], and HCV escape variants resistant to SPC3649 could potentially occur[142].
For almost two decades, major endeavors to develop nucleic acid-based therapeutics against hepatitis C virus have been undertaken. Compounds such as ribozymes, aptamers, siRNAs, and antisense oligonucleotides have been shown to perturb various steps in the HCV life cycle (Figure 1 and Table 1). However, clinical application of these nucleic acid-based therapeutics has been hampered by their low efficiency, off-target effects, toxicity, inefficient delivery, and the lack of cell culture and animal models. These limitations have been gradually overcome with recently improved delivery carriers (viral and synthetic) and chemical modifications of nucleic acids that can ameliorate the efficiency and bioavailability, while also reducing the toxicity and off-target effects[11,12]. Moreover, great efforts have been made to establish HCV cell culture systems[66,67,76] and small animal models[143,144], which are highly useful for the evaluation of anti-viral efficacy, and thus, for the realization of effective nucleic acid-based anti-HCV drugs in the future.
P- Reviewers: Costantini S, McConnell MR, Seron K S- Editor: Qi Y L- Editor: A E- Editor: Ma Ss
| 1. | Lauer GM, Walker BD. Hepatitis C virus infection. N Engl J Med. 2001;345:41-52. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 2042] [Cited by in F6Publishing: 1987] [Article Influence: 86.4] [Reference Citation Analysis (0)] |
| 2. | Alter MJ. Epidemiology of hepatitis C virus infection. World J Gastroenterol. 2007;13:2436-2441. [PubMed] [Cited in This Article: ] |
| 3. | Perz JF, Armstrong GL, Farrington LA, Hutin YJ, Bell BP. The contributions of hepatitis B virus and hepatitis C virus infections to cirrhosis and primary liver cancer worldwide. J Hepatol. 2006;45:529-538. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1764] [Cited by in F6Publishing: 1760] [Article Influence: 97.8] [Reference Citation Analysis (0)] |
| 4. | Gottwein JM, Scheel TK, Jensen TB, Lademann JB, Prentoe JC, Knudsen ML, Hoegh AM, Bukh J. Development and characterization of hepatitis C virus genotype 1-7 cell culture systems: role of CD81 and scavenger receptor class B type I and effect of antiviral drugs. Hepatology. 2009;49:364-377. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 296] [Cited by in F6Publishing: 299] [Article Influence: 19.9] [Reference Citation Analysis (0)] |
| 5. | Houghton M. Prospects for prophylactic and therapeutic vaccines against the hepatitis C viruses. Immunol Rev. 2011;239:99-108. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 120] [Cited by in F6Publishing: 119] [Article Influence: 9.2] [Reference Citation Analysis (0)] |
| 6. | Bartenschlager R, Lohmann V, Penin F. The molecular and structural basis of advanced antiviral therapy for hepatitis C virus infection. Nat Rev Microbiol. 2013;11:482-496. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 287] [Cited by in F6Publishing: 276] [Article Influence: 25.1] [Reference Citation Analysis (0)] |
| 7. | Fried MW, Shiffman ML, Reddy KR, Smith C, Marinos G, Gonçales FL, Häussinger D, Diago M, Carosi G, Dhumeaux D. Peginterferon alfa-2a plus ribavirin for chronic hepatitis C virus infection. N Engl J Med. 2002;347:975-982. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 4847] [Cited by in F6Publishing: 4689] [Article Influence: 213.1] [Reference Citation Analysis (0)] |
| 8. | Manns MP, Wedemeyer H, Cornberg M. Treating viral hepatitis C: efficacy, side effects, and complications. Gut. 2006;55:1350-1359. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 470] [Cited by in F6Publishing: 479] [Article Influence: 26.6] [Reference Citation Analysis (0)] |
| 9. | Kieffer TL, Sarrazin C, Miller JS, Welker MW, Forestier N, Reesink HW, Kwong AD, Zeuzem S. Telaprevir and pegylated interferon-alpha-2a inhibit wild-type and resistant genotype 1 hepatitis C virus replication in patients. Hepatology. 2007;46:631-639. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 307] [Cited by in F6Publishing: 328] [Article Influence: 19.3] [Reference Citation Analysis (0)] |
| 10. | Kieffer TL, Kwong AD, Picchio GR. Viral resistance to specifically targeted antiviral therapies for hepatitis C (STAT-Cs). J Antimicrob Chemother. 2010;65:202-212. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 96] [Cited by in F6Publishing: 101] [Article Influence: 6.7] [Reference Citation Analysis (0)] |
| 11. | Burnett JC, Rossi JJ. RNA-based therapeutics: current progress and future prospects. Chem Biol. 2012;19:60-71. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 711] [Cited by in F6Publishing: 647] [Article Influence: 53.9] [Reference Citation Analysis (0)] |
| 12. | Kole R, Krainer AR, Altman S. RNA therapeutics: beyond RNA interference and antisense oligonucleotides. Nat Rev Drug Discov. 2012;11:125-140. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 815] [Cited by in F6Publishing: 844] [Article Influence: 70.3] [Reference Citation Analysis (0)] |
| 13. | Wilson RC, Doudna JA. Molecular mechanisms of RNA interference. Annu Rev Biophys. 2013;42:217-239. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 781] [Cited by in F6Publishing: 678] [Article Influence: 61.6] [Reference Citation Analysis (0)] |
| 14. | Stenvang J, Petri A, Lindow M, Obad S, Kauppinen S. Inhibition of microRNA function by antimiR oligonucleotides. Silence. 2012;3:1. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 364] [Cited by in F6Publishing: 384] [Article Influence: 32.0] [Reference Citation Analysis (0)] |
| 15. | Mulhbacher J, St-Pierre P, Lafontaine DA. Therapeutic applications of ribozymes and riboswitches. Curr Opin Pharmacol. 2010;10:551-556. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 87] [Cited by in F6Publishing: 76] [Article Influence: 5.4] [Reference Citation Analysis (0)] |
| 16. | Andrei G, De Clercq E, Snoeck R. Drug targets in cytomegalovirus infection. Infect Disord Drug Targets. 2009;9:201-222. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 62] [Cited by in F6Publishing: 68] [Article Influence: 4.5] [Reference Citation Analysis (0)] |
| 17. | Toth PP. Emerging LDL therapies: Mipomersen-antisense oligonucleotide therapy in the management of hypercholesterolemia. J Clin Lipidol. 2013;7:S6-10. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 24] [Cited by in F6Publishing: 28] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 18. | Rinaldi M, Chiosi F, dell’Omo R, Romano MR, Parmeggiani F, Semeraro F, Mastropasqua R, Costagliola C. Intravitreal pegaptanib sodium (Macugen®) for treatment of diabetic macular oedema: a morphologic and functional study. Br J Clin Pharmacol. 2012;74:940-946. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 14] [Cited by in F6Publishing: 15] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 19. | Janssen HL, Reesink HW, Lawitz EJ, Zeuzem S, Rodriguez-Torres M, Patel K, van der Meer AJ, Patick AK, Chen A, Zhou Y. Treatment of HCV infection by targeting microRNA. N Engl J Med. 2013;368:1685-1694. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1665] [Cited by in F6Publishing: 1627] [Article Influence: 147.9] [Reference Citation Analysis (0)] |
| 20. | Serganov A, Patel DJ. Ribozymes, riboswitches and beyond: regulation of gene expression without proteins. Nat Rev Genet. 2007;8:776-790. [PubMed] [Cited in This Article: ] |
| 21. | Bartenschlager R, Lohmann V. Replication of hepatitis C virus. J Gen Virol. 2000;81:1631-1648. [PubMed] [Cited in This Article: ] |
| 22. | Ohkawa K, Yuki N, Kanazawa Y, Ueda K, Mita E, Sasaki Y, Kasahara A, Hayashi N. Cleavage of viral RNA and inhibition of viral translation by hepatitis C virus RNA-specific hammerhead ribozyme in vitro. J Hepatol. 1997;27:78-84. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 9] [Cited by in F6Publishing: 12] [Article Influence: 0.4] [Reference Citation Analysis (0)] |
| 23. | Sakamoto N, Wu CH, Wu GY. Intracellular cleavage of hepatitis C virus RNA and inhibition of viral protein translation by hammerhead ribozymes. J Clin Invest. 1996;98:2720-2728. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 61] [Cited by in F6Publishing: 61] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 24. | Welch PJ, Tritz R, Yei S, Leavitt M, Yu M, Barber J. A potential therapeutic application of hairpin ribozymes: in vitro and in vivo studies of gene therapy for hepatitis C virus infection. Gene Ther. 1996;3:994-1001. [PubMed] [Cited in This Article: ] |
| 25. | Welch PJ, Yei S, Barber JR. Ribozyme gene therapy for hepatitis C virus infection. Clin Diagn Virol. 1998;10:163-171. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 31] [Cited by in F6Publishing: 31] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 26. | Puerta-Fernández E, Romero-López C, Barroso-delJesus A, Berzal-Herranz A. Ribozymes: recent advances in the development of RNA tools. FEMS Microbiol Rev. 2003;27:75-97. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 76] [Cited by in F6Publishing: 69] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 27. | Müller S, Appel B, Krellenberg T, Petkovic S. The many faces of the hairpin ribozyme: structural and functional variants of a small catalytic RNA. IUBMB Life. 2012;64:36-47. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 24] [Cited by in F6Publishing: 27] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 28. | Jarczak D, Korf M, Beger C, Manns MP, Krüger M. Hairpin ribozymes in combination with siRNAs against highly conserved hepatitis C virus sequence inhibit RNA replication and protein translation from hepatitis C virus subgenomic replicons. FEBS J. 2005;272:5910-5922. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 37] [Cited by in F6Publishing: 39] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 29. | Asif-Ullah M, Lévesque M, Robichaud G, Perreault JP. Development of ribozyme-based gene-inactivations; the example of the hepatitis delta virus ribozyme. Curr Gene Ther. 2007;7:205-216. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 18] [Cited by in F6Publishing: 21] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 30. | Yu YC, Mao Q, Gu CH, Li QF, Wang YM. Activity of HDV ribozymes to trans-cleave HCV RNA. World J Gastroenterol. 2002;8:694-698. [PubMed] [Cited in This Article: ] |
| 31. | Lévesque MV, Lévesque D, Brière FP, Perreault JP. Investigating a new generation of ribozymes in order to target HCV. PLoS One. 2010;5:e9627. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 15] [Cited by in F6Publishing: 18] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 32. | Gonzalez-Carmona MA, Schüssler S, Serwe M, Alt M, Ludwig J, Sproat BS, Steigerwald R, Hoffmann P, Quasdorff M, Schildgen O. Hammerhead ribozymes with cleavage site specificity for NUH and NCH display significant anti-hepatitis C viral effect in vitro and in recombinant HepG2 and CCL13 cells. J Hepatol. 2006;44:1017-1025. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 10] [Cited by in F6Publishing: 11] [Article Influence: 0.6] [Reference Citation Analysis (0)] |
| 33. | Macejak DG, Jensen KL, Jamison SF, Domenico K, Roberts EC, Chaudhary N, von Carlowitz I, Bellon L, Tong MJ, Conrad A. Inhibition of hepatitis C virus (HCV)-RNA-dependent translation and replication of a chimeric HCV poliovirus using synthetic stabilized ribozymes. Hepatology. 2000;31:769-776. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 80] [Cited by in F6Publishing: 90] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 34. | Macejak DG, Jensen KL, Pavco PA, Phipps KM, Heinz BA, Colacino JM, Blatt LM. Enhanced antiviral effect in cell culture of type 1 interferon and ribozymes targeting HCV RNA. J Viral Hepat. 2001;8:400-405. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 18] [Cited by in F6Publishing: 22] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 35. | Lee PA, Blatt LM, Blanchard KS, Bouhana KS, Pavco PA, Bellon L, Sandberg JA. Pharmacokinetics and tissue distribution of a ribozyme directed against hepatitis C virus RNA following subcutaneous or intravenous administration in mice. Hepatology. 2000;32:640-646. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 37] [Cited by in F6Publishing: 40] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 36. | Swan T, Raymond D. Hepatitis C virus (HCV) and HIV/HCV coinfection: A critical review of research and treatment. New York: Treatment Action Group 2004; . [Cited in This Article: ] |
| 37. | Kashani-Sabet M. Ribozyme therapeutics. J Investig Dermatol Symp Proc. 2002;7:76-78. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 27] [Cited by in F6Publishing: 29] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 38. | Steele D, Kertsburg A, Soukup GA. Engineered catalytic RNA and DNA : new biochemical tools for drug discovery and design. Am J Pharmacogenomics. 2003;3:131-144. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 16] [Cited by in F6Publishing: 18] [Article Influence: 0.9] [Reference Citation Analysis (0)] |
| 39. | Trepanier J, Tanner JE, Momparler RL, Le ON, Alvarez F, Alfieri C. Cleavage of intracellular hepatitis C RNA in the virus core protein coding region by deoxyribozymes. J Viral Hepat. 2006;13:131-138. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 27] [Cited by in F6Publishing: 28] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 40. | Kumar D, Chaudhury I, Kar P, Das RH. Site-specific cleavage of HCV genomic RNA and its cloned core and NS5B genes by DNAzyme. J Gastroenterol Hepatol. 2009;24:872-878. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 15] [Cited by in F6Publishing: 16] [Article Influence: 1.1] [Reference Citation Analysis (0)] |
| 41. | Roy S, Gupta N, Subramanian N, Mondal T, Banerjea AC, Das S. Sequence-specific cleavage of hepatitis C virus RNA by DNAzymes: inhibition of viral RNA translation and replication. J Gen Virol. 2008;89:1579-1586. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 23] [Cited by in F6Publishing: 23] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 42. | Lee B, Kim KB, Oh S, Choi JS, Park JS, Min DH, Kim DE. Suppression of hepatitis C virus genome replication in cells with RNA-cleaving DNA enzymes and short-hairpin RNA. Oligonucleotides. 2010;20:285-296. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 11] [Cited by in F6Publishing: 12] [Article Influence: 0.9] [Reference Citation Analysis (0)] |
| 43. | James HA, Gibson I. The therapeutic potential of ribozymes. Blood. 1998;91:371-382. [PubMed] [Cited in This Article: ] |
| 44. | Yuan Y, Altman S. Selection of guide sequences that direct efficient cleavage of mRNA by human ribonuclease P. Science. 1994;263:1269-1273. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 80] [Cited by in F6Publishing: 84] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 45. | Nadal A, Martell M, Lytle JR, Lyons AJ, Robertson HD, Cabot B, Esteban JI, Esteban R, Guardia J, Gómez J. Specific cleavage of hepatitis C virus RNA genome by human RNase P. J Biol Chem. 2002;277:30606-30613. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 33] [Cited by in F6Publishing: 37] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 46. | Nadal A, Robertson HD, Guardia J, Gómez J. Characterization of the structure and variability of an internal region of hepatitis C virus RNA for M1 RNA guide sequence ribozyme targeting. J Gen Virol. 2003;84:1545-1548. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 13] [Cited by in F6Publishing: 16] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 47. | Sabariegos R, Nadal A, Beguiristain N, Piron M, Gómez J. Catalytic RNase P RNA from Synechocystis sp. cleaves the hepatitis C virus RNA near the AUG start codon. FEBS Lett. 2004;577:517-522. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 11] [Cited by in F6Publishing: 13] [Article Influence: 0.7] [Reference Citation Analysis (0)] |
| 48. | Been MD, Cech TR. One binding site determines sequence specificity of Tetrahymena pre-rRNA self-splicing, trans-splicing, and RNA enzyme activity. Cell. 1986;47:207-216. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 150] [Cited by in F6Publishing: 178] [Article Influence: 4.7] [Reference Citation Analysis (0)] |
| 49. | Sullenger BA, Cech TR. Ribozyme-mediated repair of defective mRNA by targeted, trans-splicing. Nature. 1994;371:619-622. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 192] [Cited by in F6Publishing: 179] [Article Influence: 6.0] [Reference Citation Analysis (0)] |
| 50. | Jones JT, Lee SW, Sullenger BA. Tagging ribozyme reaction sites to follow trans-splicing in mammalian cells. Nat Med. 1996;2:643-648. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 87] [Cited by in F6Publishing: 93] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 51. | Ryu KJ, Kim JH, Lee SW. Ribozyme-mediated selective induction of new gene activity in hepatitis C virus internal ribosome entry site-expressing cells by targeted trans-splicing. Mol Ther. 2003;7:386-395. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 55] [Cited by in F6Publishing: 58] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 52. | Bergeron LJ, Reymond C, Perreault JP. Functional characterization of the SOFA delta ribozyme. RNA. 2005;11:1858-1868. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 19] [Cited by in F6Publishing: 17] [Article Influence: 0.9] [Reference Citation Analysis (0)] |
| 53. | Bergeron LJ, Perreault JP. Target-dependent on/off switch increases ribozyme fidelity. Nucleic Acids Res. 2005;33:1240-1248. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 24] [Cited by in F6Publishing: 28] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 54. | Lee CH, Kim JH, Kim HW, Myung H, Lee SW. Hepatitis C virus replication-specific inhibition of microRNA activity with self-cleavable allosteric ribozyme. Nucleic Acid Ther. 2012;22:17-29. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 9] [Cited by in F6Publishing: 11] [Article Influence: 0.9] [Reference Citation Analysis (0)] |
| 55. | Krützfeldt J, Rajewsky N, Braich R, Rajeev KG, Tuschl T, Manoharan M, Stoffel M. Silencing of microRNAs in vivo with ‘antagomirs’. Nature. 2005;438:685-689. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 3005] [Cited by in F6Publishing: 2983] [Article Influence: 157.0] [Reference Citation Analysis (0)] |
| 56. | Esau C, Davis S, Murray SF, Yu XX, Pandey SK, Pear M, Watts L, Booten SL, Graham M, McKay R. miR-122 regulation of lipid metabolism revealed by in vivo antisense targeting. Cell Metab. 2006;3:87-98. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1612] [Cited by in F6Publishing: 1588] [Article Influence: 88.2] [Reference Citation Analysis (0)] |
| 57. | Elmén J, Lindow M, Silahtaroglu A, Bak M, Christensen M, Lind-Thomsen A, Hedtjärn M, Hansen JB, Hansen HF, Straarup EM. Antagonism of microRNA-122 in mice by systemically administered LNA-antimiR leads to up-regulation of a large set of predicted target mRNAs in the liver. Nucleic Acids Res. 2008;36:1153-1162. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 520] [Cited by in F6Publishing: 509] [Article Influence: 29.9] [Reference Citation Analysis (0)] |
| 58. | Gramantieri L, Ferracin M, Fornari F, Veronese A, Sabbioni S, Liu CG, Calin GA, Giovannini C, Ferrazzi E, Grazi GL. Cyclin G1 is a target of miR-122a, a microRNA frequently down-regulated in human hepatocellular carcinoma. Cancer Res. 2007;67:6092-6099. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 614] [Cited by in F6Publishing: 621] [Article Influence: 36.5] [Reference Citation Analysis (0)] |
| 59. | Tsai WC, Hsu PW, Lai TC, Chau GY, Lin CW, Chen CM, Lin CD, Liao YL, Wang JL, Chau YP. MicroRNA-122, a tumor suppressor microRNA that regulates intrahepatic metastasis of hepatocellular carcinoma. Hepatology. 2009;49:1571-1582. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 450] [Cited by in F6Publishing: 465] [Article Influence: 31.0] [Reference Citation Analysis (0)] |
| 60. | Tsai WC, Hsu SD, Hsu CS, Lai TC, Chen SJ, Shen R, Huang Y, Chen HC, Lee CH, Tsai TF. MicroRNA-122 plays a critical role in liver homeostasis and hepatocarcinogenesis. J Clin Invest. 2012;122:2884-2897. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 612] [Cited by in F6Publishing: 632] [Article Influence: 52.7] [Reference Citation Analysis (0)] |
| 61. | Kaur G, Roy I. Therapeutic applications of aptamers. Expert Opin Investig Drugs. 2008;17:43-60. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 62] [Cited by in F6Publishing: 63] [Article Influence: 3.9] [Reference Citation Analysis (0)] |
| 62. | Que-Gewirth NS, Sullenger BA. Gene therapy progress and prospects: RNA aptamers. Gene Ther. 2007;14:283-291. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 166] [Cited by in F6Publishing: 160] [Article Influence: 9.4] [Reference Citation Analysis (0)] |
| 63. | Tuerk C, Gold L. Systematic evolution of ligands by exponential enrichment: RNA ligands to bacteriophage T4 DNA polymerase. Science. 1990;249:505-510. [PubMed] [Cited in This Article: ] |
| 64. | Ellington AD, Szostak JW. In vitro selection of RNA molecules that bind specific ligands. Nature. 1990;346:818-822. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 7090] [Cited by in F6Publishing: 6659] [Article Influence: 195.9] [Reference Citation Analysis (0)] |
| 65. | Nakamura Y, Ishiguro A, Miyakawa S. RNA plasticity and selectivity applicable to therapeutics and novel biosensor development. Genes Cells. 2012;17:344-364. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 22] [Cited by in F6Publishing: 24] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 66. | Lohmann V, Körner F, Koch J, Herian U, Theilmann L, Bartenschlager R. Replication of subgenomic hepatitis C virus RNAs in a hepatoma cell line. Science. 1999;285:110-113. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 2294] [Cited by in F6Publishing: 2224] [Article Influence: 89.0] [Reference Citation Analysis (0)] |
| 67. | Wakita T, Pietschmann T, Kato T, Date T, Miyamoto M, Zhao Z, Murthy K, Habermann A, Kräusslich HG, Mizokami M. Production of infectious hepatitis C virus in tissue culture from a cloned viral genome. Nat Med. 2005;11:791-796. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 2241] [Cited by in F6Publishing: 2239] [Article Influence: 117.8] [Reference Citation Analysis (0)] |
| 68. | Nishikawa F, Kakiuchi N, Funaji K, Fukuda K, Sekiya S, Nishikawa S. Inhibition of HCV NS3 protease by RNA aptamers in cells. Nucleic Acids Res. 2003;31:1935-1943. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 34] [Cited by in F6Publishing: 38] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 69. | Nishikawa F, Funaji K, Fukuda K, Nishikawa S. In vitro selection of RNA aptamers against the HCV NS3 helicase domain. Oligonucleotides. 2004;14:114-129. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 38] [Cited by in F6Publishing: 40] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 70. | Hwang B, Cho JS, Yeo HJ, Kim JH, Chung KM, Han K, Jang SK, Lee SW. Isolation of specific and high-affinity RNA aptamers against NS3 helicase domain of hepatitis C virus. RNA. 2004;10:1277-1290. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 40] [Cited by in F6Publishing: 39] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 71. | Umehara T, Fukuda K, Nishikawa F, Sekiya S, Kohara M, Hasegawa T, Nishikawa S. Designing and analysis of a potent bi-functional aptamers that inhibit protease and helicase activities of HCV NS3. Nucleic Acids Symp Ser (Oxf). 2004;195-196. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 10] [Cited by in F6Publishing: 13] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 72. | Blight KJ, Kolykhalov AA, Rice CM. Efficient initiation of HCV RNA replication in cell culture. Science. 2000;290:1972-1974. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1154] [Cited by in F6Publishing: 1139] [Article Influence: 47.5] [Reference Citation Analysis (0)] |
| 73. | Grobler JA, Markel EJ, Fay JF, Graham DJ, Simcoe AL, Ludmerer SW, Murray EM, Migliaccio G, Flores OA. Identification of a key determinant of hepatitis C virus cell culture adaptation in domain II of NS3 helicase. J Biol Chem. 2003;278:16741-16746. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 20] [Cited by in F6Publishing: 22] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 74. | Wu JZ, Yao N, Walker M, Hong Z. Recent advances in discovery and development of promising therapeutics against hepatitis C virus NS5B RNA-dependent RNA polymerase. Mini Rev Med Chem. 2005;5:1103-1112. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 37] [Cited by in F6Publishing: 40] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 75. | Jones LA, Clancy LE, Rawlinson WD, White PA. High-affinity aptamers to subtype 3a hepatitis C virus polymerase display genotypic specificity. Antimicrob Agents Chemother. 2006;50:3019-3027. [PubMed] [Cited in This Article: ] |
| 76. | Saeed M, Gondeau C, Hmwe S, Yokokawa H, Date T, Suzuki T, Kato T, Maurel P, Wakita T. Replication of hepatitis C virus genotype 3a in cultured cells. Gastroenterology. 2013;144:56-58.e7. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 42] [Cited by in F6Publishing: 44] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 77. | Bellecave P, Cazenave C, Rumi J, Staedel C, Cosnefroy O, Andreola ML, Ventura M, Tarrago-Litvak L, Astier-Gin T. Inhibition of hepatitis C virus (HCV) RNA polymerase by DNA aptamers: mechanism of inhibition of in vitro RNA synthesis and effect on HCV-infected cells. Antimicrob Agents Chemother. 2008;52:2097-2110. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 31] [Cited by in F6Publishing: 35] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 78. | Lee CH, Lee YJ, Kim JH, Lim JH, Kim JH, Han W, Lee SH, Noh GJ, Lee SW. Inhibition of hepatitis C virus (HCV) replication by specific RNA aptamers against HCV NS5B RNA replicase. J Virol. 2013;87:7064-7074. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 40] [Cited by in F6Publishing: 43] [Article Influence: 3.9] [Reference Citation Analysis (0)] |
| 79. | Yang D, Meng X, Yu Q, Xu L, Long Y, Liu B, Fang X, Zhu H. Inhibition of hepatitis C virus infection by DNA aptamer against envelope protein. Antimicrob Agents Chemother. 2013;57:4937-4944. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 29] [Cited by in F6Publishing: 34] [Article Influence: 3.1] [Reference Citation Analysis (0)] |
| 80. | Kikuchi K, Umehara T, Fukuda K, Kuno A, Hasegawa T, Nishikawa S. A hepatitis C virus (HCV) internal ribosome entry site (IRES) domain III-IV-targeted aptamer inhibits translation by binding to an apical loop of domain IIId. Nucleic Acids Res. 2005;33:683-692. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 68] [Cited by in F6Publishing: 61] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 81. | Marton S, Romero-López C, Berzal-Herranz A. RNA aptamer-mediated interference of HCV replication by targeting the CRE-5BSL3.2 domain. J Viral Hepat. 2013;20:103-112. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 15] [Cited by in F6Publishing: 17] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 82. | Fire A, Xu S, Montgomery MK, Kostas SA, Driver SE, Mello CC. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature. 1998;391:806-811. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 10522] [Cited by in F6Publishing: 9712] [Article Influence: 373.5] [Reference Citation Analysis (1)] |
| 83. | Kim DH, Rossi JJ. Strategies for silencing human disease using RNA interference. Nat Rev Genet. 2007;8:173-184. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 788] [Cited by in F6Publishing: 758] [Article Influence: 44.6] [Reference Citation Analysis (0)] |
| 84. | Bernstein E, Caudy AA, Hammond SM, Hannon GJ. Role for a bidentate ribonuclease in the initiation step of RNA interference. Nature. 2001;409:363-366. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 3476] [Cited by in F6Publishing: 3191] [Article Influence: 138.7] [Reference Citation Analysis (0)] |
| 85. | Zamore PD, Tuschl T, Sharp PA, Bartel DP. RNAi: double-stranded RNA directs the ATP-dependent cleavage of mRNA at 21 to 23 nucleotide intervals. Cell. 2000;101:25-33. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1876] [Cited by in F6Publishing: 1751] [Article Influence: 73.0] [Reference Citation Analysis (0)] |
| 86. | Matranga C, Tomari Y, Shin C, Bartel DP, Zamore PD. Passenger-strand cleavage facilitates assembly of siRNA into Ago2-containing RNAi enzyme complexes. Cell. 2005;123:607-620. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 785] [Cited by in F6Publishing: 775] [Article Influence: 40.8] [Reference Citation Analysis (0)] |
| 87. | Rand TA, Petersen S, Du F, Wang X. Argonaute2 cleaves the anti-guide strand of siRNA during RISC activation. Cell. 2005;123:621-629. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 540] [Cited by in F6Publishing: 516] [Article Influence: 27.2] [Reference Citation Analysis (0)] |
| 88. | Elbashir SM, Lendeckel W, Tuschl T. RNA interference is mediated by 21- and 22-nucleotide RNAs. Genes Dev. 2001;15:188-200. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 2287] [Cited by in F6Publishing: 2246] [Article Influence: 97.7] [Reference Citation Analysis (0)] |
| 89. | Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281-297. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 25833] [Cited by in F6Publishing: 26782] [Article Influence: 1339.1] [Reference Citation Analysis (0)] |
| 90. | Elbashir SM, Harborth J, Lendeckel W, Yalcin A, Weber K, Tuschl T. Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nature. 2001;411:494-498. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 6971] [Cited by in F6Publishing: 6870] [Article Influence: 298.7] [Reference Citation Analysis (0)] |
| 91. | Hannon GJ, Rossi JJ. Unlocking the potential of the human genome with RNA interference. Nature. 2004;431:371-378. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 789] [Cited by in F6Publishing: 781] [Article Influence: 39.1] [Reference Citation Analysis (0)] |
| 92. | Mittal V. Improving the efficiency of RNA interference in mammals. Nat Rev Genet. 2004;5:355-365. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 202] [Cited by in F6Publishing: 192] [Article Influence: 9.6] [Reference Citation Analysis (0)] |
| 93. | Yokota T, Sakamoto N, Enomoto N, Tanabe Y, Miyagishi M, Maekawa S, Yi L, Kurosaki M, Taira K, Watanabe M. Inhibition of intracellular hepatitis C virus replication by synthetic and vector-derived small interfering RNAs. EMBO Rep. 2003;4:602-608. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 235] [Cited by in F6Publishing: 250] [Article Influence: 11.9] [Reference Citation Analysis (0)] |
| 94. | Motavaf M, Safari S, Alavian SM. Therapeutic potential of RNA interference: a new molecular approach to antiviral treatment for hepatitis C. J Viral Hepat. 2012;19:757-765. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 20] [Cited by in F6Publishing: 22] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 95. | Seo MY, Abrignani S, Houghton M, Han JH. Small interfering RNA-mediated inhibition of hepatitis C virus replication in the human hepatoma cell line Huh-7. J Virol. 2003;77:810-812. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 108] [Cited by in F6Publishing: 112] [Article Influence: 5.3] [Reference Citation Analysis (0)] |
| 96. | Prabhu R, Garry RF, Dash S. Small interfering RNA targeted to stem-loop II of the 5’ untranslated region effectively inhibits expression of six HCV genotypes. Virol J. 2006;3:100. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 34] [Cited by in F6Publishing: 39] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 97. | Brummelkamp TR, Bernards R, Agami R. A system for stable expression of short interfering RNAs in mammalian cells. Science. 2002;296:550-553. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 3486] [Cited by in F6Publishing: 3442] [Article Influence: 156.5] [Reference Citation Analysis (0)] |
| 98. | Paddison PJ, Caudy AA, Bernstein E, Hannon GJ, Conklin DS. Short hairpin RNAs (shRNAs) induce sequence-specific silencing in mammalian cells. Genes Dev. 2002;16:948-958. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1123] [Cited by in F6Publishing: 1097] [Article Influence: 49.9] [Reference Citation Analysis (0)] |
| 99. | Kim DH, Behlke MA, Rose SD, Chang MS, Choi S, Rossi JJ. Synthetic dsRNA Dicer substrates enhance RNAi potency and efficacy. Nat Biotechnol. 2005;23:222-226. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 649] [Cited by in F6Publishing: 637] [Article Influence: 31.9] [Reference Citation Analysis (0)] |
| 100. | Watanabe T, Sudoh M, Miyagishi M, Akashi H, Arai M, Inoue K, Taira K, Yoshiba M, Kohara M. Intracellular-diced dsRNA has enhanced efficacy for silencing HCV RNA and overcomes variation in the viral genotype. Gene Ther. 2006;13:883-892. [PubMed] [Cited in This Article: ] |
| 101. | Kanda T, Steele R, Ray R, Ray RB. Small interfering RNA targeted to hepatitis C virus 5’ nontranslated region exerts potent antiviral effect. J Virol. 2007;81:669-676. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 61] [Cited by in F6Publishing: 68] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 102. | Ray RB, Kanda T. Inhibition of HCV replication by small interfering RNA. Methods Mol Biol. 2009;510:251-262. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 10] [Cited by in F6Publishing: 12] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 103. | Randall G, Grakoui A, Rice CM. Clearance of replicating hepatitis C virus replicon RNAs in cell culture by small interfering RNAs. Proc Natl Acad Sci USA. 2003;100:235-240. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 259] [Cited by in F6Publishing: 273] [Article Influence: 13.0] [Reference Citation Analysis (0)] |
| 104. | Liu M, Ding H, Zhao P, Qin ZL, Gao J, Cao MM, Luan J, Wu WB, Qi ZT. RNA interference effectively inhibits mRNA accumulation and protein expression of hepatitis C virus core and E2 genes in human cells. Biosci Biotechnol Biochem. 2006;70:2049-2055. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 23] [Cited by in F6Publishing: 25] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 105. | Kim M, Shin D, Kim SI, Park M. Inhibition of hepatitis C virus gene expression by small interfering RNAs using a tri-cistronic full-length viral replicon and a transient mouse model. Virus Res. 2006;122:1-10. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 33] [Cited by in F6Publishing: 36] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 106. | Ansar M, Ashfaq UA, Shahid I, Sarwar MT, Javed T, Rehman S, Hassan S, Riazuddin S. Inhibition of full length hepatitis C virus particles of 1a genotype through small interference RNA. Virol J. 2011;8:203. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 13] [Cited by in F6Publishing: 15] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 107. | Ali Ashfaq U, Ansar M, Sarwar MT, Javed T, Rehman S, Riazuddin S. Post-transcriptional inhibition of hepatitis C virus replication through small interference RNA. Virol J. 2011;8:112. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 9] [Cited by in F6Publishing: 12] [Article Influence: 0.9] [Reference Citation Analysis (0)] |
| 108. | Kapadia SB, Brideau-Andersen A, Chisari FV. Interference of hepatitis C virus RNA replication by short interfering RNAs. Proc Natl Acad Sci USA. 2003;100:2014-2018. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 310] [Cited by in F6Publishing: 327] [Article Influence: 15.6] [Reference Citation Analysis (0)] |
| 109. | Wilson JA, Jayasena S, Khvorova A, Sabatinos S, Rodrigue-Gervais IG, Arya S, Sarangi F, Harris-Brandts M, Beaulieu S, Richardson CD. RNA interference blocks gene expression and RNA synthesis from hepatitis C replicons propagated in human liver cells. Proc Natl Acad Sci USA. 2003;100:2783-2788. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 227] [Cited by in F6Publishing: 240] [Article Influence: 11.4] [Reference Citation Analysis (0)] |
| 110. | Randall G, Panis M, Cooper JD, Tellinghuisen TL, Sukhodolets KE, Pfeffer S, Landthaler M, Landgraf P, Kan S, Lindenbach BD. Cellular cofactors affecting hepatitis C virus infection and replication. Proc Natl Acad Sci USA. 2007;104:12884-12889. [PubMed] [Cited in This Article: ] |
| 111. | Ng TI, Mo H, Pilot-Matias T, He Y, Koev G, Krishnan P, Mondal R, Pithawalla R, He W, Dekhtyar T. Identification of host genes involved in hepatitis C virus replication by small interfering RNA technology. Hepatology. 2007;45:1413-1421. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 103] [Cited by in F6Publishing: 107] [Article Influence: 6.3] [Reference Citation Analysis (0)] |
| 112. | Supekova L, Supek F, Lee J, Chen S, Gray N, Pezacki JP, Schlapbach A, Schultz PG. Identification of human kinases involved in hepatitis C virus replication by small interference RNA library screening. J Biol Chem. 2008;283:29-36. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 88] [Cited by in F6Publishing: 87] [Article Influence: 5.1] [Reference Citation Analysis (0)] |
| 113. | Tai AW, Benita Y, Peng LF, Kim SS, Sakamoto N, Xavier RJ, Chung RT. A functional genomic screen identifies cellular cofactors of hepatitis C virus replication. Cell Host Microbe. 2009;5:298-307. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 348] [Cited by in F6Publishing: 350] [Article Influence: 23.3] [Reference Citation Analysis (0)] |
| 114. | Zeisel MB, Koutsoudakis G, Schnober EK, Haberstroh A, Blum HE, Cosset FL, Wakita T, Jaeck D, Doffoel M, Royer C. Scavenger receptor class B type I is a key host factor for hepatitis C virus infection required for an entry step closely linked to CD81. Hepatology. 2007;46:1722-1731. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 202] [Cited by in F6Publishing: 211] [Article Influence: 12.4] [Reference Citation Analysis (0)] |
| 115. | Jahan S, Samreen B, Khaliq S, Ijaz B, Khan M, Siddique MH, Ahmad W, Hassan S. HCV entry receptors as potential targets for siRNA-based inhibition of HCV. Genet Vaccines Ther. 2011;9:15. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 12] [Cited by in F6Publishing: 13] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 116. | Jahan S, Khaliq S, Samreen B, Ijaz B, Khan M, Ahmad W, Ashfaq UA, Hassan S. Effect of combined siRNA of HCV E2 gene and HCV receptors against HCV. Virol J. 2011;8:295. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 16] [Cited by in F6Publishing: 17] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 117. | Xue Q, Ding H, Liu M, Zhao P, Gao J, Ren H, Liu Y, Qi ZT. Inhibition of hepatitis C virus replication and expression by small interfering RNA targeting host cellular genes. Arch Virol. 2007;152:955-962. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 15] [Cited by in F6Publishing: 17] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 118. | Korf M, Jarczak D, Beger C, Manns MP, Krüger M. Inhibition of hepatitis C virus translation and subgenomic replication by siRNAs directed against highly conserved HCV sequence and cellular HCV cofactors. J Hepatol. 2005;43:225-234. [PubMed] [Cited in This Article: ] |
| 119. | Randall G, Chen L, Panis M, Fischer AK, Lindenbach BD, Sun J, Heathcote J, Rice CM, Edwards AM, McGilvray ID. Silencing of USP18 potentiates the antiviral activity of interferon against hepatitis C virus infection. Gastroenterology. 2006;131:1584-1591. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 132] [Cited by in F6Publishing: 128] [Article Influence: 7.1] [Reference Citation Analysis (0)] |
| 120. | Nakagawa S, Umehara T, Matsuda C, Kuge S, Sudoh M, Kohara M. Hsp90 inhibitors suppress HCV replication in replicon cells and humanized liver mice. Biochem Biophys Res Commun. 2007;353:882-888. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 79] [Cited by in F6Publishing: 74] [Article Influence: 4.1] [Reference Citation Analysis (0)] |
| 121. | Henry SD, van der Wegen P, Metselaar HJ, Tilanus HW, Scholte BJ, van der Laan LJ. Simultaneous targeting of HCV replication and viral binding with a single lentiviral vector containing multiple RNA interference expression cassettes. Mol Ther. 2006;14:485-493. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 78] [Cited by in F6Publishing: 85] [Article Influence: 4.7] [Reference Citation Analysis (0)] |
| 122. | Khaliq S, Khaliq SA, Zahur M, Ijaz B, Jahan S, Ansar M, Riazuddin S, Hassan S. RNAi as a new therapeutic strategy against HCV. Biotechnol Adv. 2010;28:27-34. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 22] [Cited by in F6Publishing: 24] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 123. | Ashfaq UA, Yousaf MZ, Aslam M, Ejaz R, Jahan S, Ullah O. siRNAs: potential therapeutic agents against hepatitis C virus. Virol J. 2011;8:276. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 17] [Cited by in F6Publishing: 22] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 124. | Wilson JA, Richardson CD. Hepatitis C virus replicons escape RNA interference induced by a short interfering RNA directed against the NS5b coding region. J Virol. 2005;79:7050-7058. [PubMed] [Cited in This Article: ] |
| 125. | Lavender H, Brady K, Burden F, Delpuech-Adams O, Denise H, Palmer A, Perkins H, Savic B, Scott S, Smith-Burchnell C. In vitro characterization of the activity of PF-05095808, a novel biological agent for hepatitis C virus therapy. Antimicrob Agents Chemother. 2012;56:1364-1375. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 9] [Cited by in F6Publishing: 10] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 126. | Suhy DA, Kao SC, Mao T, Whiteley L, Denise H, Souberbielle B, Burdick AD, Hayes K, Wright JF, Lavender H. Safe, long-term hepatic expression of anti-HCV shRNA in a nonhuman primate model. Mol Ther. 2012;20:1737-1749. [DOI] [Cited in This Article: ] [Cited by in Crossref: 40] [Cited by in F6Publishing: 41] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 127. | Yang X, Haurigot V, Zhou S, Luo G, Couto LB. Inhibition of hepatitis C virus replication using adeno-associated virus vector delivery of an exogenous anti-hepatitis C virus microRNA cluster. Hepatology. 2010;52:1877-1887. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 34] [Cited by in F6Publishing: 37] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 128. | Grimm D, Streetz KL, Jopling CL, Storm TA, Pandey K, Davis CR, Marion P, Salazar F, Kay MA. Fatality in mice due to oversaturation of cellular microRNA/short hairpin RNA pathways. Nature. 2006;441:537-541. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1240] [Cited by in F6Publishing: 1305] [Article Influence: 72.5] [Reference Citation Analysis (0)] |
| 129. | Gupta S, Schoer RA, Egan JE, Hannon GJ, Mittal V. Inducible, reversible, and stable RNA interference in mammalian cells. Proc Natl Acad Sci USA. 2004;101:1927-1932. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 155] [Cited by in F6Publishing: 169] [Article Influence: 8.5] [Reference Citation Analysis (0)] |
| 130. | Sakamoto N, Tanabe Y, Yokota T, Satoh K, Sekine-Osajima Y, Nakagawa M, Itsui Y, Tasaka M, Sakurai Y, Cheng-Hsin C. Inhibition of hepatitis C virus infection and expression in vitro and in vivo by recombinant adenovirus expressing short hairpin RNA. J Gastroenterol Hepatol. 2008;23:1437-1447. [PubMed] [Cited in This Article: ] |
| 131. | Chandra PK, Kundu AK, Hazari S, Chandra S, Bao L, Ooms T, Morris GF, Wu T, Mandal TK, Dash S. Inhibition of hepatitis C virus replication by intracellular delivery of multiple siRNAs by nanosomes. Mol Ther. 2012;20:1724-1736. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 56] [Cited by in F6Publishing: 54] [Article Influence: 4.5] [Reference Citation Analysis (0)] |
| 132. | Bennett CF, Swayze EE. RNA targeting therapeutics: molecular mechanisms of antisense oligonucleotides as a therapeutic platform. Annu Rev Pharmacol Toxicol. 2010;50:259-293. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 952] [Cited by in F6Publishing: 974] [Article Influence: 69.6] [Reference Citation Analysis (0)] |
| 133. | Chan JH, Lim S, Wong WS. Antisense oligonucleotides: from design to therapeutic application. Clin Exp Pharmacol Physiol. 2006;33:533-540. [PubMed] [Cited in This Article: ] |
| 134. | Soler M, McHutchison JG, Kwoh TJ, Dorr FA, Pawlotsky JM. Virological effects of ISIS 14803, an antisense oligonucleotide inhibitor of hepatitis C virus (HCV) internal ribosome entry site (IRES), on HCV IRES in chronic hepatitis C patients and examination of the potential role of primary and secondary HCV resistance in the outcome of treatment. Antivir Ther. 2004;9:953-968. [PubMed] [Cited in This Article: ] |
| 135. | McHutchison JG, Patel K, Pockros P, Nyberg L, Pianko S, Yu RZ, Dorr FA, Kwoh TJ. A phase I trial of an antisense inhibitor of hepatitis C virus (ISIS 14803), administered to chronic hepatitis C patients. J Hepatol. 2006;44:88-96. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 52] [Cited by in F6Publishing: 57] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 136. | McCaffrey AP, Meuse L, Karimi M, Contag CH, Kay MA. A potent and specific morpholino antisense inhibitor of hepatitis C translation in mice. Hepatology. 2003;38:503-508. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 63] [Cited by in F6Publishing: 68] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 137. | González-Carmona MA, Vogt A, Heinicke T, Quasdorff M, Hoffmann P, Yildiz Y, Schneider C, Serwe M, Bartenschlager R, Sauerbruch T. Inhibition of hepatitis C virus gene expression by adenoviral vectors encoding antisense RNA in vitro and in vivo. J Hepatol. 2011;55:19-28. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 9] [Cited by in F6Publishing: 10] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 138. | Jopling CL, Yi M, Lancaster AM, Lemon SM, Sarnow P. Modulation of hepatitis C virus RNA abundance by a liver-specific MicroRNA. Science. 2005;309:1577-1581. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1993] [Cited by in F6Publishing: 1929] [Article Influence: 101.5] [Reference Citation Analysis (0)] |
| 139. | Henke JI, Goergen D, Zheng J, Song Y, Schüttler CG, Fehr C, Jünemann C, Niepmann M. microRNA-122 stimulates translation of hepatitis C virus RNA. EMBO J. 2008;27:3300-3310. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 492] [Cited by in F6Publishing: 513] [Article Influence: 32.1] [Reference Citation Analysis (0)] |
| 140. | Jangra RK, Yi M, Lemon SM. Regulation of hepatitis C virus translation and infectious virus production by the microRNA miR-122. J Virol. 2010;84:6615-6625. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 246] [Cited by in F6Publishing: 254] [Article Influence: 18.1] [Reference Citation Analysis (0)] |
| 141. | Lanford RE, Hildebrandt-Eriksen ES, Petri A, Persson R, Lindow M, Munk ME, Kauppinen S, Ørum H. Therapeutic silencing of microRNA-122 in primates with chronic hepatitis C virus infection. Science. 2010;327:198-201. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1284] [Cited by in F6Publishing: 1262] [Article Influence: 84.1] [Reference Citation Analysis (0)] |
| 142. | Li YP, Gottwein JM, Scheel TK, Jensen TB, Bukh J. MicroRNA-122 antagonism against hepatitis C virus genotypes 1-6 and reduced efficacy by host RNA insertion or mutations in the HCV 5’ UTR. Proc Natl Acad Sci USA. 2011;108:4991-4996. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 145] [Cited by in F6Publishing: 146] [Article Influence: 11.2] [Reference Citation Analysis (0)] |
| 143. | Bukh J. Animal models for the study of hepatitis C virus infection and related liver disease. Gastroenterology. 2012;142:1279-1287.e3. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 101] [Cited by in F6Publishing: 104] [Article Influence: 8.7] [Reference Citation Analysis (0)] |
| 144. | Dorner M, Horwitz JA, Donovan BM, Labitt RN, Budell WC, Friling T, Vogt A, Catanese MT, Satoh T, Kawai T. Completion of the entire hepatitis C virus life cycle in genetically humanized mice. Nature. 2013;501:237-241. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 178] [Cited by in F6Publishing: 175] [Article Influence: 15.9] [Reference Citation Analysis (0)] |