Published online Dec 21, 2013. doi: 10.3748/wjg.v19.i47.8924
Revised: October 30, 2013
Accepted: November 28, 2013
Published online: December 21, 2013
Single nucleotide polymorphisms near the interleukin 28B (IL-28B) gene have been identified as strong predictors of both spontaneous or Peg-interferon (Peg-IFN) and ribavirin (RBV) induced clearance of hepatitis C virus (HCV). Several studies have shown that, in patients with genotype 1 (GT-1), rs12979860 C/C and rs8099917 T/T substitutions are associated with a more than twofold increase in sustained virological response rate to Peg-IFN and RBV treatment. Although new treatment regimens based on combination of DAA with or without IFN are in the approval phase, until combination regimens with a backbone of Peg-IFN will be used, we can expect that IL28B holds its importance. The clinical relevance of IL28B genotyping in treatment of patients infected with HCV genotype 2 (GT-2) and 3 (GT-3) remains controversial. Therefore, after a careful examination of the available literature, we analyzed the impact of IL28B in GT-2 and -3. Simple size of the studies and GT-2 and GT-3 proportion were discussed. An algorithm for the practical use of IL28B in these patients was suggested at the aim of optimizing treatment.
Core tip: The clinical relevance of interleukin 28B genotyping in patients with hepatitis C virus genotype 2 and 3 infection is debated. In this critical analysis of studies performed so far, it was shown that this genetic tool may help in optimizing treatment of genotype 3 patients, whilst it plays a marginal role in genotype 2 infected patients management.
- Citation: Mangia A, Mottola L, Santoro R. Interleukin 28B polymorphisms as predictor of response in hepatitis C virus genotype 2 and 3 infected patients. World J Gastroenterol 2013; 19(47): 8924-8928
- URL: https://www.wjgnet.com/1007-9327/full/v19/i47/8924.htm
- DOI: https://dx.doi.org/10.3748/wjg.v19.i47.8924
In patients with hepatitis C virus (HCV) genotype 1 (GT-1), the standard treatment based on dual combination of Pegylated interferon (Peg-IFN) and ribavirin (RBV) has been replaced by the triple combination regimen including a protease inhibitor; on the contrary, in patients with HCV genotype 2 (GT-2) and genotype 3 (GT-3), it continues to represent the standard of care[1,2].
Interleukin28B (IL-28B) genotype is a strong predictor of response to IFN-based treatment in GT-1[3-6], but at a first glance, genetic analyses so far conducted in GT-2 and -3 patients provided ambiguous results[7-14]. The studies published so far may have bias but we should also bear in mind that identification of response predictors assumes a different relevance in different HCV genotypes. Indeed, high rates of sustained virologic response (SVR) achieved in GT-2 infected patients make response predictors of marginal interest and limit their use to the identification of patients who may take profit from a treatment of short duration whilst. On the contrary, in GT-3 patients, the unsatisfactory rate of SVR reported even with IFN-free regimens, induce a continuous search of predictors of response[15].
A detailed examination of the studies on IL28B in GT-2 and GT-3 patients suggests that there are valid explanations for the contrasting results on SVR association. In our opinion, the analysis of genetic predictors performed in mixed cohorts, incorporating GT-2 or -3 in different proportions may be the first responsible of these contrasting results[10,11]. Additional confounders are the different treatment regimens of the patients included in these studies, heterogeneous in terms of duration and intensity[11,14], and, more importantly, the different populations of patients evaluated in these retrospective genetic analyses that combines naive and previous treatment-experienced[16].
Finally, the use of either rs8099917 SNP located 7.6 kb upstream the IL-28B gene or rs12979860, located 3.2 kb upstream the open reading frame of IL28B gene may be an additional source of confusion. Indeed, these two SNPs showed similar distribution in Caucasian patients but different frequency and, consequently, lower strength of association in races other than Caucasian.
To overcome some of these issues, two meta-analyses on the role of IL28B in GT-2 and 3 have been recently published[17,18]. However, as happened with (the) single studies, these meta-analyses reached contrasting results. In the study by Chen et al[17] no association between SVR and IL-28B CC was found in the subgroup of GT-2 and 3, although it was shown that TT at rs8099917 SNP is associated with a favorable response in GT-2 Asian subjects. In the second meta-analysis, the Authors reached the conclusion that the favorable IL-28B CC genotype is a statistically significant predictor of SVR and rapid virological response (RVR) in Caucasian patients treated with Peg-IFN and ribavirin for 24 wk, with the exception of Asian patients with GT-2 achieving higher rates of RVR when carrying the favorable IL-28B genotype[18].
Data summarizing the results of the studies on rs1297860 in HCV mono-infected patients are reported in Table 1. Studies including GT-2 and -3 lumped together[10,11,13,19] and studies investigating cohorts of patients with GT-3 alone were separately analyzed[12,14,20,21]. Results by genotype were provided by a large study from our group investigating 710 patients[21]. Another study focused on viral kinetics of IL28B polymorphisms by GT-1 vs GT-2 and -3[20].
The results of the largest studies on IL28B treatment response prediction in GT-2 and GT-3 are here analyzed. Combined results for GT-2 and GT-3 together are generally provided. Stored DNA samples from 268 Caucasian patients enrolled in a multicenter controlled trial from Italy were tested for rs12979860. Patients were randomized to Peg-IFN and RBV for standard (24 wk) or variable (12/24 wk) treatment duration on the basis of RVR. Two hundred and thirteen patients were GT-2 and 55 GT-3 infected[10]. IL-28B CC-type was present in 37% of patients. Rates of SVR were 82% in patients with CC-type, 75% in CT and 58% in TT. The CC-type resulted an independent predictor of SVR, but the predictive role was largely driven by the capability of predicting SVR among patients without RVR. Like in GT-1 (22), among the 165 (61%) patients with HCV RNA undetectable at week 4, IL-28B genotype was not predictive of SVR[10].
These findings reinforce the concept that a week 4 undetectable HCV RNA is the strongest predictor of SVR to Peg-IFN and RBV treatment; at the same time, they suggest that the clinical relevance of IL-28B genotype for GT-2 and -3 is far from being borderline, as it can help in selecting patients that may be interferon insensitive at baseline.
The results reported by Sarrazin et al[11] regarding 267 patients (GT-2 = 77, GT-3 = 190), among which only 205 received treatment, are apparently in contrast with our conclusions. The Authors showed an association between IL-28B CC-type and SVR in the subgroup of patients with RVR. No association was observed in patients without RVR[11]. However, despite a lower rate of RVR in observed in this study than in others (40%), only 11 patients without RVR were analyzed (CC = 3, CT = 4 TT = 3). Therefore, a type II error cannot be ruled out. Moreover, if we analyze CC vs CT plus TT patients who completed the treatment, we can observe a trend toward a statistically significant association with SVR in the subgroup of patients without RVR (P = 0.08).
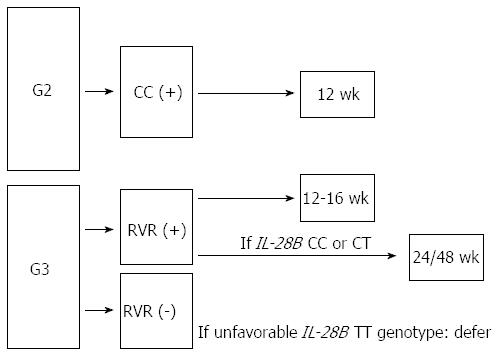
If we consider the results of the previously reported studies on GT-2 and -3 in comparison with those attained in patients with G1 infection[22], we could hypothesize that the lower the rate of RVR, the stronger the association between SVR and CC-type (Figure 1).
Similar considerations apply to the study by Lindh et al[13]. In this study, 341 White patients with GT-2 and -3 were evaluated. The RVR rate was 61%; 134 patients did not achieve RVR. Patients were subdivided according to a treatment duration of 12 or 24 wk (n = 166 and n = 175, respectively). When patients were considered overall, a significant association between IL28B and RVR was shown. However, association between CC and SVR was significant for patients treated for 24 wk (P = 0.02), but not for those receiving a short course of treatment with Peg-IFN alpha-2a and fixed 800 mg doses of RBV. Among patients without RVR, CC genotype was represented in 35%, CT in 47% and TT in 15%. In the subgroup of patients without RVR treated for 24 wk, SVR rates were higher for patients with CC as compared to CT and TT (74% vs 59% and 29%, respectively).
The study by Bitetto et al[19] evaluated 101 patients with GT-2 and -3 as part of a larger cohort of patients with different HCV genotypes. In this study no association with either RVR or SVR was demonstrated among GT-2 and -3. In particular, 78% of IL28B CC infected with GT-2 and GT-3 combined together and 88% of IL28B CT/TT achieved viral clearance after treatment.
Two studies focused on GT-3 only; Scherzer et al[12], evaluated a small cohort of 71 patients, while Moghaddam et al[14] studied 281 patients (Table 2)[20]. Data on 475 GT-3 were separately available also in the study by Fattovich et al[20] and in our prospective cohort of 710 patients with GT-2 and GT-3[21]. These studies are analysed below.
Scherzer et al[12] investigated both rs12979860 and rs8099917 SNPs in a cohort of patients originally randomized 1:1 to 800 or 400 mg of RBV in combination with Peg-IFN alpha-2a. The results of this study might be limited by the small sample size, moreover the low dosages of RBV (used) may impact the generalizability of the conclusions. In the final analysis, only patients who completed the treatment were considered, they were 37 and 31 in each arm, respectively. A CC genotype was identified in 38% of patients, but no association with SVR was observed. Indeed, 19/25 (76%) CC and 34/44 CT and TT combined together (77.3%) achieved SVR[12]. As shown in patients with genotype 1 infection[22], higher levels of baseline HCV RNA were associated in this study with CC genotype as compared to CT or TT[12].
A larger study investigating IL28B in GT-3 has been published by Moghaddam et al[14]. The Authors evaluated both rs12979860 and rs8099917 SNPs in DNA extracted from plasma of 281 GT-3 patients representing 51% of patients enrolled into two previous clinical trials, a non-randomised and a randomised one[23,24]. Authors demonstrated rates of SVR comparable between CC, CT and TT (77% vs 81% and 96%), whereas a statistically significant association between RVR and favorable genotypes was shown. Indeed, 84% of CC-type, 62% of CT and 56% of TT achieved RVR (OR = 3.3, 95%CI: 1.9-5.8). Genotyping rs8099917 SNP, the results were not different (OR = 2.7, 95%CI: 1.6-4.7). In this study, the exclusion of a number of patients who did not fit the inclusion criteria may have represented a bias. Moreover, the association analysis between the different host genotypes and SVR should have been adjusted for confounders, as for example an uneven distribution of patients with favourable genotypes across the different treatment arms. Strikingly, in this study the frequency of CC genotype was 8%-9% higher than in other studies[10].
Negative results were reported also by Fattovich et al[20] in a retrospective cohort study on Italian patients. This study offered the possibility to a separate analysis of GT-2 and GT-3 but the overall number was not higher than 159 including 104 GT-2 and 55 GT-3. No association with RVR was reported in 24 of 28 patients with IL28CC and GT-3 in comparison to 20 of 27 CT/TT (P = 0.31). Similarly, the results were not different for 20 of 23 GT-3 CC and 28 of 32 subjects with CT/TT who achieved SVR (P = 0.79).
The Write study with IL28B available in 93.7% of 710 GT-2 and GT-3 patients is the largest series prospectively evaluating for IL28B. Results of this study including 475 GT-3 showed that while within GT-2 no association between IL28B CC and SVR (90.3% for CC vs 82.0% for non CC, P = 0.15) can be observed, within GT-3, the association between IL-28B CC and SVR is highly significant (84% vs 60%, P < 0.001).
These results demonstrate that when the sample size is adequate, the association between IL28B and RVR or SVR can be appreciated. Therefore, it may be rational to evaluate IL-28B genotyping in patients with GT-3, unless further evidence suggest otherwise. Despite the occurrence of side effects or poor tolerability, patients who bear a favorable IL-28B genotype should not discontinue treatment. At the same time, an unfavorable IL-28B genotype in patients with GT-3 infection may suggest to defer treatment in waiting for more efficacious drugs.
After a careful analysis of the available data, a few aspects deserve consideration. Sample size is one of the most relevant issues in genetic studies as the power of the single study is influenced by the effect size and by the frequency of the minor allele[25]. The effect size of IL28B is large, yet the frequency of the minor allele for SNP rs1297860 ranges between 8% and 16% across the studies evaluating GT-2 and -3[10,12]. Although with such variability, it might be difficult to establish a minimum sample size valid across the studies, the risk for many of them to be underpowered is not negligible[26]. With the assumption of a 0.37 frequency of CC-type and an expected rate of SVR of 0.68 in CT patients, more than 520 patients are required to detect an odds ratio of at least 1.8. Therefore, study’s conclusions should be based on studies with large sample size.
A further consideration owes to be made, the role of predictive factors is not absolute, but it depends on the efficacy of treatment. With about 80% of SVR attained in patients with GT-2 treated with P/R combination it is easy to understand that the sensitivity of the IL-28B genotype for the prediction of SVR in patients with GT-2 is limited and it is easy to understand that combining together GT-2 and GT-3 the sensitivity of the IL-28B genotype for the prediction of SVR is no higher than 40%-47%[10,11]. The lower rate of SVR in patients with GT-3 only put things in a different context suggesting that the combination of unfavorable IL28B and advanced fibrosis may represent a good reason to defer treatment based on Peg-IFN backbone due to the expectancy of a very poor response. Waiting for alternative treatment may be in these case a more reasonable choice.
Based on these considerations, we have imagined an algorithm for the management of patients with chronic GT-2 and GT-3 infection including IL28B and on treatment response (Figure 1). Our proposal is to perform IL-28B genotyping in patients with GT-3 at the aim of encouraging them to treatment, when undecided, to establish the duration of treatment and to decide not to treat those with very poor likelihood of SVR.
In conclusion, in easy to treat GT-2 patients IL28B may be considered as an additional not essential predictor of shortened treatment duration, while in GT-3, genotyping of IL-28B polymorphisms may be used to convince skeptical patients, to maintain on treatment those who are at risk of withdrawing because of side effects and to defer treatment in patients with low likelihood of response.
P- Reviewers: Grizzi F, Sanefuji K, Takami T S- Editor: Qi Y L- Editor: A E- Editor: Wang CH
| 1. | Ghany MG, Nelson DR, Strader DB, Thomas DL, Seeff LB. An update on treatment of genotype 1 chronic hepatitis C virus infection: 2011 practice guideline by the American Association for the Study of Liver Diseases. Hepatology. 2011;54:1433-1444. [PubMed] [DOI] [Cited in This Article: ] |
| 2. | European Association for the Study of the Liver. EASL Clinical Practice Guidelines: management of hepatitis C virus infection. J Hepatol. 2011;55:245-264. [PubMed] [DOI] [Cited in This Article: ] |
| 3. | Thomas DL, Thio CL, Martin MP, Qi Y, Ge D, O’Huigin C, Kidd J, Kidd K, Khakoo SI, Alexander G. Genetic variation in IL28B and spontaneous clearance of hepatitis C virus. Nature. 2009;461:798-801. [PubMed] [DOI] [Cited in This Article: ] |
| 4. | Ge D, Fellay J, Thompson AJ, Simon JS, Shianna KV, Urban TJ, Heinzen EL, Qiu P, Bertelsen AH, Muir AJ. Genetic variation in IL28B predicts hepatitis C treatment-induced viral clearance. Nature. 2009;461:399-401. [PubMed] [DOI] [Cited in This Article: ] |
| 5. | Tanaka Y, Nishida N, Sugiyama M, Kurosaki M, Matsuura K, Sakamoto N, Nakagawa M, Korenaga M, Hino K, Hige S. Genome-wide association of IL28B with response to pegylated interferon-alpha and ribavirin therapy for chronic hepatitis C. Nat Genet. 2009;41:1105-1109. [PubMed] [DOI] [Cited in This Article: ] |
| 6. | Suppiah V, Moldovan M, Ahlenstiel G, Berg T, Weltman M, Abate ML, Bassendine M, Spengler U, Dore GJ, Powell E. IL28B is associated with response to chronic hepatitis C interferon-alpha and ribavirin therapy. Nat Genet. 2009;41:1100-1104. [PubMed] [DOI] [Cited in This Article: ] |
| 7. | Rauch A, Kutalik Z, Descombes P, Cai T, Di Iulio J, Mueller T, Bochud M, Battegay M, Bernasconi E, Borovicka J. Genetic variation in IL28B is associated with chronic hepatitis C and treatment failure: a genome-wide association study. Gastroenterology. 2010;138:1338-1345, 1345.e1-7. [PubMed] [DOI] [Cited in This Article: ] |
| 8. | Rallón NI, Naggie S, Benito JM, Medrano J, Restrepo C, Goldstein D, Shianna KV, Vispo E, Thompson A, McHutchison J. Association of a single nucleotide polymorphism near the interleukin-28B gene with response to hepatitis C therapy in HIV/hepatitis C virus-coinfected patients. AIDS. 2010;24:F23-F29. [PubMed] [DOI] [Cited in This Article: ] |
| 9. | Bochud PY, Bibert S, Negro F, Haagmans B, Soulier A, Ferrari C, Missale G, Zeuzem S, Pawlotsky JM, Schalm S. IL28B polymorphisms predict reduction of HCV RNA from the first day of therapy in chronic hepatitis C. J Hepatol. 2011;55:980-988. [PubMed] [DOI] [Cited in This Article: ] |
| 10. | Mangia A, Thompson AJ, Santoro R, Piazzolla V, Tillmann HL, Patel K, Shianna KV, Mottola L, Petruzzellis D, Bacca D. An IL28B polymorphism determines treatment response of hepatitis C virus genotype 2 or 3 patients who do not achieve a rapid virologic response. Gastroenterology. 2010;139:821-827, 827.e1. [PubMed] [DOI] [Cited in This Article: ] |
| 11. | Sarrazin C, Susser S, Doehring A, Lange CM, Müller T, Schlecker C, Herrmann E, Lötsch J, Berg T. Importance of IL28B gene polymorphisms in hepatitis C virus genotype 2 and 3 infected patients. J Hepatol. 2011;54:415-421. [PubMed] [DOI] [Cited in This Article: ] |
| 12. | Scherzer TM, Hofer H, Staettermayer AF, Rutter K, Beinhardt S, Steindl-Munda P, Kerschner H, Kessler HH, Ferenci P. Early virologic response and IL28B polymorphisms in patients with chronic hepatitis C genotype 3 treated with peginterferon alfa-2a and ribavirin. J Hepatol. 2011;54:866-871. [PubMed] [DOI] [Cited in This Article: ] |
| 13. | Lindh M, Lagging M, Färkkilä M, Langeland N, Mørch K, Nilsson S, Norkrans G, Pedersen C, Buhl MR, Westin J. Interleukin 28B gene variation at rs12979860 determines early viral kinetics during treatment in patients carrying genotypes 2 or 3 of hepatitis C virus. J Infect Dis. 2011;203:1748-1752. [PubMed] [DOI] [Cited in This Article: ] |
| 14. | Moghaddam A, Melum E, Reinton N, Ring-Larsen H, Verbaan H, Bjøro K, Dalgard O. IL28B genetic variation and treatment response in patients with hepatitis C virus genotype 3 infection. Hepatology. 2011;53:746-754. [PubMed] [DOI] [Cited in This Article: ] |
| 15. | Lawitz E, Mangia A, Wyles D, Rodriguez-Torres M, Hassanein T, Gordon SC, Schultz M, Davis MN, Kayali Z, Reddy KR. Sofosbuvir for previously untreated chronic hepatitis C infection. N Engl J Med. 2013;368:1878-1887. [PubMed] [DOI] [Cited in This Article: ] |
| 16. | Yu ML, Huang CF, Huang JF, Chang NC, Yang JF, Lin ZY, Chen SC, Hsieh MY, Wang LY, Chang WY. Role of interleukin-28B polymorphisms in the treatment of hepatitis C virus genotype 2 infection in Asian patients. Hepatology. 2011;53:7-13. [PubMed] [DOI] [Cited in This Article: ] |
| 17. | Chen Y, Xu HX, Wang LJ, Liu XX, Mahato RI, Zhao YR. Meta-analysis: IL28B polymorphisms predict sustained viral response in HCV patients treated with pegylated interferon-α and ribavirin. Aliment Pharmacol Ther. 2012;36:91-103. [PubMed] [DOI] [Cited in This Article: ] |
| 18. | Rangnekar AS, Fontana RJ. IL-28B polymorphisms and the response to antiviral therapy in HCV genotype 2 and 3 varies by ethnicity: a meta-analysis. J Viral Hepat. 2013;20:377-384. [PubMed] [DOI] [Cited in This Article: ] |
| 19. | Bitetto D, Fattovich G, Fabris C, Ceriani E, Falleti E, Fornasiere E, Pasino M, Ieluzzi D, Cussigh A, Cmet S. Complementary role of vitamin D deficiency and the interleukin-28B rs12979860 C/T polymorphism in predicting antiviral response in chronic hepatitis C. Hepatology. 2011;53:1118-1126. [PubMed] [DOI] [Cited in This Article: ] |
| 20. | Fattovich G, Covolo L, Bibert S, Askarieh G, Lagging M, Clément S, Malerba G, Pasino M, Guido M, Puoti M. IL28B polymorphisms, IP-10 and viral load predict virological response to therapy in chronic hepatitis C. Aliment Pharmacol Ther. 2011;33:1162-1172. [PubMed] [DOI] [Cited in This Article: ] |
| 21. | Mangia A, Francavilla R, Merola V, Barbarini G, Russello M, Lorenzo Nosotti L, Nicola Minerva N, Bacca D, Romano M, Cuccorese G. Benefit of weight based dosages of Ribavirin (RBV) in combination with PegInterferon (PegIFN) α2a in naïve patients with HCV2 and HCV3: results of an Italian Randomized Controlled Trial, the WRITE study. Hepatology. 2012;56:1737. [Cited in This Article: ] |
| 22. | Mangia A, Thompson AJ, Santoro R, Piazzolla V, Copetti M, Minerva N, Petruzzellis D, Mottola L, Bacca D, McHutchison JG. Limited use of interleukin 28B in the setting of response-guided treatment with detailed on-treatment virological monitoring. Hepatology. 2011;54:772-780. [PubMed] [DOI] [Cited in This Article: ] |
| 23. | Dalgard O, Bjøro K, Hellum KB, Myrvang B, Ritland S, Skaug K, Raknerud N, Bell H. Treatment with pegylated interferon and ribavarin in HCV infection with genotype 2 or 3 for 14 weeks: a pilot study. Hepatology. 2004;40:1260-1265. [PubMed] [DOI] [Cited in This Article: ] |
| 24. | Dalgard O, Bjøro K, Ring-Larsen H, Bjornsson E, Holberg-Petersen M, Skovlund E, Reichard O, Myrvang B, Sundelöf B, Ritland S. Pegylated interferon alfa and ribavirin for 14 versus 24 weeks in patients with hepatitis C virus genotype 2 or 3 and rapid virological response. Hepatology. 2008;47:35-42. [PubMed] [DOI] [Cited in This Article: ] |
| 25. | Thio CL. Host genetic factors and antiviral immune responses to hepatitis C virus. Clin Liver Dis. 2008;12:713-726, xi. [PubMed] [DOI] [Cited in This Article: ] |
| 26. | Mosbruger TL, Duggal P, Goedert JJ, Kirk GD, Hoots WK, Tobler LH, Busch M, Peters MG, Rosen HR, Thomas DL. Large-scale candidate gene analysis of spontaneous clearance of hepatitis C virus. J Infect Dis. 2010;201:1371-1380. [PubMed] [DOI] [Cited in This Article: ] |