Published online Apr 21, 2013. doi: 10.3748/wjg.v19.i15.2331
Revised: January 13, 2013
Accepted: February 2, 2013
Published online: April 21, 2013
Processing time: 138 Days and 19.1 Hours
AIM: To evaluate the expression of special AT-rich sequence-binding protein 1 (SATB1) gene in colorectal cancer and its role in colorectal cancer cell proliferation and invasion.
METHODS: Immunohistochemistry was used to detect the protein expression of SATB1 in 30 colorectal cancer (CRC) tissue samples and pair-matched adjacent non-tumor samples. Cell growth was investigated after enhancing expression of SATB1. Wound-healing assay and Transwell assay were used to investigate the impact of SATB1 on migratory and invasive abilities of SW480 cells in vitro. Nude mice that received subcutaneous implantation or lateral tail vein were used to study the effects of SATB1 on tumor growth or metastasis in vivo.
RESULTS: SATB1 was over-expressed in CRC tissues and CRC cell lines. SATB1 promotes cell proliferation and cell cycle progression in CRC SW480 cells. SATB1 overexpression could promote cell growth in vivo. In addition, SATB1 could significantly raise the ability of cell migration and invasion in vitro and promote the ability of tumor metastasis in vivo. SATB1 could up-regulate matrix metalloproteases 2, 9, cyclin D1 and vimentin, meanwhile SATB1 could down-regulate E-cadherin in CRC.
CONCLUSION: SATB1 acts as a potential growth and metastasis promoter in CRC. SATB1 may be useful as a therapeutic target for CRC.
- Citation: Fang XF, Hou ZB, Dai XZ, Chen C, Ge J, Shen H, Li XF, Yu LK, Yuan Y. Special AT-rich sequence-binding protein 1 promotes cell growth and metastasis in colorectal cancer. World J Gastroenterol 2013; 19(15): 2331-2339
- URL: https://www.wjgnet.com/1007-9327/full/v19/i15/2331.htm
- DOI: https://dx.doi.org/10.3748/wjg.v19.i15.2331
The special AT-rich sequence-binding protein 1 (SATB1), which locates at human chromosome 3p23, is a thymocyte-specific matrix association region-binding protein that links specific DNA elements to its unique cage-like network[1]. Phosphorylation of SATB1 serves as a molecular switch in determining whether it acts as a transcriptional activator or repressor[2]. SATB1 is predominantly expressed in thymocytes and regulates the spatiotemporal expression of numerous genes that involved in T cell proliferation, development, and differentiation[3]. SATB1 has recently attracted considerable attention in cancer research and its overexpression is a frequent event in various cancers, such as breast cancer, laryngeal cancer, gastric cancer and liver cancer[4-9]. Furthermore, accumulating evidence showed that SATB1 is also associated with tumor growth and metastasis[4-6,8-10]. Han et al[5] found that SATB1 up-regulated the expression of matrix metalloproteases (MMP)2, MMP9 and down-regulated E-cadherin in breast cancer. On the other hand, SATB1 depletion blocks the up-regulation of E-cadherin and extracellular matrix (ECM) protein vimentin. Meng et al[11] showed that SATB1 plays a pivotal role in epithelial to mesenchymal transition (EMT) process and promotes liver cancer invasion. Only one study suggested that SATB1 is over-expressed in human rectal cancer and the expression of SATB1 is associated with clinicopathological parameters, including invasive depth and tumor-node-metastasis (TNM) stage in rectal cancer. Despite its importance, the roles and mechanisms of SATB1 in growth and metastasis of human colorectal cancer (CRC) remain poorly understood.
CRC is the third most common malignancy and the fourth cause of cancer mortality in the world[12-14]. Although novel molecule-based therapies including monoclonal antibodies are currently widely used in the treatment of CRC, many patients with CRC still die from disease recurrence and metastasis[15,16]. Consequently, further elucidation of the molecular mechanisms of CRC will be beneficial for developing novel therapeutic strategies to conquer this disease.
In this study, we analyzed the expression of SATB1 in CRC tissues and found that it was over-expressed in the cancerous tissue samples compared with the normal adjacent tissue samples. We also carried out in vitro and in vivo functional analysis of SATB1 by ectopical SATB1 expression in SW480 CRC cells. Further investigations focused on the regulation of SATB1 in potential downstream molecules MMPs (MMP2, MMP9), cyclin D1 (CCND1), E-cadherin and vimentin.
Human CRC cell lines SW480, SW620, RKO, HT29, HCT116 and Lovo were obtained from Shanghai Institute of Cell Biology (Shanghai, China) and were cultured in RPMI 1640 medium (Invitrogen, Carlsbad, CA, United States), supplemented with 10% fetal bovine serum. The pcDNA3.1 (Invitrogen, Carlsbad, CA, United States) was used to construct a SATB1 over-expressing plasmid. DNA fragment with mature SATB1 or a negative control sequence was inserted to this vector. Stable transfection of the plasmids was carried out using Lipofectamine2000 (Invitrogen, Carlsbad, CA, United States) according to the manufacturer’s instruction.
Paraffin-embedded tumors and paired normal tissue samples were obtained from 30 CRC patients with the approval from the Ethics Committee of the Second Hospital of Zhejiang University Medical College. Immunohistochemical (IHC) analyses were performed on 3-μm, formalin-fixed and paraffin-embedded sections. Primary antibodies for SATB1 were diluted at 1:250 (BD Biosciences, California, United States) for IHC[17,18]. For immunoblot analysis, 20 g total cellular protein was loaded per lane, separated by 4%-12% SDS-polyacrylamide gel electrophoresis, and then transferred to nitrocellulose (Invitrogen, Carlsbad, CA, United States) by electroblotting. The membranes were incubated with either SATB1 antibody (diluted 1:1000; BD Biosciences, California, United States) or α-tubulin antibody (diluted 1:200; Santa Cruz Biotechnology) at 4 °C overnight[19].
Cell proliferation assay was determined by standard 3-(4,5-cimethylthiazol-2-yl)-2,5-diphenyl tetrazolium bromide (MTT) assays. Briefly, the cells were seeded at a density of 2 × 103 cells per well in 96-well culture plates (Costar). Cell proliferation was assessed 24, 48 and 72 h later. One-tenth volume of 5 mg/mL MTT was added to each well, and the plate was further incubated at 37 °C for another 4 h; thereafter, the medium was replaced and the formazan crystals formed were dissolved in 150 μL dimethyl suophoxide with oscillation for 10 min. The optical density was determined with a multiwell spectrophotometer (BioTek, VT, United States) at 570 nm. Absorbance values were presented as percentages relative to untreated controls. The MTT assays were repeated at least three times[20]. For colony formation assay, cells were trypsinized and counted. One hundred cells were seeded in six-well plates. After 2 wk of growth, colonies with a diameter greater than 4 mm were counted. Experiments were performed in quadruplicate[21].
Scratch wound healing assay was performed as previously described. Briefly, transfected cells in 6-well plates were cultured until cells reached confluence and starved overnight. Cell layers were wounded using a 200 μL pipette tip and cultured for another 48 h. Photographs were taken at time 0, 48 and 72 h[22].
A transwell cell migration and Matrigel invasion assay was used to investigate the impact of SATB1 on migratory and invasive ability of SW480 cells. For migration detection, transfected cells were placed in transwell Chamber at 2 × 104 cells/well. The lower transwell chamber contained 10% fetal bovine serum for use as a chemoattractant. For invasion assay, the bottom of the culture inserts (8-mm pores) were coated with 30 μL of the mixture containing serum-free RPMI-1640 and Matrigel (1:8; BD Biosciences, Bedford, MA, United States). The Matrigel was allowed to solidify at 37 °C overnight. After solidification, cells (2 × 104 cells/well) were reseeded onto the upper chamber. Twenty-four hours later, the cells that had migrated or invaded through the membrane were fixed with 95% alcohol and stained with crystal violet. The number of migrated cells or invaded cells was quantified by counting 5 independent symmetrical visual fields under microscope[23].
Cells of 2 × 104 were harvested, washed and resuspended in 200 mL phosphate-buffered saline, and was subcutaneously injected into the flanks of 5-wk-old female nude mice. Animal experimental procedures were performed strictly in accordance with the related ethics regulations of our university. Tumor sizes were measured in two dimensions with calipers every week. Tumor volumes (mm3) were calculated using the following formula: V = (length × width2)/2[24]. For in vivo metastasis assays, SW480-SATB1 cells or SW480-negtive control (SW480-NC) cells were transplanted into nude mice (5-wk-old BALB/c-nu/nu, ten per group, 1 × 106 cells for each mice) through the lateral tail vein. Mice were killed after 10 wk. The lungs were dissected and subjected to hematoxylin and eosin staining. The numbers of metastases in the lungs were examined histologically.
Total RNA was extracted from cells expressing SATB1 and negative control cells with Trizol (Invitrogen, Carlsbad, CA, United States). The expression of CCND1, E-cadherin, vimentin, MMP2 and MMP9 was detected by quantitative real-time polymerase chain reaction (PCR). The primers are as follows: CCND1, the forward primer 5’-TATTGCGCTGCTACCGTTGA-3’ and the reverse primer 5’-CCAATAGCAGCAAACAATGTGAAA-3’; MMP2, the forward primer TCTTCAAGGACCGGTTCATTTG and the reverse primer GATGCTTCCAAACTTCACGCTC; MMP9, the forward primer CACTGTCCACCCCTCAGAGC and the reverse primer GCCACTTGTCGGCGATAAGG; E-cadherin, the forward primer 5’-TGCCCAGAAAATGAAAAAGG-3’ and the reverse primer 5’-GTGTATGTGGCAATGCGTTC-3’; Vimentin, the forward primer 5’-TGGCCGACGCCATCAACACC-3’ and the reverse primer 5’-CACCTCGACGCGGGCTTTGT-3’; β-actin was used as an internal control. The primers for β-actin were 5’-TGACGGGGTCACCCACACTGTGCCCATCT-3’ and 5’-GAAGTAGTAAGTGGGAACCGTGT-3’. Real-time PCR was performed using the SYBR® Green (Invitrogen) dye detection method on ABI PRISM 7900 HT Sequence Detection System under default conditions: 95 °C for 10 min, and 35 cycles of 95 °C for 15 s and 55 °C for 1 min. Comparative Ct method was used for quantification of the transcripts[25].
Each experiment was repeated at least 3 times. All results were expressed as mean ± SD. The difference between means was analyzed with Student’s t test or the χ2 test. All statistical analysis were performed using SPSS 16.0 software (Chicago, IL, United States). Differences were considered significant when P < 0.05[26].
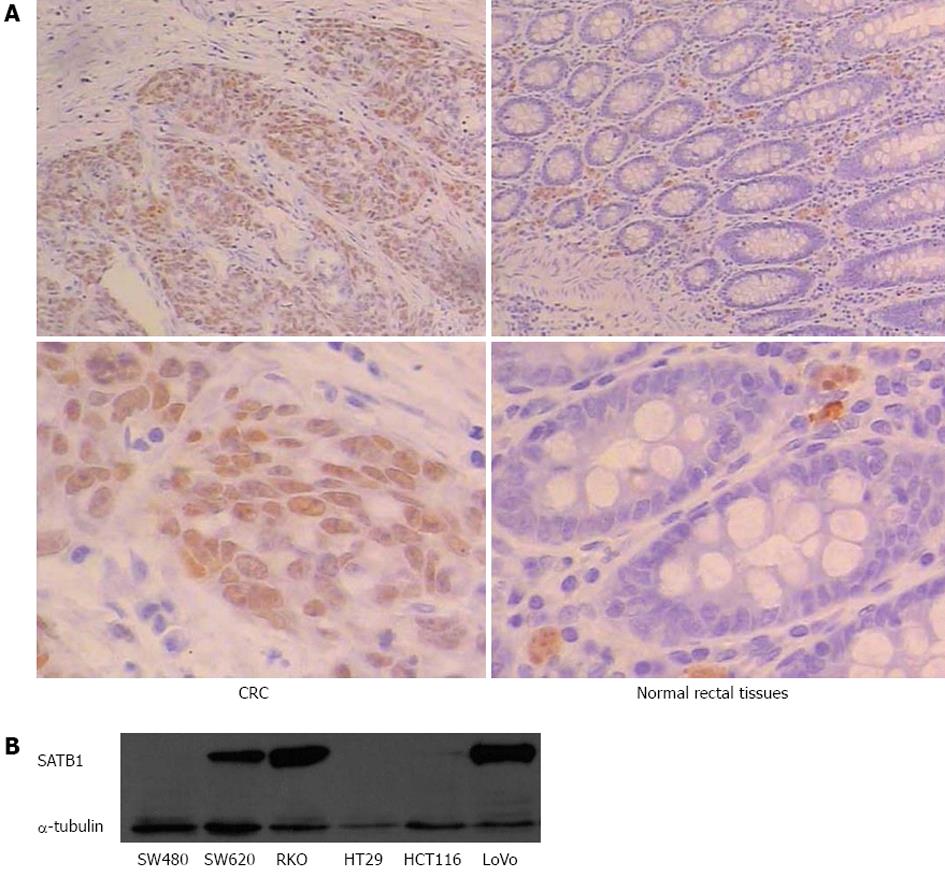
To assess the role of SATB1 in CRC, we examined the protein expression of SATB1 in 30 human CRC tissue samples and pair-matched adjacent non-tumor tissue samples by IHC. We observed positive immunoreactivities in CRC in 53% (16 of 30) of cancer tissue samples, compared with only 10% (3 of 30) in the adjacent mucosa tissue cells. The representative examples of IHC staining results are shown in Figure 1A. Statistical analysis using Pearson χ2 (df = 1, two-sided) indicates that the difference in SATB1 expression between cancer and adjacent tissues was significant (P < 0.01) (Table 1). Western blot analysis of SATB1 in established CRC cell lines showed that the expression level of SATB1 in SW620 was higher in the SW480 (Figure 1B). SW480 and SW620 were a matched pair of primary and metastatic population of cells from the same patient[27]. SW620 cells were derived from the metastasis lymph node of Dukes’ type C colorectal adenocarcinoma.
| Cancer tissues | Normal tissues | |
| Total number of samples | 30 | 30 |
| Samples with SATB1 expression in nucleus | 16 | 3 |
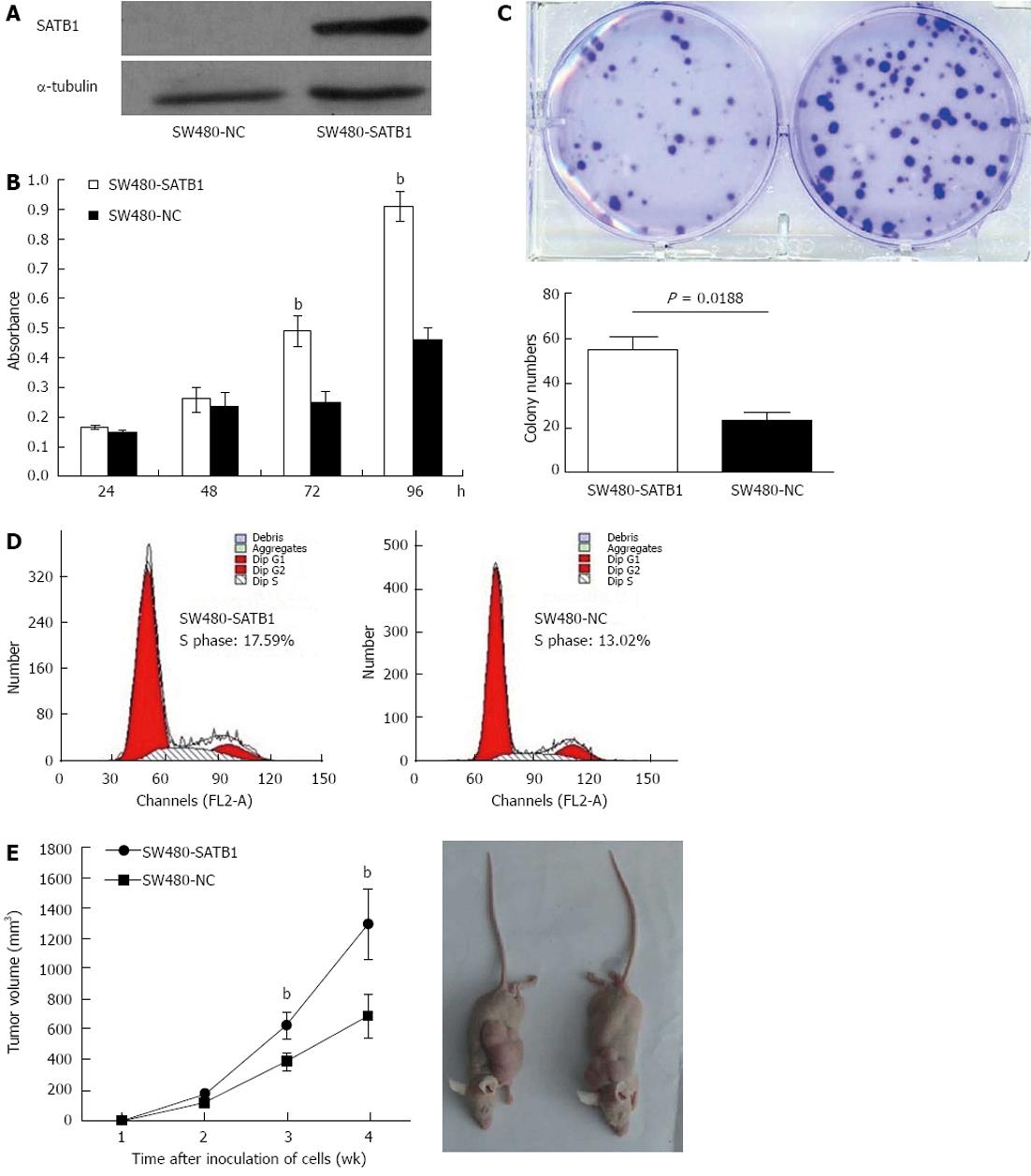
In order to investigate the role of SATB1 in CRC carcinogenesis, we tested the effect of SATB1 on the proliferation of SW480 cells. We established stable SATB1 expressing CRC cells. As shown in Figure 2A, SATB1 levels were higher in cells stably expressing SATB1 than the negative control cells. MTT assay showed that introduction of SATB1 caused a remarkable promotion of cell proliferation in SW480 cells (P < 0.05; Figure 2B). Furthermore, expression of SATB1 in SW480 cells significantly enhanced the numbers of colony formation. As shown in Figure 2C, the colony number for the negative control cells was 55.3 ± 5.0, while that for the SATB1 overexpression was 23.7 ± 3.2 (P = 0.018). To clarify the mechanisms underlying growth promotion by SATB1 in CRC cell lines, we performed cell-cycle analysis using flow cytometry on the cells stained with propidium iodide. SW480-SATB1 cells showed a higher proportion of cells in S phase (17.59%), compared with the control cells (13.02% for SW480-NC cells) (Figure 2D).
In order to assess the role of SATB1 on CRC tumorigenesis in vivo , equal numbers of SW480-SATB1 cells and SW480-NC cells were implanted onto flanks of 5-wk-old female nude mice, and the growth of the implanted tumors was measured at weeks 1-4. The results indicated that SW480 cells with enhanced SATB1 expression could promote the growth of subcutaneous tumors (Figure 2E) (P < 0.01).
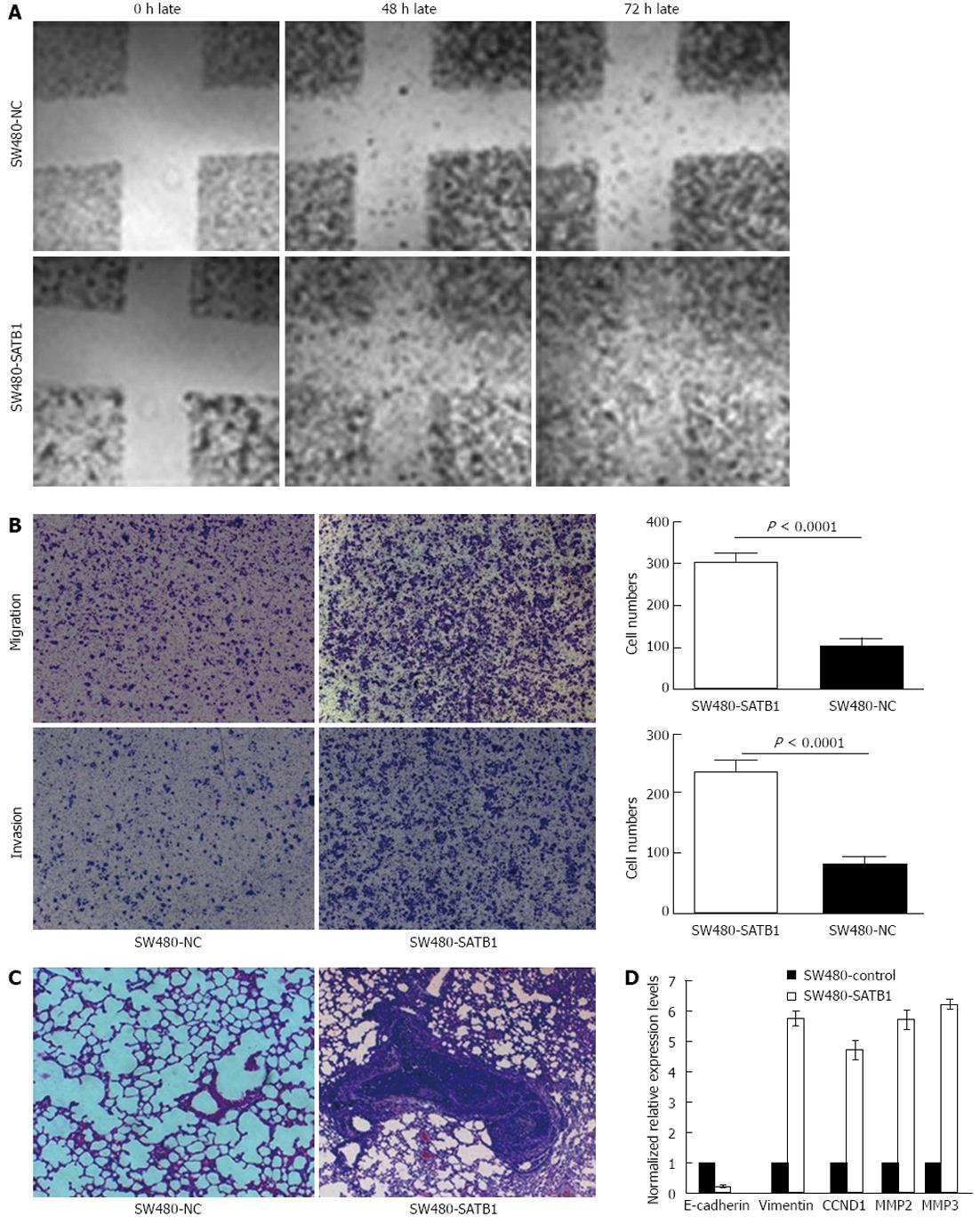
Wound-healing assay was performed to examine the effect of SATB1 expression on cell migration. We found that SW480-SATB1 cells healed the scratch wound earlier than negative controls cells (Figure 3A). We also estimated the effects of SATB1 on the migration and invasion of SW480 cells using a Transwell cell migration and Matrigel invasion assay. The data demonstrated that the overexpression of SATB1 markedly promoted the migration and invasion of SW480 cells. The number of SW480-SATB1 cells (307 ± 20, P < 0.0001) that had migrated through the membrane without Matrigel was significantly higher than that of SW480-NC cells (104 ± 20) (Figure 3B). A similar result was found with the invaded cells; the number of SW480-SATB1 cells (237 ± 19, P < 0.0001) passing through the Matrigel was significantly higher than that of SW480-NC cells (82 ± 10) (Figure 3B). To further explore the effects of SATB1 on tumor metastasis in vivo, SW480-SATB1 cells or SW480-NC cells were transplanted into nude mice through the lateral tail vein. Histological analysis of the lung of mice confirmed that SATB1 could promote lung metastasis formation. Lung metastasis of SW480 cells was apparent in mice injected with SW480-SATB1 cells (Figure 3C). In contrast, few metastatic tumors were detected in mice injected with SW480-NC cells (Figure 3C). Our results indicate that SATB1 could promote CRC cell metastasis in vivo.
We detected the potential downstream molecules regulated by SATB1 via real-time PCR analysis to probe into the possible mechanism that SATB1 promotes CRC cell proliferation and metastasis. The results showed that the expression of CCND1 was up-regulated in SW480 cells with enhanced SATB1 expression. We also found that MMP2 and MMP9, the major MMPs that have a key role in the proteolytic cascade-leading ECM cleavage during metastasis in colon carcinoma, were up-regulated in SW480-SATB1 cells (Figure 3D). In addition, the expression of EMT related gene vimentin was increased and E-cadherin was decreased in SW480-SATB1 cells (Figure 3D).
Previous studies have suggested the important role of SATB1 in tumor growth and metastasis. But there have been few researches on the relationship between SATB1 and CRC. More recently, Meng et al[11] reported that high level of SATB1 expression was closely correlated with invasive depth and TNM stage in 93 paired samples of human rectal cancer. However, the effects of SATB1 on CRC remain poorly understood. In this study, we showed that overexpression of SATB1 in CRC tissue samples and cell lines could promote cell growth in vitro and in vivo. In addition, SATB1 could significantly increase the ability of cell migration and invasion in vitro and promote the ability of tumor metastasis in vivo. We further showed that SATB1 could up-regulate MMPs 2, 9 and vimentin, meanwhile SATB1 could down-regulate E-cadherin in CRC. The data from the current study suggested that SATB1 acts as a potential growth and metastasis promoter in CRC.
Immunohistochemical results showed that the SATB1 protein was overexpressed in CRC tissues and was localized in the nuclei of cancer cells. We also found that SATB1 was overexpressed in cell lines derived from CRC. Our finding is consistent with a recent report showing that the expression of SATB1 was increased in rectal cancer and cell lines[11]. These data prompted us to analyze the functional effects of SATB1 in CRC cells. We found that SATB1 promotes cell proliferation and cell cycle progression in CRC SW480 cells. In addition, SATB1 expression could promote cell growth in vivo. These results suggested that SATB1 may play a tumor promoter role in CRC carcinogenesis.
Invasion and metastasis are the most influential factors for clinical outcome of CRC. Recent studies have shown that SATB1 contributes to tumor metastasis in many types of tumors, such as breast cancer, gastric cancer, and liver cancer[5,7-9]. Up to date, there was only one report about SATB1 expression and clinical feature in rectal cancer which found that high levels of SATB1 expression were closely correlated with invasive depth and TNM stage in human rectal cancer samples[11]. This report suggested that SATB1 may facilitate CRC metastasis. In this study, we found that ectopical SATB1 expression endows the non-aggressive SW480 cells with a capability of migration and invasion in vitro and metastasis in vivo. So we consider that SATB1 may play a crucial role in promoting cancer invasion and metastasis in CRC.
MMP2 and 9, which degrade ECM and promote tumor invasion[28,29], were up-regulated in the SW480-SATB1 cells that ectopically expressed SATB1. We also found up-regulation of vimentin and down-regulation of E-cadherin in mRNA level in the SW480-SATB1 cells. As a genome organizer, SATB1 recruits chromatin remodeling factors and regulates the spatiotemporal expression of numerous genes involving tumor growth and metastasis. SATB1 has been found to promote breast tumor metastasis and reprograms the genome to change the expression profiles consistent with invasive tumors[5]. MMP2 and MMP9 are gelatinases that belong to multigene family of proteolytic enzymes[30]. MMP2 and MMP9 are capable of degrading essentially all the ECM components and the basal membrane, both of which play an crucial role in preventing the migration of cancer cells[31]. In this sense, MMP2 and MMP9 play an important role in the proteolytic cascade-leading ECM degradation during metastasis in colon carcinoma[32,33]. E-cadherin is an adherent junction protein and tumor suppressor. Low E-cadherin and high vimentin are traditional markers currently accustomed to discern cells that have undergone a EMT process[34]. EMT is a process that epithelial cells lose polarity, cell-to-cell contacts, and cytoskeletal integrity contributing to the dissemination of carcinoma cells from epithelial tumors[35,36]. EMT is thought to be responsible for seeding distant dissemination, eventually leading to cancer-related mortality[34]. Han et al[5] firstly reported that SATB1 regulated EMT related gene such as E-cadherin, vimentin, fibronectin, N-cadherin, SNAIL and SIP1, and SATB1 depletion restores cell polarity and reduces aggressive phenotypes of breast cancer MDA-MB-231 cells in vitro. Another link between SATB1 and EMT was emphasized by Tu et al[9], suggesting that SATB1 mainly induces EMT concomitant with increased expression of Snail1, Slug, Twist and vimentin and decreased expression of E-cadherin, tight junction protein ZO-1 and desmoplakin in liver cancer cell lines. The data from the current study suggest that SATB1 can promote CRC metastasis by degrading ECM and inducing EMT in part.
In conclusion, this study demonstrated that SATB1 is over-expressed in CRC and can promote the growth and metastasis of CRC cells in vitro and in vivo. We thus have found a new potential promoting factor for the development and progression of CRC.
Elucidation of the molecular mechanisms of colorectal cancer (CRC) is beneficial for developing novel therapeutic strategies to conquer this disease and in this study the authors analyzed the role of special AT-rich sequence-binding protein 1 (SATB1) in CRC carcinogenesis.
Only one study suggested that the expression of SATB1 is associated with clinicopathological parameters in CRC. But the roles and mechanisms of SATB1 in growth and metastasis of human CRC remain poorly understood.
SATB1 promotes CRC cell growth, migration and invasion in vitro and the ability of tumor metastasis in vivo.
SATB1 acts as a potential growth and metastasis promoter in CRC. SATB1 may be useful as a therapeutic target for CRC.
SATB1 gene locates at human chromosome 3p23, and is a thymocyte-specific matrix association region-binding protein. SATB1 is predominantly expressed in thymocytes and could regulate the spatiotemporal expression of numerous genes that involved in T cell proliferation, development, and differentiation. SATB1 has recently attracted considerable attention in cancer research and its overexpression is a frequent event in various cancers. Furthermore, accumulating evidence showed that SATB1 is also associated with tumor growth and metastasis.
The manuscript is well written and the findings are important in its field of investigation.
P- Reviewer Fabio G S- Editor Jiang L L- Editor Ma JY E- Editor Li JY
| 1. | Dickinson LA, Joh T, Kohwi Y, Kohwi-Shigematsu T. A tissue-specific MAR/SAR DNA-binding protein with unusual binding site recognition. Cell. 1992;70:631-645. [PubMed] |
| 2. | Pavan Kumar P, Purbey PK, Sinha CK, Notani D, Limaye A, Jayani RS, Galande S. Phosphorylation of SATB1, a global gene regulator, acts as a molecular switch regulating its transcriptional activity in vivo. Mol Cell. 2006;22:231-243. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 127] [Cited by in RCA: 139] [Article Influence: 7.3] [Reference Citation Analysis (0)] |
| 3. | Alvarez JD, Yasui DH, Niida H, Joh T, Loh DY, Kohwi-Shigematsu T. The MAR-binding protein SATB1 orchestrates temporal and spatial expression of multiple genes during T-cell development. Genes Dev. 2000;14:521-535. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 179] [Cited by in RCA: 206] [Article Influence: 8.2] [Reference Citation Analysis (0)] |
| 4. | Iorns E, Hnatyszyn HJ, Seo P, Clarke J, Ward T, Lippman M. The role of SATB1 in breast cancer pathogenesis. J Natl Cancer Inst. 2010;102:1284-1296. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 55] [Cited by in RCA: 57] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 5. | Han HJ, Russo J, Kohwi Y, Kohwi-Shigematsu T. SATB1 reprogrammes gene expression to promote breast tumour growth and metastasis. Nature. 2008;452:187-193. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 377] [Cited by in RCA: 396] [Article Influence: 23.3] [Reference Citation Analysis (0)] |
| 6. | Zhao XD, Ji WY, Zhang W, He LX, Yang J, Liang HJ, Wang LL. Overexpression of SATB1 in laryngeal squamous cell carcinoma. ORL J Otorhinolaryngol Relat Spec. 2010;72:1-5. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 46] [Cited by in RCA: 50] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 7. | Lu X, Cheng C, Zhu S, Yang Y, Zheng L, Wang G, Shu X, Wu K, Liu K, Tong Q. SATB1 is an independent prognostic marker for gastric cancer in a Chinese population. Oncol Rep. 2010;24:981-987. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1] [Cited by in RCA: 2] [Article Influence: 0.1] [Reference Citation Analysis (0)] |
| 8. | Cheng C, Lu X, Wang G, Zheng L, Shu X, Zhu S, Liu K, Wu K, Tong Q. Expression of SATB1 and heparanase in gastric cancer and its relationship to clinicopathologic features. APMIS. 2010;118:855-863. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 71] [Cited by in RCA: 77] [Article Influence: 5.1] [Reference Citation Analysis (0)] |
| 9. | Tu W, Luo M, Wang Z, Yan W, Xia Y, Deng H, He J, Han P, Tian D. Upregulation of SATB1 promotes tumor growth and metastasis in liver cancer. Liver Int. 2012;32:1064-1078. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 70] [Cited by in RCA: 84] [Article Influence: 6.5] [Reference Citation Analysis (0)] |
| 10. | Patani N, Jiang W, Mansel R, Newbold R, Mokbel K. The mRNA expression of SATB1 and SATB2 in human breast cancer. Cancer Cell Int. 2009;9:18. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 72] [Cited by in RCA: 78] [Article Influence: 4.9] [Reference Citation Analysis (0)] |
| 11. | Meng WJ, Yan H, Zhou B, Zhang W, Kong XH, Wang R, Zhan L, Li Y, Zhou ZG, Sun XF. Correlation of SATB1 overexpression with the progression of human rectal cancer. Int J Colorectal Dis. 2012;27:143-150. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 59] [Cited by in RCA: 63] [Article Influence: 4.8] [Reference Citation Analysis (0)] |
| 12. | Tenesa A, Dunlop MG. New insights into the aetiology of colorectal cancer from genome-wide association studies. Nat Rev Genet. 2009;10:353-358. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 308] [Cited by in RCA: 302] [Article Influence: 18.9] [Reference Citation Analysis (0)] |
| 13. | Zheng X, Wang L, Zhu Y, Guan Q, Li H, Xiong Z, Deng L, Lu J, Miao X, Cheng L. The SNP rs961253 in 20p12.3 is associated with colorectal cancer risk: a case-control study and a meta-analysis of the published literature. PLoS One. 2012;7:e34625. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 11] [Cited by in RCA: 13] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 14. | Gezen C, Kement M, Altuntas YE, Okkabaz N, Seker M, Vural S, Gumus M, Oncel M. Results after multivisceral resections of locally advanced colorectal cancers: an analysis on clinical and pathological t4 tumors. World J Surg Oncol. 2012;10:39. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 48] [Cited by in RCA: 53] [Article Influence: 4.1] [Reference Citation Analysis (0)] |
| 15. | Meyerhardt JA, Li L, Sanoff HK, Carpenter W, Schrag D. Effectiveness of bevacizumab with first-line combination chemotherapy for Medicare patients with stage IV colorectal cancer. J Clin Oncol. 2012;30:608-615. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 100] [Cited by in RCA: 112] [Article Influence: 8.6] [Reference Citation Analysis (0)] |
| 16. | Van Cutsem E, Köhne CH, Láng I, Folprecht G, Nowacki MP, Cascinu S, Shchepotin I, Maurel J, Cunningham D, Tejpar S. Cetuximab plus irinotecan, fluorouracil, and leucovorin as first-line treatment for metastatic colorectal cancer: updated analysis of overall survival according to tumor KRAS and BRAF mutation status. J Clin Oncol. 2011;29:2011-2019. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1314] [Cited by in RCA: 1452] [Article Influence: 103.7] [Reference Citation Analysis (0)] |
| 17. | Fang X, Yu W, Li L, Shao J, Zhao N, Chen Q, Ye Z, Lin SC, Zheng S, Lin B. ChIP-seq and functional analysis of the SOX2 gene in colorectal cancers. OMICS. 2010;14:369-384. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 46] [Cited by in RCA: 57] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 18. | Lin B, Madan A, Yoon JG, Fang X, Yan X, Kim TK, Hwang D, Hood L, Foltz G. Massively parallel signature sequencing and bioinformatics analysis identifies up-regulation of TGFBI and SOX4 in human glioblastoma. PLoS One. 2010;5:e10210. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 64] [Cited by in RCA: 74] [Article Influence: 4.9] [Reference Citation Analysis (0)] |
| 19. | Foltz G, Ryu GY, Yoon JG, Nelson T, Fahey J, Frakes A, Lee H, Field L, Zander K, Sibenaller Z. Genome-wide analysis of epigenetic silencing identifies BEX1 and BEX2 as candidate tumor suppressor genes in malignant glioma. Cancer Res. 2006;66:6665-6674. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 101] [Cited by in RCA: 118] [Article Influence: 6.2] [Reference Citation Analysis (0)] |
| 20. | Hou Z, Xie L, Yu L, Qian X, Liu B. MicroRNA-146a is down-regulated in gastric cancer and regulates cell proliferation and apoptosis. Med Oncol. 2012;29:886-892. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 64] [Cited by in RCA: 75] [Article Influence: 5.4] [Reference Citation Analysis (0)] |
| 21. | Fang X, Yoon JG, Li L, Yu W, Shao J, Hua D, Zheng S, Hood L, Goodlett DR, Foltz G. The SOX2 response program in glioblastoma multiforme: an integrated ChIP-seq, expression microarray, and microRNA analysis. BMC Genomics. 2011;12:11. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 111] [Cited by in RCA: 125] [Article Influence: 8.9] [Reference Citation Analysis (0)] |
| 22. | Hou Z, Yin H, Chen C, Dai X, Li X, Liu B, Fang X. microRNA-146a targets the L1 cell adhesion molecule and suppresses the metastatic potential of gastric cancer. Mol Med Rep. 2012;6:501-506. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 36] [Cited by in RCA: 40] [Article Influence: 3.1] [Reference Citation Analysis (0)] |
| 23. | Zhou HM, Dong TT, Wang LL, Feng B, Zhao HC, Fan XK, Zheng MH. Suppression of colorectal cancer metastasis by nigericin through inhibition of epithelial-mesenchymal transition. World J Gastroenterol. 2012;18:2640-2648. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 29] [Cited by in RCA: 29] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 24. | Chen Y, Shi L, Zhang L, Li R, Liang J, Yu W, Sun L, Yang X, Wang Y, Zhang Y. The molecular mechanism governing the oncogenic potential of SOX2 in breast cancer. J Biol Chem. 2008;283:17969-17978. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 257] [Cited by in RCA: 289] [Article Influence: 17.0] [Reference Citation Analysis (0)] |
| 25. | Ozden SA, Ozyurt H, Ozgen Z, Kilinc O, Oncel M, Gul AE, Karadayi N, Serakinci N, Kan B, Orun O. Prognostic role of sensitive-to-apoptosis gene expression in rectal cancer. World J Gastroenterol. 2011;17:4905-4910. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 6] [Cited by in RCA: 8] [Article Influence: 0.6] [Reference Citation Analysis (0)] |
| 26. | Shen H, Yuan Y, Hu HG, Zhong X, Ye XX, Li MD, Fang WJ, Zheng S. Clinical significance of K-ras and BRAF mutations in Chinese colorectal cancer patients. World J Gastroenterol. 2011;17:809-816. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 27] [Cited by in RCA: 36] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 27. | Kubens BS, Zänker KS. Differences in the migration capacity of primary human colon carcinoma cells (SW480) and their lymph node metastatic derivatives (SW620). Cancer Lett. 1998;131:55-64. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 41] [Cited by in RCA: 45] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 28. | Jimenez RE, Hartwig W, Antoniu BA, Compton CC, Warshaw AL, Fernández-Del Castillo C. Effect of matrix metalloproteinase inhibition on pancreatic cancer invasion and metastasis: an additive strategy for cancer control. Ann Surg. 2000;231:644-654. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 39] [Cited by in RCA: 51] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 29. | Horikawa T, Yoshizaki T, Sheen TS, Lee SY, Furukawa M. Association of latent membrane protein 1 and matrix metalloproteinase 9 with metastasis in nasopharyngeal carcinoma. Cancer. 2000;89:715-723. [PubMed] |
| 30. | Egeblad M, Werb Z. New functions for the matrix metalloproteinases in cancer progression. Nat Rev Cancer. 2002;2:161-174. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4447] [Cited by in RCA: 4487] [Article Influence: 195.1] [Reference Citation Analysis (0)] |
| 31. | Deryugina EI, Quigley JP. Matrix metalloproteinases and tumor metastasis. Cancer Metastasis Rev. 2006;25:9-34. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1413] [Cited by in RCA: 1499] [Article Influence: 78.9] [Reference Citation Analysis (0)] |
| 32. | Yoon SO, Park SJ, Yun CH, Chung AS. Roles of matrix metalloproteinases in tumor metastasis and angiogenesis. J Biochem Mol Biol. 2003;36:128-137. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 143] [Cited by in RCA: 174] [Article Influence: 7.9] [Reference Citation Analysis (0)] |
| 33. | John A, Tuszynski G. The role of matrix metalloproteinases in tumor angiogenesis and tumor metastasis. Pathol Oncol Res. 2001;7:14-23. [PubMed] |
| 34. | Roussos ET, Keckesova Z, Haley JD, Epstein DM, Weinberg RA, Condeelis JS. AACR special conference on epithelial-mesenchymal transition and cancer progression and treatment. Cancer Res. 2010;70:7360-7364. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 62] [Cited by in RCA: 71] [Article Influence: 4.7] [Reference Citation Analysis (0)] |
| 35. | Thiery JP. Epithelial-mesenchymal transitions in tumour progression. Nat Rev Cancer. 2002;2:442-454. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4877] [Cited by in RCA: 5106] [Article Influence: 222.0] [Reference Citation Analysis (0)] |
| 36. | Guarino M, Rubino B, Ballabio G. The role of epithelial-mesenchymal transition in cancer pathology. Pathology. 2007;39:305-318. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 538] [Cited by in RCA: 616] [Article Influence: 34.2] [Reference Citation Analysis (0)] |