Published online Jan 28, 2011. doi: 10.3748/wjg.v17.i4.526
Revised: November 2, 2010
Accepted: November 9, 2010
Published online: January 28, 2011
AIM: To clarify the role of high in normal-1 (HIN-1) gene promoter methylation during gastric cancer development.
METHODS: Gastric cancer cell lines and tissue specimens were analyzed for expression of HIN-1 mRNA and protein using the semi-quantitative reverse transcription polymerase chain reaction and immunohistochemistry. The methylation of the HIN-1 gene promoter was detected in gastric carcinoma cells and tissues using methylation-specific polymerase chain reaction. The 3-(4,5-dimethylthiazol-2yl)-5-(3-carboxymethoxyphenyl)-2-(4-sulfophenyl)-2H-tetrazolium cell viability assay and flow cytometry were used to assess the changes in behaviors of gastric cancer cells with or without 5-aza-2’-deoxycytidine treatment.
RESULTS: HIN-1 was not expressed in 4 of 5 gastric cancer cell lines. The demethylation reagent 5-aza-2’-deoxycytidine was able to induce or upregulate HIN-1 expression in gastric cancer cell lines, which is associated with reduction of tumor cell viability. Furthermore, methylation of the HIN-1 gene promoter was shown in 57.8% (26/45) of the primary gastric cancer and 42.1% (17/38) of adjacent tissue samples, but was not shown in normal gastric mucosa (0/10). From the clinicopathological data of the patients, methylation of the HIN-1 gene promoter was found to be associated with tumor differentiation (P = 0.000).
CONCLUSION: High methylation of HIN-1 gene promoter results in silence of HIN-1 expression in gastric cancer. 5-aza-2’-deoxycytidine reverses HIN-1 methylation and reduces viability of gastric cancer cells.
- Citation: Gong Y, Guo MZ, Ye ZJ, Zhang XL, Zhao YL, Yang YS. Silence of HIN-1 expression through methylation of its gene promoter in gastric cancer. World J Gastroenterol 2011; 17(4): 526-533
- URL: https://www.wjgnet.com/1007-9327/full/v17/i4/526.htm
- DOI: https://dx.doi.org/10.3748/wjg.v17.i4.526
Gastric cancer is the second most common cause of cancer death worldwide after lung cancer[1,2]. Gastric carcinogenesis, like all other cancers, is a multistep process, involving numerous genetic and epigenetic alterations, such as abnormalities in growth factors/receptors, angiogenic factors, cell cycle regulators, and DNA mismatch repair genes. These abnormalities also define biological characteristics of gastric cancer cells, which can serve as therapeutic targets for gastric cancer[3,4]. Although genetic abnormalities including gene mutation and deletion are prominent in causing oncogene activation and tumor suppressor gene inactivation, epigenetic silence of tumor suppressor genes via aberrant promoter hypermethylation have also been shown to be frequent events in gastric carcinoma[5,6]. DNA high methylation of tumor suppressor genes frequently occurs in the early stage of human carcinogenesis, and investigating the methylation of these gene promoters may contribute to the diagnosis, prognosis and target therapy in gastric carcinoma[7,8].
High in normal-1 (HIN-1) gene was originally isolated through a serial analysis of gene expression from normal and ductal carcinoma in situ luminal mammary epithelial cells. The latter is believed to be the precursor of invasive ductal carcinoma[9]. HIN-1 is highly expressed in normal luminal mammary epithelial cells but lost in the majority of breast cancers. Restoration of HIN-1 expression suppressed growth of breast cancer cells[10]. HIN-1 can also regulate cell-cycle reentry, suppresses tumor cell migration and invasion, and induces apoptosis in breast cancer cell lines[10]. Although HIN-1 processes the putative tumor suppressor function, no somatically genetic changes of HIN-1 gene were found in breast cancer[9]. Previous studies demonstrated frequent methylation of HIN-1 gene promoter in breast cancer, prostate cancer, malignant mesotheliomas, non-small cell lung cancer, lymphoma, retinoblastoma, Wilms’ tumor, and rhabdomyosarcoma[11-14].
However, expression of this putative tumor suppressor gene in gastric cancer has not been fully studied. Therefore, in this study, we first confirmed the methylation of HIN-1 gene promoter in human gastric cancer cell lines and determined the role of 5-aza-2’-deoxycytidine [5-aza-dc, a drug that inhibits the DNA methyltransferase (DNMT)-mediated hypermethylation of promoter region CpG islands] in regulation of HIN-1 expression in gastric cancer cells. We also detected the methylation of HIN-1 gene promoter in tissue specimens and found the association between HIN-1 gene promoter methylation and clinicopathologic characteristics of gastric cancer.
Gastric carcinoma cell lines KATOIII, AGS, PHM82, NUGC3, and BCG823 were obtained from American Type Culture Collection (Manassas, VA) and cultured in either RPMI 1640 medium or RPMI 1640/Ham’s F-12 medium (all from Invitrogen, Carlsbad, CA) supplemented with 10% fetal bovine serum in a humidified incubator with 5% CO2 and 95% air at 37°C. These cells were passaged at a ratio of 1:3 with trypsin once they reached confluence (approximately 106 cells) into 75 cm2 culture flasks (Sarstedt, Newton, NC). For treatment with 5-aza-2’-deoxycytidine, these cell lines were split and cultured at a low density (30% confluence) overnight and then treated with 5-aza-2’-deoxycytidine (Sigma, St. Louis, MO) at a concentration of 1 μmol/L for up to 96 h. The growth medium was refreshed every 24 h, and at the end of the treatment, DNA and RNA from these cells were isolated as described below.
In the current study, 45 surgically resected and pathologically confirmed gastric tumors and 38 adjacent non-tumor tissues were obtained from the PLA General Hospital, Beijing, China between January 2009 and January 2010 and stored in liquid nitrogen until use. Ten cases of normal gastric mucosa were also obtained from the gastric endoscopic biopsies of tumor-free patients. This study was approved by our hospital’s Institutional Review Board.
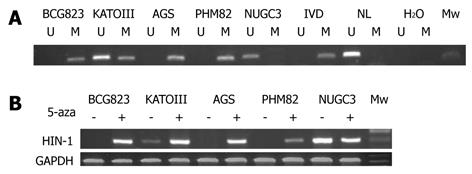
Genomic DNA from these cell lines and tissue specimens were extracted using a proteinase-K method described previously[15]. The extracted DNA was then dissolved in Tris-EDTA (TE) buffer and stored at -20°C. To assess the methylation levels of the HIN-1 gene promoter, genomic DNA from gastric cancer cell lines and tissue specimens were first subjected to bisulfite treatment and then methylation-specific polymerase chain reaction (MSP) as described previously[16]. The MSP primers for HIN-1 were designed and synthesized according to genomic sequences skirting the presumed transcription start sites for HIN-1. The HIN-1 MSP primers spanned a region of 92 base pairs for unmethylation (location is from +128 to +41) and 88 base pairs for methylation (location is from +131 to +40). The primer sequences were: HIN-1-UN 5'-GAAGTTTTGTGGTTTTGTTTGGGTAGTT-3', HIN-1-UN-AS 5'-CACACAAAACCCCAAAAAAACAACA-3', HIN-1-ME-S 5'-GTTTCGTGGTTTTGTTCGGGTAGTC-3' and HIN-1-ME-AS 5'-GCAAAACCCCAAAAAAACGACG-3'. Each MSP reaction incorporated approximately 100 ng of bisulfite-treated DNA, 25 picomoles of each primer, 100 pmoles dNTPs, 2.5 μL 10 × PCR buffer, and 1 unit of JumpStart Red Taq Polymerase (Sigma) in a final reaction volume of 25 μL. The PCR amplification conditions were an initial 95°C for 5 min and then 35 cycles of 95°C for 30 s, 60°C for 30 s, and 72°C for 30 s and a final extension at 72°C for 5 min and then stored at 4°C. The MSP products were separated on 2% agarose gel electrophoresis and visualized under the ultraviolet (UV) light.
Total cellular RNA from the cell lines was isolated using the TRIzol reagent (Invitrogen) according to the manufacturer’s instructions. RNA quality and quantity were assessed using agarose gel electrophoresis (1%) and spectrophotometric analysis of 260/280 ratios. The RNA was stored at -70°C prior to use. The first strand cDNA was synthesized with oligo-(dT) primer using a reverse transcriptase kit from Invitrogen.
Two micrograms RNA was subjected to the first strand cDNA synthesis, and 1 μL cDNA from RT reaction was subjected to PCR amplification of gene expression in a total 25 μL reaction volume. The PCR amplification was carried out using primer sets derived from the published HIN-1 gene sequences: HIN-1 primers were 5'-TCTGCGTGGCCCTGTCCTG-3' (sense) and 5'-GCTCAGCCAAACACTGTCAG-3' (antisense)[14]. This primer set, designed to cross the intronic sequences, can prevent from amplification of genomic DNA for control of genomic DNA contamination during RNA isolation. A total of 32 cycles of PCR amplification were performed based on our pre-experiment for semi-quantitative measurement of HIN-1 gene expression levels. Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was amplified for 25 cycles as an internal control of equal loading and cDNA quality and quantity. The sequence of GAPDH primers: 5'-GACCACAGTCCATGCCATCAC-3' (sense) and 5'-GTCCACCACCCTGTTGCTGTA-3' (antisense). The PCR products were then electrophoresed in 1.5% agarose gels containing ethidium bromide and reviewed under the UV light.
The cells were grown and treated with or without 5-aza-2’-deoxycytidine for 6 d and total cellular protein was then extracted from these cells in 200 μL ice-cold mild lysis buffer containing 10 μL nonidet P-40, 0.15 mol/L NaCl, 0.01 mol/L sodium phosphate (pH 7.2), 2 mmol/L EDTA, 50 mmol/L sodium fluoride, 0.2 mmol/L sodium vanadate, and 1 μg/mL aprotinin. The cell mixture was centrifuged at 20 000 r/min for 15 min and supernatants were then collected. The concentration of protein was quantified by the BCA protein assay from Pierce (Rockford, IL, USA) and an equal amount of protein was separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and then transferred onto PDVF membranes (Millipore, Billerica, USA). Western blotting analyses were then carried out using an anti-HIN-1 (Novus Biologicals, Littleton, USA) or an anti-β-actin antibody (Boster, Wuhan, China). The blots were developed with chemiluminescence substrate solution from Pierce and exposed to X-ray film.
Gastric cancer cells were grown in 96-well plates and treated with or without 5-aza-2’-deoxycytidine for up to 6 d, and then cell proliferation was determined using CCK-8 solution (Beyotime, China) according to the manufacturer’s instructions. The optical density was measured at 492 nm using an ELISA plate reader (TECAN, Switzerland). The experiments were performed in triplicate and repeated three times.
Gastric cancer cells were treated with or without 5-aza-2’-deoxycytidine for up to 6 d. Both attached and floating cells were harvested and fixed with 70% ethanol for at least 48 h. After resuspension in 50 μg/mL, the cells were treated with 100 μg/mL RNase for 30 min and stained with propidium iodide and then analyzed by flow cytometry (FACscalibur; Becton Dickinson, Franklin Lakes, NJ).
Sections 5 μm thick were formalin-fixed and paraffin-embedded in xylene and rehydrated through an ethanol series. Antigen retrieval was carried out at this stage in a microwave oven. Sections were then blocked with 3% hydrogen peroxidase followed by incubation with a 50% protein blocking agent. Fetal bovine serum (10%), with or without HIN-1 antibody (1:60), was applied to each slide, and the slides were incubated for 30 min, and counterstained with hematoxylin. Tissues without the specific antibody were used as negative controls. Anti-HIN-1 (Novus Biologicals, Littleton, USA) and PV-6000-G Kit (Beijing Zhongshan Jinqiao Biotechnology, Beijing, China) were used for the immunohistochemical (IHC) staining. HIN-1 expression was regarded as positive when 10% or more cancer cells exhibited HIN-1 expression.
The statistical analyses of the experimental data were carried out using SPSS 13.0 software for Windows (Chicago, IL). P values for dichotomous variables were two-tailed and based on the Pearson χ2 test or the Pearson χ2 test with continuity correction. Continuous variables were analyzed with Student’s t test. A value of P < 0.05 was considered statistically significant.
Silence of HIN-1 expression through methylation of HIN-1 gene promoter and 5-aza-2’-deoxycytidine induction of HIN-1 gene expression in gastric cancer cell lines
To find out whether the silence of HIN-1 gene expression is caused by methylation of the HIN-1 gene promoter, we first detected the methylation status of HIN-1 in 5 gastric cancer cell lines. The MSP analysis showed that HIN-1 gene promoter was highly methylated in AGS, PHM82, and BCG-823 cells, but not methylated or partially methylated in NUGC 3 and KATOIII cell lines (Figure 1A). We detected HIN-1 expression in five gastric cancer cell lines and found that HIN-1 mRNA was not expressed in AGS, PHM82, and BCG 823 cells, but expressed in NUGC 3 and weakly expressed in KATOIII cells. HIN-1 expression was induced or upregulated in these cell lines after we treated them with 5-aza-2’-deoxycytidine (Figure 1B).
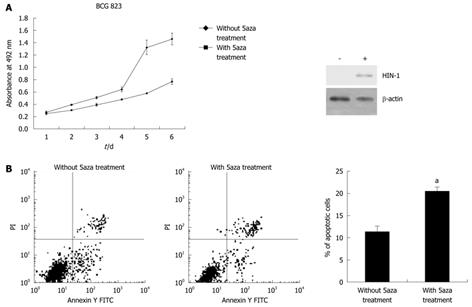
We determined the ability of 5-aza-2’-deoxycytidine to regulate gastric cancer cell viability using BCG-823 cells treated with 5-aza-2’-deoxycytidine. The results showed that treatment with 1 μmol/L of 5-aza-2’-deoxycytidine for up to 6 d significantly upregulated expression of HIN-1 but reduced the number of the viable cells (Figure 2A) and induced them to undergo apoptosis compared with the untreated tumor cells (20.46% ± 1.24% vs 11.28% ± 1.01%, P = 0.001, Figure 2B). These data were associated with HIN-1 expression induced by 5-aza-2’-deoxycytidine (Figure 1B and Figure 2A).
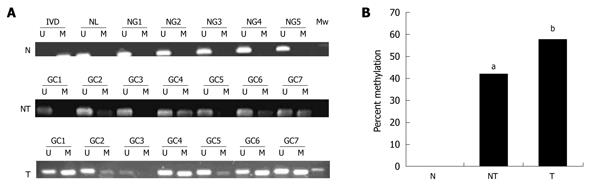
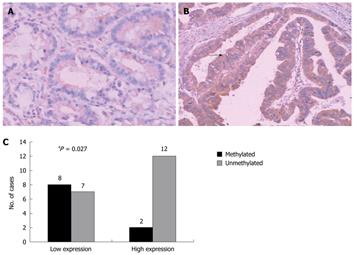
To translate this in vitro finding into ex vivo tissue specimens, MSP analysis of HIN-1 gene promoter methylation was conducted in 45 patients with human gastric carcinoma (32 male and 13 female). The patients’ average age was 55 ± 13 years and other clinicopathological data are listed in Table 1. MSP analysis showed that methylation of the HIN-1 gene promoter was frequently detected in gastric cancer (57.78%, 26/45) and adjacent non-tumor tissues (42.1%, 17/38), but not in normal gastric mucosa. Statistically, there was no difference in methylation of the HIN-1 gene promoter between gastric cancer and adjacent non-tumor tissues. However, there were statistically significant differences between gastric cancer and normal gastric mucosa, and between adjacent non-tumor tissues and normal mucosa (Figure 3A and B, P = 0.002 and P = 0.005, respectively). To correlate HIN-1 gene promoter methylation with HIN-1 expression, 29 gastric cancer tissues (GCs) were subjected to immunohistochemistry analysis. Representative immunostaining is shown in Figure 4A and B. GC cases with low HIN-1 immunostaining had more frequent DNA methylation than GCs with high immunostaining (53.33% vs 14.29%, P = 0.027, Figure 4C). These data demonstrate that DNA methylation contributes to the decreased expression of HIN-1 in GCs.
| Variable | Patients | HIN-1 methylation | P value |
| Sex | 0.161 | ||
| Male | 32 | 17 | |
| Female | 13 | 9 | |
| Age (yr) | 0.401 | ||
| ≤ 50 | 14 | 9 | |
| > 50 | 31 | 17 | |
| Tumor size (cm) | 0.283 | ||
| < 5 | 25 | 16 | |
| ≥ 5 | 19 | 10 | |
| Tumor differentiation | 0.000a | ||
| Moderate/poor | 21 | 17 | |
| Well | 23 | 8 | |
| Stage | 0.683 | ||
| I-II | 13 | 7 | |
| III-IV | 29 | 17 | |
| Nodal status | 0.903 | ||
| - | 9 | 5 | |
| + | 35 | 20 |
Methylation status of HIN-1 gene promoter was associated with tumor differentiation. The methylation frequency in well-differentiated and moderately/poorly-differentiated tumors was 34.78% (8/23) and 80.95% (17/21), respectively, indicating that HIN-1 was more frequently methylated in poorly-differentiated gastric cancer than that in well-differentiated gastric cancer (P = 0.000, Table 1). However, there was no correlation between HIN-1 methylation and other parameters (such as age, tumor size, and lymph node metastasis) (Table 1).
In the current study, we determined HIN-1 gene expression and the methylation status of the HIN-1 gene promoter in gastric cancer cells. We found that the expression of HIN-1 mRNA was lost in gastric cancer cells. MSP analysis revealed high methylation of the HIN-1 gene promoter in these tumor cells. 5-aza-2’-deoxycytidine treatment induced HIN-1 expression, but reduced viability of gastric cancer cells. Furthermore, ex vivo data demonstrated that the HIN-1 gene promoter is frequently methylated in gastric cancer and the adjacent non-tumor tissues, but not in normal gastric mucosae. HIN-1 gene promoter methylation was associated with differentiation of gastric cancer. This study demonstrated frequent methylation of the HIN-1 gene promoter in gastric cancer. Therefore, the HIN-1 gene promoter methylation may be further evaluated as a biomarker for early detection of gastric cancer.
Inactivation of tumor suppressor genes contributes to cancer development. Such inactivation may be caused by genetic or epigenetic alterations, including gene mutation, deletion, promoter methylation, abnormal splicing, deregulation of imprinting and haploinsufficiency[4]. Among these abnormalities, loss of heterozygosity (LOH) was shown to cause inactivation of most candidate tumor suppressor genes in the critical regions of chromosomes 3p, 5q, 8p and 9p[17-20]. However, changes in methylation status of these genes also frequently occur. The HIN-1 gene is located at 5q35 and plays a role in epithelial cell differentiation. HIN-1 can also regulate cell-cycle reentry, suppresses tumor cell migration and invasion, and induces apoptosis in breast cancer cell lines[10]. The HIN-1 gene is frequently methylated in different cancers, but not by mutation[11]. For example, HIN-1 gene promoter hypermethylation was found in the majority (70%) of breast cancer and pre-invasive lesions. Hypermethylation of the HIN-1 promoter region also occurs in cancer and the adjacent tissues of the lung, prostate, pancreas, and esophagus, but not in normal tissues[21]. Methylation of the HIN-1 gene promoter was associated with esophageal squamous carcinoma progression[14]. Our current data demonstrated aberrant methylation of HIN-1 gene promoter regions and subsequent loss of HIN-1 expression in gastric cancer cell lines and tumor tissue specimens. These results are consistent with previous studies on other cancers[9,12]. HIN-1 methylation existed in 57.78% (26/45) of gastric cancer and 42.1% (17/38) of adjacent non-tumor tissues, which indicated that it is a common feature of gastric cancer and may be the early stage accident in gastric carcinogenesis.
The pathogenesis of intestinal-type gastric cancer is usually initiated or caused by Helicobacter pylori (H. pylori) infection[22]. However, the underlying mechanism remains to be defined, and a better understanding of pathogenesis of gastric cancer could help develop molecular diagnostic and patient-tailored therapeutic targets[23]. In the previous studies, we reported that field defect, an area of abnormal tissue that precedes and is predisposed to the development of cancer, could be predicted by detection of gene promoter methylation[24]. Such abnormal fields are of interest because they give insight into the early stages of carcinogenesis and may provide biomarkers of cancer risk[25,26]. Aberrant promoter hypermethylation has been shown to be a common event in human cancer mainly due to the loss of function of tumor suppressor. This neoplasia-related event is thought to occur early in carcinogenesis, and hence, promoter hypermethylation is being widely studied as a biomarker for the diagnosis and detection of early lesions. In this context, HIN-1 was frequently methylated in gastric carcinoma adjacent tissues but not in normal gastric mucosa. It suggests that HIN-1 methylation may represent the field defect of gastric carcinoma. HIN-1 gene promoter methylation may be an early event in gastric cancer. However, further studies are required to determine whether H. pylori infection is responsible for this.
Our current data showed a statistical difference between methylation of the HIN-1 gene promoter and gastric cancer differentiation, HIN-1 was more frequently methylated in poorly-differentiated gastric carcinomas than in well-differentiated ones, which may suggest the role of HIN-1 in regulation of cell differentiation.
We also found that methylation of HIN-1 gene promoter only occurred in gastric cancer but not in normal gastric mucosa. 5-aza-2’-deoxycytidine induced expression of HIN-1, which is associated with reduced viability of gastric cells, indicating that HIN-1 plays an important role in suppressing gastric carcinogenesis. However, we cannot rule out whether other tumor suppressor genes are also induced and restored by 5-aza-2’-deoxycytidine, which plays a role in regulation of tumor cell viability. The latter warrants further studies because some other studies showed that epigenetic modification of pro-apoptotic genes is one of the mechanisms by which the tumor cells are resistant to chemotherapy[27,28]. Therefore, treatment with a demethylating agent like 5-aza-2’-deoxycytidine prior to chemotherapy may help improve the therapeutic efficacy for gastric cancer.
In summary, silence of HIN-1 expression is achieved through the gene methylation in gastric cancer. Methylation of HIN-1 is correlated with tumor differentiation. Future studies will evaluate whether HIN-1 gene promoter methylation can be used as a biomarker for the early detection of gastric cancer.
Gastric cancer is the second most common cause of cancer death worldwide. However, the cause of gastric cancer development remains to be determined. Lost expression of tumor suppressor genes, such as high in normal-1 (HIN-1), may contribute to the development of gastric cancer. This study determined the cause of HIN-1 gene inactivation: epigenetic silence through methylation of the gene promoter.
Silence of HIN-1 gene through hypermethylation of the gene promoter is a common event in different cancers including breast, prostate, and non-small cell lung cancers and malignant mesotheliomas, lymphoma, retinoblastoma, Wilms’ tumor, and rhabdomyosarcoma. This study investigated the role of HIN-1 in gastric cancer and showed for the first time that the hypermethylation of HIN-1 gene promoter was the mechanism for HIN-1 gene silence in gastric cancer.
The authors confirmed the methylation of HIN-1 gene promoter in human gastric cancer cell lines and determined the role of 5-aza-2’-deoxycytidine in regulation of HIN-1 expression in gastric cancer cells.
The HIN-1 gene promoter methylation may be further evaluated as a biomarker for early detection of gastric cancer.
HIN-1 gene was originally isolated through a serial analysis of gene expression from normal and ductal carcinoma in situ luminal mammary epithelial cells. HIN-1 gene promoter is frequently methylated in gastric cancer and the adjacent non-tumor tissues, but not in normal gastric mucosa.
This manuscript demonstrated promising data illustrating the methylation status of H1N-1 gene promoter and its potential role in suppression of gastric carcinoma development.
Peer reviewer: Huanbiao Mo, PhD, Associate Professor, Department of Nutrition and Food Sciences, Texas Woman’s University, PO Box 425888, Denton, TX 76204, United States
S- Editor Sun H L- Editor Ma JY E- Editor Zheng XM
| 1. | Herszényi L, Tulassay Z. Epidemiology of gastrointestinal and liver tumors. Eur Rev Med Pharmacol Sci. 2010;14:249-258. [Cited in This Article: ] |
| 2. | Brenner H, Rothenbacher D, Arndt V. Epidemiology of stomach cancer. Methods Mol Biol. 2009;472:467-477. [Cited in This Article: ] |
| 3. | Arkenau HT. Gastric cancer in the era of molecularly targeted agents: current drug development strategies. J Cancer Res Clin Oncol. 2009;135:855-866. [Cited in This Article: ] |
| 4. | Yasui W, Sentani K, Motoshita J, Nakayama H. Molecular pathobiology of gastric cancer. Scand J Surg. 2006;95:225-231. [Cited in This Article: ] |
| 5. | Herman JG, Baylin SB. Gene silencing in cancer in association with promoter hypermethylation. N Engl J Med. 2003;349:2042-2054. [Cited in This Article: ] |
| 6. | Wang JF, Dai DQ. Metastatic suppressor genes inactivated by aberrant methylation in gastric cancer. World J Gastroenterol. 2007;13:5692-5698. [Cited in This Article: ] |
| 7. | Tokugawa T, Sugihara H, Tani T, Hattori T. Modes of silencing of p16 in development of esophageal squamous cell carcinoma. Cancer Res. 2002;62:4938-4944. [Cited in This Article: ] |
| 8. | Huang KH, Huang SF, Chen IH, Liao CT, Wang HM, Hsieh LL. Methylation of RASSF1A, RASSF2A, and HIN-1 is associated with poor outcome after radiotherapy, but not surgery, in oral squamous cell carcinoma. Clin Cancer Res. 2009;15:4174-4180. [Cited in This Article: ] |
| 9. | Krop IE, Sgroi D, Porter DA, Lunetta KL, LeVangie R, Seth P, Kaelin CM, Rhei E, Bosenberg M, Schnitt S. HIN-1, a putative cytokine highly expressed in normal but not cancerous mammary epithelial cells. Proc Natl Acad Sci USA. 2001;98:9796-9801. [Cited in This Article: ] |
| 10. | Krop I, Parker MT, Bloushtain-Qimron N, Porter D, Gelman R, Sasaki H, Maurer M, Terry MB, Parsons R, Polyak K. HIN-1, an inhibitor of cell growth, invasion, and AKT activation. Cancer Res. 2005;65:9659-9669. [Cited in This Article: ] |
| 11. | Krop I, Player A, Tablante A, Taylor-Parker M, Lahti-Domenici J, Fukuoka J, Batra SK, Papadopoulos N, Richards WG, Sugarbaker DJ. Frequent HIN-1 promoter methylation and lack of expression in multiple human tumor types. Mol Cancer Res. 2004;2:489-494. [Cited in This Article: ] |
| 12. | Wong TS, Kwong DL, Sham JS, Tsao SW, Wei WI, Kwong YL, Yuen AP. Promoter hypermethylation of high-in-normal 1 gene in primary nasopharyngeal carcinoma. Clin Cancer Res. 2003;9:3042-3046. [Cited in This Article: ] |
| 13. | Lehmann U, Berg-Ribbe I, Wingen LU, Brakensiek K, Becker T, Klempnauer J, Schlegelberger B, Kreipe H, Flemming P. Distinct methylation patterns of benign and malignant liver tumors revealed by quantitative methylation profiling. Clin Cancer Res. 2005;11:3654-3660. [Cited in This Article: ] |
| 14. | Guo M, Ren J, Brock MV, Herman JG, Carraway HE. Promoter methylation of HIN-1 in the progression to esophageal squamous cancer. Epigenetics. 2008;3:336-341. [Cited in This Article: ] |
| 15. | Blin N, Stafford DW. A general method for isolation of high molecular weight DNA from eukaryotes. Nucleic Acids Res. 1976;3:2303-2308. [Cited in This Article: ] |
| 16. | Herman JG, Graff JR, Myöhänen S, Nelkin BD, Baylin SB. Methylation-specific PCR: a novel PCR assay for methylation status of CpG islands. Proc Natl Acad Sci USA. 1996;93:9821-9826. [Cited in This Article: ] |
| 17. | Gorringe KL, Ramakrishna M, Williams LH, Sridhar A, Boyle SE, Bearfoot JL, Li J, Anglesio MS, Campbell IG. Are there any more ovarian tumor suppressor genes? A new perspective using ultra high-resolution copy number and loss of heterozygosity analysis. Genes Chromosomes Cancer. 2009;48:931-942. [Cited in This Article: ] |
| 18. | Franko J, Krasinskas AM, Nikiforova MN, Zarnescu NO, Lee KK, Hughes SJ, Bartlett DL, Zeh HJ 3rd, Moser AJ. Loss of heterozygosity predicts poor survival after resection of pancreatic adenocarcinoma. J Gastrointest Surg. 2008;12:1664-1672; discussion 1672-1673. [Cited in This Article: ] |
| 19. | Chang YC, Yeh KT, Liu TC, Chang JG. Molecular cytogenetic characterization of esophageal cancer detected by comparative genomic hybridization. J Clin Lab Anal. 2010;24:167-174. [Cited in This Article: ] |
| 20. | Ye Y, McDevitt MA, Guo M, Zhang W, Galm O, Gore SD, Karp JE, Maciejewski JP, Kowalski J, Tsai HL. Progressive chromatin repression and promoter methylation of CTNNA1 associated with advanced myeloid malignancies. Cancer Res. 2009;69:8482-8490. [Cited in This Article: ] |
| 21. | Shigematsu H, Suzuki M, Takahashi T, Miyajima K, Toyooka S, Shivapurkar N, Tomlinson GE, Mastrangelo D, Pass HI, Brambilla E. Aberrant methylation of HIN-1 (high in normal-1) is a frequent event in many human malignancies. Int J Cancer. 2005;113:600-604. [Cited in This Article: ] |
| 22. | Hamilton JP, Meltzer SJ. A review of the genomics of gastric cancer. Clin Gastroenterol Hepatol. 2006;4:416-425. [Cited in This Article: ] |
| 23. | Chen J, Röcken C, Malfertheiner P, Ebert MP. Recent advances in molecular diagnosis and therapy of gastric cancer. Dig Dis. 2004;22:380-385. [Cited in This Article: ] |
| 24. | Guo M, House MG, Hooker C, Han Y, Heath E, Gabrielson E, Yang SC, Baylin SB, Herman JG, Brock MV. Promoter hypermethylation of resected bronchial margins: a field defect of changes? Clin Cancer Res. 2004;10:5131-5136. [Cited in This Article: ] |
| 25. | Bernstein C, Bernstein H, Payne CM, Dvorak K, Garewal H. Field defects in progression to gastrointestinal tract cancers. Cancer Lett. 2008;260:1-10. [Cited in This Article: ] |
| 26. | Belshaw NJ, Pal N, Tapp HS, Dainty JR, Lewis MP, Williams MR, Lund EK, Johnson IT. Patterns of DNA methylation in individual colonic crypts reveal aging and cancer-related field defects in the morphologically normal mucosa. Carcinogenesis. 2010;31:1158-1163. [Cited in This Article: ] |
| 27. | Nyce J, Leonard S, Canupp D, Schulz S, Wong S. Epigenetic mechanisms of drug resistance: drug-induced DNA hypermethylation and drug resistance. Proc Natl Acad Sci USA. 1993;90:2960-2964. [Cited in This Article: ] |
| 28. | Tian K, Jurukovski V, Wang XP, Kaplan MH, Xu H. Epigenetic regulation of WTH3 in primary and cultured drug-resistant breast cancer cells. Cancer Res. 2005;65:10024-10031. [Cited in This Article: ] |