Published online Nov 7, 2010. doi: 10.3748/wjg.v16.i41.5233
Revised: August 2, 2010
Accepted: August 9, 2010
Published online: November 7, 2010
AIM: To investigate variants of immunity-related GTPase family M (IRGM) and NKX2-3 genes and genotype-phenotype in Eastern European patients with inflammatory bowel disease (IBD).
METHODS: We analyzed 1707 Hungarian and Czech subjects with Crohn’s disease (CD) (n = 810, age: 37.1 ± 12.6 years, duration: 10.7 ± 8.4 years) and ulcerative colitis (UC) (n = 428, age: 43.7 ± 15.0 years, duration: 12.6 ± 9.9 years), as well as 469 healthy controls. IRGM rs13361189, NKX2-3 rs10883365 and ECM1 rs13294 polymorphisms were tested by LightCycler allele discrimination. Detailed clinical phenotypes were determined by reviewing the medical charts.
RESULTS: NKX2-3 rs10883365 variant allele was associated with increased risk for CD (P = 0.009, OR = 1.24, 95% CI = 1.06-1.48) and UC (P = 0.001, OR = 1.36, 95% CI = 1.13-1.63), whereas variant IRGM allele increased risk for CD (P = 0.029, OR = 1.36, 95% CI = 1.03-1.79). In contrast, ECM1 rs13294 was not associated with either CD or UC. In CD, the variant IRGM allele was associated with a colon-only location (P = 0.02, OR = 1.62, 95% CI = 1.07-2.44), whereas in UC, the ECM1 variant was associated with cutaneous manifestations (P = 0.002, OR = 3.36, 95% CI = 1.48-7.63). Variant alleles did not predict resistance to steroids or azathioprine, efficacy of infliximab, or need for surgery.
CONCLUSION: NKX2-3 and IRGM are susceptibility loci for IBD in Eastern European patients. Further studies are needed to confirm the reported phenotype-genotype associations.
- Citation: Meggyesi N, Kiss LS, Koszarska M, Bortlik M, Duricova D, Lakatos L, Molnar T, Leniček M, Vítek L, Altorjay I, Papp M, Tulassay Z, Miheller P, Papp J, Tordai A, Andrikovics H, Lukas M, Lakatos PL. NKX2-3 and IRGM variants are associated with disease susceptibility to IBD in Eastern European patients. World J Gastroenterol 2010; 16(41): 5233-5240
- URL: https://www.wjgnet.com/1007-9327/full/v16/i41/5233.htm
- DOI: https://dx.doi.org/10.3748/wjg.v16.i41.5233
Inflammatory bowel diseases (IBDs) are multifactorial with both environmental and genetic components; the latter displaying heterogeneity in terms of disease presentation as well as response to treatment[1]. Crohn’s disease (CD) has a strong genetic component, and to date, at least nine susceptibility loci have been identified[2]. The first, and most consistently replicated critical mutations have been found in the NOD2/CARD15 gene on chromosome 16 (IBD1). Other loci encode genes that are involved in a number of homeostatic mechanisms: innate pattern recognition receptors (including NOD2/CARD15, TLR4 and CARD9), differentiation of Th17-lymphocytes (IL-23R, JAK2, STAT3, CCR6 and ICOSLG), autophagy [ATG16L1, immunity-related GTPase family M (IRGM) and LRRK2], maintenance of epithelial barrier integrity (IBD5, DLG5, PTGER4, ITLN1, DMBT1 and XBP1), and orchestration of the secondary immune response (HLA-region, TNFSF15/TL1A, IRF5, PTPN2, PTPN22, NKX2-3, IL-12B, IL-18RAP and MST1)[3-6]. As the incidence of IBD is rapidly increasing in some parts of Eastern Europe[1], it is of great importance to study social and environmental, as well as host-genetic factors that might underlie this trend.
The IRGM gene is located on chromosome 5q33.1, and encodes an autophagy-inducing protein and belongs to the immunity-related guanosine triphosphatases (IRGs), also known as p47 guanosine triphosphatases. IRGs play an important role in host defense against intracellular pathogens[7]. Genetic association of the autophagy genes, autophagy-related 16-like 1 (ATG16L1) and IRGM has been suggested in adult-onset CD by genome-wide association scan studies (GWASs), but not in UC[5,6,8-10]. Although a meta-analysis of the three index GWASs has implicated a single nucleotide polymorphism (SNP; rs11747270) based on imputed data with an OR of 1.33[10], other studies including a new meta-analysis[11] have confirmed the IRGM signal on rs13361189 and rs4958847 immediately flanking IRGM.
Recently, the rs13361189 variant, upstream of IRGM, has been shown to be in perfect linkage disequilibrium with a 20-kb deletion polymorphism that affects the expression of IRGM (and cellular autophagy) in a tissue-specific manner[9]. These results suggest that the CD association at IRGM arises from an alteration in IRGM regulation that affects the efficacy of autophagy, and identifies rs13361189 and its strongly correlated neighbors at the 5’ end of IRGM, including the 20-kb deletion polymorphism as a likely causal variant.
Association between the rs10883365 variant of NK2 transcription factor related, locus 3 (NKX2-3) gene on chromosome 10q24.2 and susceptibility of CD has been reported in Western Europe, including The Wellcome Trust Case Control Consortium (WTCCC) GWA and a replication study from The Netherlands, and in Japan, with an OR of 1.2-1.6[12-14]. In addition, a modest association (P = 3.3 × 10-4 in the ulcerative colitis panel and P = 2.4 × 10-6 using the expanded WTCCC control panel) has also been reported between rs10883365 and UC in a non-synonymous SNP scan by Fisher et al[15]. NKX2-3-deficient mice show severe defects in gut development; primarily in the epithelium of the small intestine[16]. In addition, the lymphoid organs of these mice, including the spleen and Peyer’s patches, have abnormal tissue architecture and abnormalities in the migration and segregation of B and T cells[17], however, the exact mechanism is unknown.
ECM1, on chromosome 1q21.2, is also a plausible candidate gene for UC; it encodes extracellular matrix protein 1, a glycoprotein expressed in small and large intestine, and it interacts with the basement membrane and inhibits matrix metalloproteinase[18]. Notably, ECM1 strongly activates nuclear factor-κB signaling, a key immune regulator. Expression is upregulated in colorectal cancer and metastases, which implicates ECM1 in epithelial-stromal interaction[18]. Of note, the WTCCC observed modest association between these ECM1 SNPs and ankylosing spondylitis, a related inflammatory disorder (P = 0.0041 and 0.0044), respectively[13]. More recently, an association between the rs3737240 and rs13294 variants of ECM1 and UC susceptibility was reported in a GWAS by Fisher et al[15], with an OR of 1.3-1.4 for the homozygous carriage of the variant allele. rs3737240 and rs13294 encode substitutions T130M and G290S: Thr130, residing within a collagen IV binding domain, is conserved in primates, whereas Gly290 is not.
Finally, data about the pharmacogenetics of IBD are still limited. Resistance to steroids is associated with high expression of β-glucocorticoid receptors (hGRβ) and overexpression of MDR1 has been found in patients who fail steroid therapy and require surgery[19]. In contrast, according to our group’s earlier results, the presence of the DLG5 Arg30Gln variant allele, but not variants in ABCG2 or MDR1 genes, predicted resistance to steroids[20,21]. Vermeire et al[22] have reported a lack of association between the presence of NOD2 mutations and response to infliximab therapy, and later, the same group reported an association between the presence of Fas ligand -843C/T variant and response to infliximab[23].
In light of the lack of data in Eastern European countries, our aim was to investigate the prevalence of IRGM rs13361189, NKX2-3 rs10883365 and ECM1 rs13294 variants in large independent cohorts of Czech and Hungarian IBD patients. We also aimed to investigate a possible association between genotype and clinical phenotype, need for surgery, and response to medical therapy.
One thousand seven hundred and seven unrelated IBD patients (CD: n = 810, age: 37.1 ± 12.6 years, duration: 10.7 ± 8.4 years and UC: n = 428, age: 43.7 ± 15.0 years, duration: 12.6 ± 9.9 years) and 469 healthy subjects (blood donors) from Hungary and the Czech Republic were investigated. The clinical data of the CD and UC patients are presented in Table 1. A detailed clinical phenotype was available in 789 CD and 422 UC patients.
| CD (n = 810) | UC (n = 428) | |
| Male/female (n) | 434/376 | 202/226 |
| Age (yr) | 37.1 ± 12.6 | 43.7 ± 15.0 |
| Age at presentation (yr)1 | 26.5 ± 10.6 | 31.3 ± 13.4 |
| Duration (yr)1 | 10.7 ± 8.4 | 12.6 ± 9.9 |
| Familial IBD1 | 11.8% | 7.6% |
| Location in CD1 | ||
| L1 | 28.2% | |
| L2 | 18.9% | - |
| L3 | 51.2% | |
| L4 only | 1.7% | |
| All L4 | 7.9% | |
| Maximum extent in UC1 | ||
| Proctitis | - | 7.8% |
| Left-sided | 52.3% | |
| Extensive | 39.9% | |
| Behavior in CD1 | ||
| B1 | 34.9% | |
| B2 | 34.5% | - |
| B3 | 30.7% | |
| Perianal disease1 | 41.2% | - |
| Frequent relapse in CD/chronic continuous in UC1 | 39.9% | 27.6% |
| Arthritis1 | 38.6% | 27.1% |
| Cutaneous1 | 7.8% | 5.9% |
| Occular1 | 7.7% | 3.8% |
| PSC1 | 2.4% | 3.8% |
| Steroid use/refractory1 | 81.1%/11.6% | 66.2%/12.3% |
| Azathioprine use1 | 68.3% | 29.3% |
| Anti-TNF use1 | 33.2% | 10.4% |
| Surgery in CD/colectomy in UC1 | 50.9% | 10.0% |
| Smoking habits2 | ||
| No | 55.1% | 71.6% |
| Yes | 31.3% | 12.4% |
| Previous | 13.6% | 16.0% |
The diagnosis was based on the Lennard-Jones criteria[24]. Age; age at onset; presence of extraintestinal manifestations [arthritis: peripheral and axial; ocular manifestations: conjunctivitis, uveitis and iridocyclitis; skin lesions: erythema nodosum and pyoderma gangrenosum; and hepatic manifestations: primary sclerosing cholangitis (PSC)]; frequency of flare-ups (frequent relapses: > 1 clinical relapse/year); therapeutic effectiveness (e.g. steroid and/or immunosuppressive use, steroid resistance); need for surgery (resections); the presence of familial IBD; smoking habits; and in CD, perianal involvement, were investigated by reviewing the medical charts by the physician and completing a questionnaire. The disease phenotype (age at onset, duration, location and behavior) was determined according to the Montreal classification[25].
The control group for mutation analysis consisted of 469 age- and sex-matched healthy blood donors (male/female: 251/218, age: 40.5 ± 11.5 years old). Control subjects did not have any gastrointestinal and/or liver diseases and were selected from consecutive blood donors in Budapest, Veszprem, Debrecen and Prague. The study protocol was approved by the Ethical and Science Committee of the Ministry of Health. Each patient was informed of the nature of the study and gave signed informed consent.
Genomic DNA was isolated from whole blood according to the manufacturers’ description with High Pure PCR Template preparation Kit (Roche, Budaors, Hungary).
Genotyping was performed using the LightCycler (Roche Diagnostics, Basel, Switzerland) allelic discrimination system. Amplification primers and hybridization probes were designed by the LightCycler Probe Design software (Roche Diagnostics). All oligonucleotides were synthesized by VBC Biotech (Vienna, Austria). The following amplification primers and hybridization probes were used for genotyping IRGM: IRGM-LCF: 5'-ATGGACAGTCAGTACCCTGCAC-3', IRGM-LCR: 5'-CTCTTTACCATTGTACTCCTTGTGCC-3', IRGM-ANC: 5'-LC Red 640-TGCTCAGCGGGTACAGTTTAGAAAGGGAA-Phosphate-3', IRGM-SENS: 5'-GAAAATCGGATGTATATTAGTAGACCC-Fluorescein-3'.
The amplification primers and hybridization probes used for genotyping of ECM1 were: ECM1-LCF: 5'-ACCCACCACCACTTGTGT-3', ECM1-LCR: 5'-TGCTTGGTGAGAACTCTTTGGTTT-3', ECM1-ANC: 5'-LC Red 640-TCAAGATGTCCCGGTCATAGTTGGGGTAAGGAG-Phosphate-3', ECM1-SENS: 5'-TGACTCGACCGATGTCAAT-Fluorescein-3'.
The amplification primers and hybridization probes used for genotyping of NKX2-3 were: NKX2-LCF: 5'-CCGCATAAGACGTTACTTAAACATGT-3', NKX2-LCR: 5'-GCTATCTACTCGAAACTGTCTGC-3', NKX2-ANC-2: 5'-TCTCCCCGGGGGTCACGTTG-Fluorescein-3', NKX2-SENS-2: 5'-LC Red 610- ACAAACACCTTCAAACCGTC-Phosphate-3'.
Polymerase chain reaction (PCR) was performed by LightCycler 480 real-time PCR System (Roche), in a reaction volume of 20 μL with 50 ng genomic DNA, 10 μL 2 × PCR Master Mix (Promega), and 5 pmol of the respective labeled oligonucleotides (sensor and anchor). For IRGM and NKX2-3, an asymmetric multiplex system was used for simultaneous amplification of the two fragments with 3:10 pmol forward:reverse (F:R) amplification oligonucleotides and 10:3 F:R amounts for IRGM and NKX2-3 SNPs, respectively. Similarly, for ECM1, an asymmetric PCR was applied with a 10:3 pmol F:R primer mix for amplification in a separate reaction. Cycling conditions were as follows: initial denaturation at 95°C for 3 min, followed by 35 cycles of denaturation at 95°C for 20 s, annealing at 55°C for 30 s, and extension at 72°C for 45 s, with a ramping rate of 4.4°C/s.
After amplification, a melting curve analysis was performed by cooling the samples to 40°C, followed by gradual heating to 80°C with a ramp rate of 0.04°C/s. The decline in fluorescence was continuously monitored. Melting curves were converted to melting peaks with wild-type and variant alleles showing distinct melting points. For the IRGM and NKX2-3 multiplex PCR, color compensation was performed to compensate for the fluorescence crosstalk between detection channels. Genotype calling was carried out by two independent investigators.
Variables were tested for normality using Shapiro Wilk’s W test. A t test with separate variance estimates, χ2 test and χ2 test with Yates correction were used to evaluate differences between IBD patients and controls, as well as within subgroups of IBD patients. Logistic regression was used to compare genetic and clinical data and results are expressed as OR with 95% CI. P < 0.05 was considered as significant. For the statistical analysis, SPSS version 15.0 (SPSS Inc., Chicago, IL, USA) was used. Statistical analysis was performed by Lakatos PL with the assistance of a statistician.
All investigated polymorphisms were in Hardy-Weinberg equilibrium (P = 0.38-0.95). The success rate of the genotyping assays was 98%-99%. The genotype and allele frequencies are presented in Table 2. NKX2-3 rs10883365 variant allele was associated with increased risk for CD (P = 0.018 after Bonferroni correction, OR = 1.24, 95% CI = 1.06-1.48) and UC (P = 0.003 after Bonferroni correction, OR = 1.36, 95% CI = 1.13-1.63), whereas the variant IRGM allele increased the risk of CD (P = 0.04 after Bonferroni correction, OR = 1.36, 95% CI = 1.03-1.79). The association between NKX2-3 rs10883365 variant and IBD was also significant in the genotypic and dominant models. A similar trend was noted between IRGM and CD. In contrast, ECM1 rs13294 was not associated with either CD or UC. The combination of IRGM carrier/NKX2-3 homozygote genotype was significantly higher in CD compared to the controls (7.2% vs 2.1%, P < 0.0001 after Bonferroni correction, OR = 3.55, 95% CI = 1.80-7.01).
| WT (%) | HET (%) | HOM (%) | P value | Carrier (%) | P value | OR (95%CI) | MAF | P value | OR (95%CI) | |
| IRGM rs13361189 | ||||||||||
| All CD (n = 808) | 636 (78.7) | 156 (19.3) | 16 (2.0) | 0.0601 | 172 (21.3) | 0.0601 | - | 188 (11.6) | 0.029 | .36 (1.03-1.79) |
| Hungarian CD (n = 456) | 355 (77.9) | 89 (19.5) | 12 (2.6) | NS12 | 101 (22.1) | NS12 | - | 113 (12.4) | NS12 | - |
| Czech CD (n = 352) | 281 (79.8) | 67 (19.0) | 4 (1.2) | NS12 | 71 (20.2) | NS12 | - | 75 (10.7) | NS12 | - |
| All UC (n = 428) | 351 (82.0) | 76 (17.8) | 1 (0.2) | NS12 | 77 (18.0) | NS12 | - | 78 (9.1) | NS12 | - |
| Hungarian UC (n = 274) | 221 (80.7) | 52 (19.0) | 1 (0.3) | NS12 | 53 (19.3) | NS12 | - | 54 (9.9) | NS12 | - |
| Czech UC (n = 154) | 130 (84.4) | 24 (15.6) | 0 | NS12 | 24 (15.6) | NS12 | - | 24 (7.8) | NS12 | - |
| All controls (n = 460) | 382 (83.0) | 75 (16.3) | 3 (0.7) | - | 78 (17.0) | - | - | 81 (8.8) | - | - |
| Hungarian controls (n = 265) | 217 (81.9) | 46 (17.4) | 2 (0.7) | - | 48 (18.1) | - | - | 50 (9.4) | - | - |
| Czech controls (n = 195) | 165 (84.6) | 29 (14.9) | 1 (0.5) | - | 30 (15.4) | - | - | 31 (7.9) | - | - |
| NKX2 rs 10883365 | ||||||||||
| All CD (n = 810) | 208 (25.7) | 389 (48.1) | 213 (26.2) | 0.0101 | 602 (74.3) | NS12 | - | 815 (50.3) | 0.009 | 1.24 (1.06-1.48) |
| Hungarian CD (n = 457) | 115 (25.2) | 220 (48.1) | 122 (26.7) | NS12 | 342 (74.8) | NS12 | - | 464 (50.8) | NS12 | - |
| Czech CD (n = 353) | 93 (26.3) | 169 (47.9) | 91 (25.8) | 0.0701 | 260 (73.7) | NS1 | - | 351 (49.7) | 0.0301 | 1.31 (1.03-1.68) |
| All UC (n = 427) | 90 (21.1) | 226 (52.9) | 111 (26.0) | 0.0041 | 337 (78.9) | 0.0061 | 1.52 (1.13-2.08) | 448 (52.5) | 0.001 | 1.36 (1.13-1.63) |
| Hungarian UC (n = 274) | 56 (20.4) | 152 (55.5) | 66 (24.1) | NS12 | 218 (79.6) | 0.0701 | 284 (51.8) | 0.060 | ||
| Czech UC (n = 153) | 34 (22.2) | 74 (48.4) | 45 (29.4) | 0.0171 | 119 (77.8) | 0.0461 | 1.63 (1.01-2.65) | 164 (53.6) | 0.0051 | 1.53 (1.14-2.07) |
| All controls (n = 469) | 136 (29.0) | 245 (52.2) | 88 (18.8) | - | 333 (71.0) | - | - | 421 (44.9) | - | - |
| Hungarian controls (n = 271) | 73 (26.9) | 145 (53.5) | 53 (19.6) | - | 198 (73.1) | - | - | 251 (46.3) | - | - |
| Czech controls (n = 198) | 63 (31.8) | 100 (50.5) | 35 (17.7) | - | 135 (68.2) | - | - | 170 (42.9) | - | - |
| ECM1 rs13294 | ||||||||||
| All CD (n = 807) | 139 (17.2) | 392 (48.6) | 276 (34.2) | NS12 | 688 (82.8) | NS12 | - | 944 (58.5) | NS12 | - |
| Hungarian CD (n = 455) | 89 (19.6) | 228 (50.1) | 138 (30.3) | NS12 | 366 (80.4) | NS12 | - | 504 (55.4) | NS12 | - |
| Czech CD (n = 352) | 50 (14.2) | 164 (46.6) | 138 (39.2) | NS12 | 302 (85.8) | NS12 | - | 440 (62.5) | NS12 | - |
| All UC (n = 428) | 77 (18.0) | 225 (52.6) | 126 (29.4) | NS12 | 351 (82.0) | NS12 | - | 477 (55.7) | NS12 | - |
| Hungarian UC (n = 275) | 44 (16.0) | 156 (56.1) | 75 (27.3) | NS12 | 231 (84.0) | NS12 | - | 306 (55.6) | NS12 | - |
| Czech UC (n = 153) | 33 (21.6) | 69 (45.1) | 51 (33.3) | NS12 | 120 (78.4) | NS12 | - | 171 (55.9) | NS12 | - |
| All controls (n = 463) | 90 (19.4) | 222 (47.9) | 151 (32.6) | - | 373 (80.6) | - | - | 524 (56.6) | - | - |
| Hungarian controls (n = 265) | 56 (21.1) | 124 (46.8) | 85 (32.1) | - | 209 (78.9) | - | - | 294 (55.5) | - | - |
| Czech controls (n = 198) | 34 (17.2) | 98 (49.5) | 66 (33.3) | - | 164 (82.8) | - | - | 230 (58.1) | - | - |
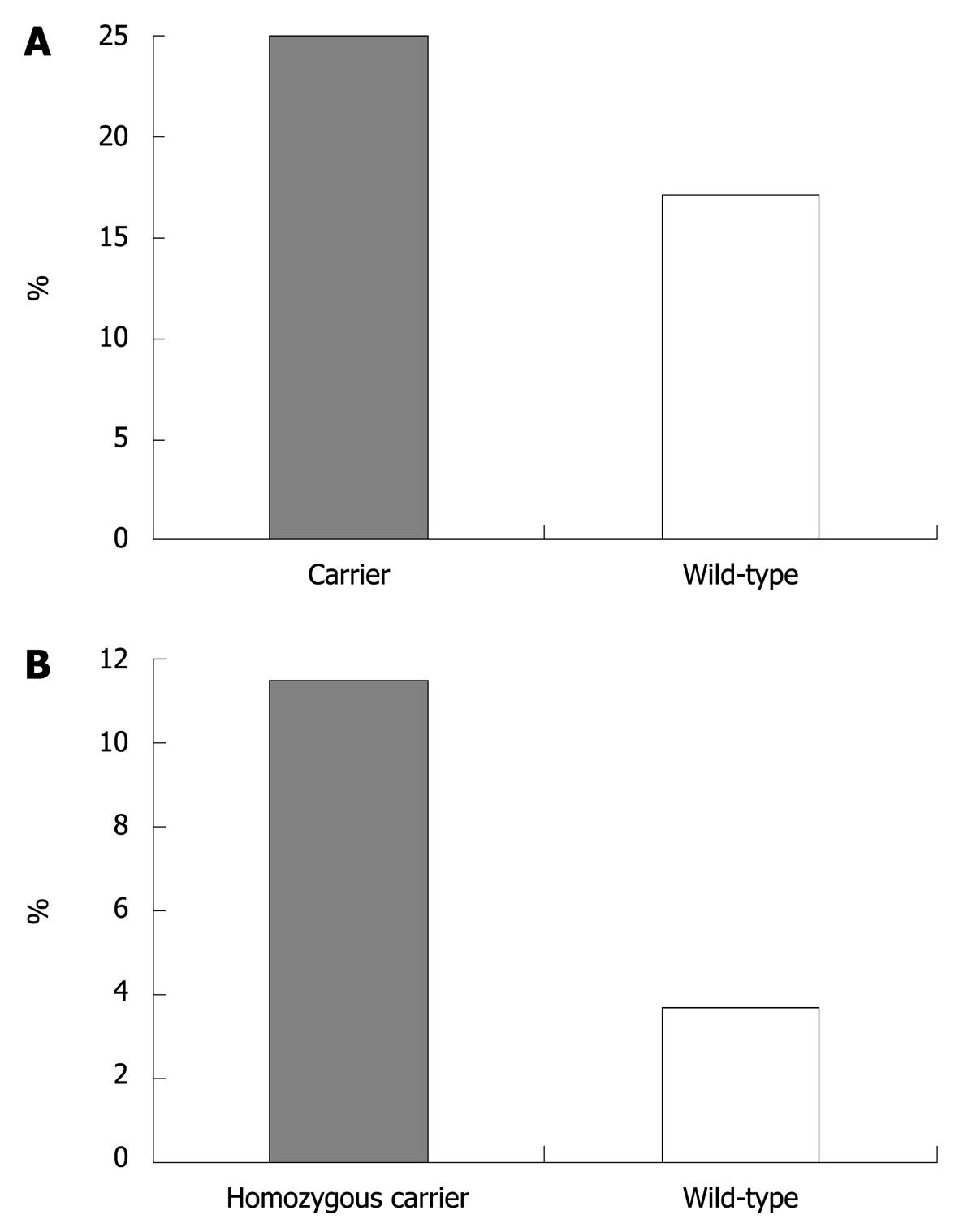
In CD, presence of the variant IRGM allele was associated with colon-only location (in carriers: 25% vs wild-type: 17.1%, P = 0.04 after Bonferroni correction, OR = 1.62, 95% CI = 1.07-2.44, Figure 1A). In UC, homozygous carriage of the ECM1 variant allele was associated with cutaneous manifestations (11.5% in homozygotes vs 3.7% in patients with other genotypes, P = 0.004 after Bonferroni correction, OR = 3.36, 95% CI = 1.48-7.63, Figure 1B). No other significant associations were found in either CD or UC patients (data not shown).
We also investigated the association between the NKX2-3, IRGM and ECM1 variants and the response to steroids, infliximab or azathioprine or the need for surgery in patients with CD. Two hundred and sixty-three unrelated CD patients were treated with anti-tumor necrosis factor (TNF): 259 received infliximab and four, adalimumab, two of whom were treated after secondary loss of response to infliximab therapy. Overall, there was no association between the presence of the above variants and short-term response (assessed at week 12) to infliximab induction therapy [5 mg/kg at weeks 0, 2 and 6; partial response: Crohn’s disease activity index (CDAI) decreased by ≥ 70 points and/or ≥ 50% decrease in the number of draining fistulas; remission: CDAI < 150 or closure of all fistulas]; steroid use/resistance; azathioprine use; or need for surgery (data not shown). Similarly, no association was found between either of the variants and steroid use/resistance, azathioprine use or need for surgery in patients with UC (data not shown).
This is the first report on the prevalence of the IRGM rs13361189, NKX2-3 rs10883365 and ECM1 rs13294 variants, in large, independent IBD cohorts from Eastern Europe. The variant NKX2-3 allele conferred a risk in both UC and CD, whereas the IRGM rs13361189 variant allele was associated with increased risk for CD. In addition, the IRGM variant was associated with disease location in CD.
The genotype and allele frequencies for IRGM and NKX2-3 variants in CD, UC and controls reported in the present study were in line with most previous reports in Caucasian populations, whereas the frequency of the variant ECM1 rs13294 allele was approximately 10% higher in both IBD and controls, compared to that reported in previous studies[7,8,12,13,15,26].
We confirmed that the IRGM rs13361189 variant was associated with disease susceptibility in CD in Eastern European populations. The allelic OR of 1.36 (95% CI = 1.03-1.79) was in the range reported in Caucasian populations (OR = 1.38-1.56)[7,13] and in a recent meta-analysis (OR = 1.34)[10]. Similar genotype distributions were observed in both the Czech and Hungarian cohort, however, due to the low variant allele frequency, the difference became significant only in the combined analysis. A similar tendency (P = 0.06) was observed in the genotypic and dominant model. In contrast, in the present study we failed to replicate the weak association between IRGM and UC (rs13361189 P = 0.0069, pooled OR = 1.16; rs4958847 P = 0.014, pooled OR = 1.13) that was reported recently in a Spanish meta-analysis[10]. Of note, the present study was underpowered to detect such small differences, however, there was even no trend for a difference between UC and controls.
In addition, in an Italian study[27], the rs4958847 polymorphism was associated with fistulizing behavior (P = 0.037, OR = 1.54, CI = 1.02-2.31) and perianal fistulas (P = 0.045, OR = 1.55, CI = 1.01-2.38) in a logistic regression analysis. This was later partially confirmed by the Leuven group. Henckaerts et al[28] have reported in a very elegant study a significant association between the IRGM rs4958847 variant, U7 gene desert rs12704036 T-allele, NOD2/CAR15 mutation, ileal involvement at diagnosis, male sex, and time to development of non-perianal fistulas in a Cox regression analysis. In a further study from New Zealand[29], rs13361189 variant increased the risk for ileal CD in 507 CD patients and 576 controls. The OR in ileal CD was 1.92 (95% CI = 1.27-2.96). Moreover, Peterson et al[30] have suggested an association between IRGM and pediatric disease onset (< 17 years) of CD in a North American cohort. However, only one of the two IRGM variants studied (rs13361189) was weakly associated with CD in their study (uncorrected P = 0.03). In the present study, we could not confirm the association between age at onset and the presence of the IRGM variant, in accordance with Canadian, Italian and Scottish results[29,31]. In contrast, we found an association between IRGM carriage and disease location in CD; colonic location was significantly more common in carriers of the variant allele (OR = 1.62, 95% CI = 1.07-2.44). Of note, however, the rs4958847 variant was not investigated in the present study.
The association between the rs10883365 variant of NKX2-3 gene and susceptibility of IBD was first reported in CD in Caucasian patients and in a Japanese study with an OR of 1.2-1.6[12-14]. In addition, using the UC panel and the expanded WTCCC control panel, Fisher et al[15] have reported a modest association (P = 3.3 × 10-4 and P = 2.4 × 10-6) between rs10883365 and UC. In the present study, we confirmed these findings; the rs10883365 variant was associated with UC and CD susceptibility in the allelic and genotypic models with an OR of 1.53 and 1.24, respectively. Based on our data, the association between NKX2-3 and IBD is stronger in UC compared to CD, at least in patients from Eastern Europe. In a recent Dutch study, the association between the rs10883365 variant of NKX2-3 and CD was linked to smoking status, with the risk being more pronounced in active and passive smokers[32]. In the present study, however, we did not find an association between NKX2-3 and smoking status, and similarly, no genotype-phenotype associations were found.
In addition, Weersma et al[13] have created genetic risk profiles by combining the presence of variant alleles in IL23R, ATG16L1, IRGM, NKX2-3, 1q24, 5p13, HERC2, CCNY, 10q21 and NOD2/CARD15, and the number of risk alleles was associated with gradually increased risk for CD. Similarly, in the present study, the combination of IRGM carrier/NKX2-3 homozygote genotype was associated with an increased risk for CD, with a much higher OR compared to either of the variant alleles alone.
Recently, an association between ECM1 rs3737240 and rs13294 variants and UC has been reported in a GWAS by Fisher et al[15], with an OR of 1.3-1.4 for the homozygous carriage of the variant allele. The association for the rs13294 variant has recently been confirmed in a Dutch study, with an OR of 1.24 in patients with UC[33]. In contrast, no association was found in CD[34], even though a different SNP (rs11205387) was investigated. Although the present study was powered to confirm a difference with an OR of 1.3-1.5 with adequate statistical power, we could not confirm an association between the ECM1 rs13294 variant and either UC or CD. The accidental association between homozygous carriage of the ECM1 variant allele and cutaneous manifestations (P = 0.002, OR = 3.36, 95% CI = 1.48-7.63, Figure 1B) requires further confirmation.
Theoretically, through influencing inflammatory responses, autophagy and epithelial-stromal interactions, polymorphisms in the above genes might be of potential importance in altering the efficacy of anti-inflammatory therapy and thereby the need for surgery. However, in the present study, none of the variant alleles was associated with the response to either steroid or infliximab therapy, or the need for surgery (resection in CD or colectomy in UC) in IBD.
In conclusion, NKX2-3 and IRGM are susceptibility loci for IBD in Eastern European patients. None of the variants investigated were associated with the need for surgery or efficacy of medical therapy. Further studies are needed to confirm the reported phenotype-genotype associations found in this study.
Sequence variants in the autophagy gene immunity-related GTPase family M (IRGM) and NKX2-3 have been reported to contribute to Crohn’s disease (CD) susceptibility, whereas ECM1 contributes to ulcerative colitis (UC) in genome-wide association scans in North America and Western Europe.
There is a lack of data in Eastern European countries, therefore, our aim was to investigate the prevalence of IRGM rs13361189, NKX2-3 rs10883365 and ECM1 rs13294 variants. In addition, the possible association between genotype and clinical phenotype and the need for surgery is conflictive, and the association between the above genetic variants and response to medical therapy was not investigated.
In the present study, the authors showed in two well-characterized, independent CD cohorts with strict clinical follow-up, that NKX2-3 rs10883365 variant allele was associated with increased susceptibility to CD and UC, whereas variant IRGM allele increased the risk for CD only. Our data suggest that the variant IRGM allele is associated with colon-only location, whereas in UC, the ECM1 variant was associated with cutaneous manifestations. None of the variants predicted resistance to steroids and azathioprine, efficacy of infliximab, or need for surgery.
The new data presented from Eastern Europe might contribute to better understanding of genetic or environmental differences between populations and the association of those differences with disease susceptibility and phenotype.
Vienna-Montreal classification: classification systems of disease phenotypes in CD. The classification assesses the age at presentation, disease location and disease behavior. The IRGM gene encodes an autophagy-inducing protein and plays an important role in host defense against intracellular pathogens. NKX2-3 is a member of the NKX family of homeodomain-containing transcription factors, which are implicated in many aspects of cell type specification and maintenance of differentiated tissue functions. ECM1 encodes extracellular matrix protein 1, a glycoprotein expressed in small and large intestine. Notably, ECM1 strongly activates nuclear-κB signaling, a key immune regulator.
The need to extend studies performed in the other parts of the world to Eastern Europe is valid, and provides a point for understanding subtle genetic or environmental differences between populations and the association of those differences with disease.
Peer reviewers: Dr. William R Parker, PhD, Assistant Professor, Department of Surgery, Duke University Medical Center, Box 2605, Durham, NC 27710, United States; Toru Hiyama, MD, PhD, Health Service Center, Hiroshima University, 1-7-1 Kagamiyama, Higashihiroshima 739-8521, Japan
S- Editor Cheng JX L- Editor Kerr C E- Editor Ma WH
| 1. | Lakatos PL. Recent trends in the epidemiology of inflammatory bowel diseases: up or down? World J Gastroenterol. 2006;12:6102-6108. [Cited in This Article: ] |
| 2. | Van Limbergen J, Wilson DC, Satsangi J. The genetics of Crohn's disease. Annu Rev Genomics Hum Genet. 2009;10:89-116. [Cited in This Article: ] |
| 3. | Lakatos PL, Lakatos L, Szalay F, Willheim-Polli C, Osterreicher C, Tulassay Z, Molnar T, Reinisch W, Papp J, Mozsik G. Toll-like receptor 4 and NOD2/CARD15 mutations in Hungarian patients with Crohn's disease: phenotype-genotype correlations. World J Gastroenterol. 2005;11:1489-1495. [Cited in This Article: ] |
| 4. | Duerr RH, Taylor KD, Brant SR, Rioux JD, Silverberg MS, Daly MJ, Steinhart AH, Abraham C, Regueiro M, Griffiths A. A genome-wide association study identifies IL23R as an inflammatory bowel disease gene. Science. 2006;314:1461-1463. [Cited in This Article: ] |
| 5. | Hampe J, Franke A, Rosenstiel P, Till A, Teuber M, Huse K, Albrecht M, Mayr G, De La Vega FM, Briggs J. A genome-wide association scan of nonsynonymous SNPs identifies a susceptibility variant for Crohn disease in ATG16L1. Nat Genet. 2007;39:207-211. [Cited in This Article: ] |
| 6. | Lakatos PL, Szamosi T, Szilvasi A, Molnar E, Lakatos L, Kovacs A, Molnar T, Altorjay I, Papp M, Tulassay Z. ATG16L1 and IL23 receptor (IL23R) genes are associated with disease susceptibility in Hungarian CD patients. Dig Liver Dis. 2008;40:867-873. [Cited in This Article: ] |
| 7. | Singh SB, Davis AS, Taylor GA, Deretic V. Human IRGM induces autophagy to eliminate intracellular mycobacteria. Science. 2006;313:1438-1441. [Cited in This Article: ] |
| 8. | Parkes M, Barrett JC, Prescott NJ, Tremelling M, Anderson CA, Fisher SA, Roberts RG, Nimmo ER, Cummings FR, Soars D. Sequence variants in the autophagy gene IRGM and multiple other replicating loci contribute to Crohn's disease susceptibility. Nat Genet. 2007;39:830-832. [Cited in This Article: ] |
| 9. | Barrett JC, Hansoul S, Nicolae DL, Cho JH, Duerr RH, Rioux JD, Brant SR, Silverberg MS, Taylor KD, Barmada MM. Genome-wide association defines more than 30 distinct susceptibility loci for Crohn's disease. Nat Genet. 2008;40:955-962. [Cited in This Article: ] |
| 10. | McCarroll SA, Huett A, Kuballa P, Chilewski SD, Landry A, Goyette P, Zody MC, Hall JL, Brant SR, Cho JH. Deletion polymorphism upstream of IRGM associated with altered IRGM expression and Crohn's disease. Nat Genet. 2008;40:1107-1112. [Cited in This Article: ] |
| 11. | Palomino-Morales RJ, Oliver J, Gómez-García M, López-Nevot MA, Rodrigo L, Nieto A, Alizadeh BZ, Martín J. Association of ATG16L1 and IRGM genes polymorphisms with inflammatory bowel disease: a meta-analysis approach. Genes Immun. 2009;10:356-364. [Cited in This Article: ] |
| 12. | Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature. 2007;447:661-678. [Cited in This Article: ] |
| 13. | Weersma RK, Stokkers PC, Cleynen I, Wolfkamp SC, Henckaerts L, Schreiber S, Dijkstra G, Franke A, Nolte IM, Rutgeerts P. Confirmation of multiple Crohn's disease susceptibility loci in a large Dutch-Belgian cohort. Am J Gastroenterol. 2009;104:630-638. [Cited in This Article: ] |
| 14. | Yamazaki K, Takahashi A, Takazoe M, Kubo M, Onouchi Y, Fujino A, Kamatani N, Nakamura Y, Hata A. Positive association of genetic variants in the upstream region of NKX2-3 with Crohn's disease in Japanese patients. Gut. 2009;58:228-232. [Cited in This Article: ] |
| 15. | Fisher SA, Tremelling M, Anderson CA, Gwilliam R, Bumpstead S, Prescott NJ, Nimmo ER, Massey D, Berzuini C, Johnson C. Genetic determinants of ulcerative colitis include the ECM1 locus and five loci implicated in Crohn's disease. Nat Genet. 2008;40:710-712. [Cited in This Article: ] |
| 16. | Pabst O, Zweigerdt R, Arnold HH. Targeted disruption of the homeobox transcription factor Nkx2-3 in mice results in postnatal lethality and abnormal development of small intestine and spleen. Development. 1999;126:2215-2225. [Cited in This Article: ] |
| 17. | Pabst O, Förster R, Lipp M, Engel H, Arnold HH. NKX2.3 is required for MAdCAM-1 expression and homing of lymphocytes in spleen and mucosa-associated lymphoid tissue. EMBO J. 2000;19:2015-2023. [Cited in This Article: ] |
| 18. | Chan I, Liu L, Hamada T, Sethuraman G, McGrath JA. The molecular basis of lipoid proteinosis: mutations in extracellular matrix protein 1. Exp Dermatol. 2007;16:881-890. [Cited in This Article: ] |
| 19. | Farrell RJ, Murphy A, Long A, Donnelly S, Cherikuri A, O'Toole D, Mahmud N, Keeling PW, Weir DG, Kelleher D. High multidrug resistance (P-glycoprotein 170) expression in inflammatory bowel disease patients who fail medical therapy. Gastroenterology. 2000;118:279-288. [Cited in This Article: ] |
| 20. | Fischer S, Lakatos PL, Lakatos L, Kovacs A, Molnar T, Altorjay I, Papp M, Szilvasi A, Tulassay Z, Osztovits J. ATP-binding cassette transporter ABCG2 (BCRP) and ABCB1 (MDR1) variants are not associated with disease susceptibility, disease phenotype response to medical therapy or need for surgeryin Hungarian patients with inflammatory bowel diseases. Scand J Gastroenterol. 2007;42:726-733. [Cited in This Article: ] |
| 21. | Lakatos PL, Fischer S, Claes K, Kovacs A, Molnar T, Altorjay I, Demeter P, Tulassay Z, Palatka K, Papp M. DLG5 R30Q is not associated with IBD in Hungarian IBD patients but predicts clinical response to steroids in Crohn's disease. Inflamm Bowel Dis. 2006;12:362-368. [Cited in This Article: ] |
| 22. | Vermeire S, Louis E, Rutgeerts P, De Vos M, Van Gossum A, Belaiche J, Pescatore P, Fiasse R, Pelckmans P, Vlietinck R. NOD2/CARD15 does not influence response to infliximab in Crohn's disease. Gastroenterology. 2002;123:106-111. [Cited in This Article: ] |
| 23. | Hlavaty T, Pierik M, Henckaerts L, Ferrante M, Joossens S, van Schuerbeek N, Noman M, Rutgeerts P, Vermeire S. Polymorphisms in apoptosis genes predict response to infliximab therapy in luminal and fistulizing Crohn's disease. Aliment Pharmacol Ther. 2005;22:613-626. [Cited in This Article: ] |
| 24. | Lennard-Jones JE. Classification of inflammatory bowel disease. Scand J Gastroenterol Suppl. 1989;170:2-6; discussion 16-19. [Cited in This Article: ] |
| 25. | Silverberg MS, Satsangi J, Ahmad T, Arnott ID, Bernstein CN, Brant SR, Caprilli R, Colombel JF, Gasche C, Geboes K. Toward an integrated clinical, molecular and serological classification of inflammatory bowel disease: Report of a Working Party of the 2005 Montreal World Congress of Gastroenterology. Can J Gastroenterol. 2005;19 Suppl A:5-36. [Cited in This Article: ] |
| 26. | Franke A, Balschun T, Karlsen TH, Hedderich J, May S, Lu T, Schuldt D, Nikolaus S, Rosenstiel P, Krawczak M. Replication of signals from recent studies of Crohn's disease identifies previously unknown disease loci for ulcerative colitis. Nat Genet. 2008;40:713-715. [Cited in This Article: ] |
| 27. | Latiano A, Palmieri O, Cucchiara S, Castro M, D'Incà R, Guariso G, Dallapiccola B, Valvano MR, Latiano T, Andriulli A. Polymorphism of the IRGM gene might predispose to fistulizing behavior in Crohn's disease. Am J Gastroenterol. 2009;104:110-116. [Cited in This Article: ] |
| 28. | Henckaerts L, Van Steen K, Verstreken I, Cleynen I, Franke A, Schreiber S, Rutgeerts P, Vermeire S. Genetic risk profiling and prediction of disease course in Crohn's disease patients. Clin Gastroenterol Hepatol. 2009;7:972-980.e2. [Cited in This Article: ] |
| 29. | Roberts RL, Hollis-Moffatt JE, Gearry RB, Kennedy MA, Barclay ML, Merriman TR. Confirmation of association of IRGM and NCF4 with ileal Crohn's disease in a population-based cohort. Genes Immun. 2008;9:561-565. [Cited in This Article: ] |
| 30. | Peterson N, Guthery S, Denson L, Lee J, Saeed S, Prahalad S, Biank V, Ehlert R, Tomer G, Grand R. Genetic variants in the autophagy pathway contribute to paediatric Crohn's disease. Gut. 2008;57:1336-1337; author reply 1337. [Cited in This Article: ] |
| 31. | Amre DK, Mack DR, Morgan K, Krupoves A, Costea I, Lambrette P, Grimard G, Dong J, Feguery H, Bucionis V. Autophagy gene ATG16L1 but not IRGM is associated with Crohn's disease in Canadian children. Inflamm Bowel Dis. 2009;15:501-507. [Cited in This Article: ] |
| 32. | van der Heide F, Nolte IM, Kleibeuker JH, Wijmenga C, Dijkstra G, Weersma RK. Differences in genetic background between active smokers, passive smokers, and non-smokers with Crohn's disease. Am J Gastroenterol. 2010;105:1165-1172. [Cited in This Article: ] |
| 33. | Festen EA, Stokkers PC, van Diemen CC, van Bodegraven AA, Boezen HM, Crusius BJ, Hommes DW, van der Woude CJ, Balschun T, Verspaget HW. Genetic analysis in a Dutch study sample identifies more ulcerative colitis susceptibility loci and shows their additive role in disease risk. Am J Gastroenterol. 2010;105:395-402. [Cited in This Article: ] |
| 34. | Anderson CA, Massey DC, Barrett JC, Prescott NJ, Tremelling M, Fisher SA, Gwilliam R, Jacob J, Nimmo ER, Drummond H. Investigation of Crohn's disease risk loci in ulcerative colitis further defines their molecular relationship. Gastroenterology. 2009;136:523-529.e3. [Cited in This Article: ] |