Published online May 14, 2009. doi: 10.3748/wjg.15.2270
Revised: March 30, 2009
Accepted: April 6, 2009
Published online: May 14, 2009
AIM: To determine if TSPAN1 overexpression is associated with clinicopathological and prognostic factors in human colorectal adenocarcinoma.
METHODS: Total RNA was extracted in 20 human adenocarcinoma tissues for TSPAN1 mRNA assay by RT-PCR. Eighty-eight specimens of human colorectal adenocarcinoma were surgically removed. TSPAN1 protein levels in cancer tissues were determined by immunohistochemistry using a polyclonal antibody against self-prepared TSPAN1. The correlation between TSPAN1 expression and the clinicopathological factors and the overall survival rate was analyzed by univariate and multivariate assay.
RESULTS: TSPAN1 mRNA was detected in 90.0% (18/20) of cancerous tissues. The light density of TSPAN1 mRNA expression levels was 0.89 ± 0.30 in adenocarcinoma by gel-image system. TSPAN1 protein expression was detected in 78.41% (69/88) and weakly expressed in 40% normal colorectal tissues. There were significant differences between colorectal adenocarcinoma and normal control epithelium (P < 0.05). TSPAN1 protein expression in colorectal cancerous tissue was significantly correlated with the histological grade, cell expression PCNA, lymph nodal metastasis and TNM staging of the disease. Patients with TSPAN1 protein overexpression had a significantly shorter survival period than that in patients with TSPAN1 protein negative or weak expression, respectively (P < 0.05). Furthermore, by multivariate analysis, TSPAN1 protein expression demonstrated an independent prognostic factor for human colorectal cancers (P < 0.05, relative risk 0.755; 95% confidence interval 0.302-1.208).
CONCLUSION: The expression of TSPAN1 gene is increased in colorectal carcinoma, suggesting that TSPAN1 might serve as an independent prognostic factor for the colorectal adenocarcinoma patients.
- Citation: Chen L, Zhu YY, Zhang XJ, Wang GL, Li XY, He S, Zhang JB, Zhu JW. TSPAN1 protein expression: A significant prognostic indicator for patients with colorectal adenocarcinoma. World J Gastroenterol 2009; 15(18): 2270-2276
- URL: https://www.wjgnet.com/1007-9327/full/v15/i18/2270.htm
- DOI: https://dx.doi.org/10.3748/wjg.15.2270
The colorectal carcinoma is one of the most common malignant neoplasms, ranking the fourth frequency in men and third in women[1]. Although the prognosis has slightly improved in the past years, colorectal cancer is still the second and third major common cause of cancer related death in men and women in the United States, respectively[2]. The incidence of colorectal cancer is the fourth in malignant tumor ranking in China, and it is increased dramatically in developing regions[34]. The colorectal cancer is thought to result from a combination of environmental factors, diet, lifestyle, chronic inflammation and accumulation of specific genetic alterations. The pathogenesis and development of colorectal cancer involve multi-genes and multi-steps. Ogino et al[5] showed the occurrence of colorectal cancer involved in a series of gene mutations, microsatellite instability (MSI) and 18q loss of heterozygosity (LOH). The other molecules studied include MST1 (Mammalian sterile 20-like kinase)[6]. Replication protein (RPA)[7], ELAV-like protein Huk and COX-2[8], α-catenin, β-catenin[9] α-ligatin, β-ligatin, Rho-a[10], etc. In fact, an established cascade of events leading to colorectal cancer development and progression is described by Vogelstein. The alteration of expression of these molecules often showed an obvious correlation with pathologic grading and clinical staging in colorectal cancer, which can be used as a biomarker for assessing prognosis. Currently, the assessment of prognosis is mainly based on pathological features of the tumor which is valuable to the triage of patients who will benefit from adjuvant therapy. The clinical pathological staging is the most popular standard prognostic approach for predicting the clinical outcome of colorectal cancer patients[1112]. The prognosis of colorectal cancer is closely related to the tumor TNM stages. However, patients with similar stages of the disease have various outcomes. Therefore, there is a need to identify useful prognostic molecular markers in guiding treatment decisions and/or in developing more effective treatments. TSPAN1 (GenBank Accession No. AF065388) is a new member of TM4SF[13], which is located at chromosome 1 p34.1. It encodes a 241 amino acid protein. TSPAN1 was reported as a tumor-related gene recently[13–17]. In several studies, TSPAN1 gene over-expression was detected in liver cancer[14], prostate cancer[15], gastric carcinoma[16] and cervix cancer[17]. It has been proposed that TSPAN1 plays a role in cell mitosis and/or cause cell abnormal differentiation. In this study, we examined fresh tumor tissues and histological sections of colorectal adenocarcinoma to determine the expression of TSPAN1 mRNA and protein, and analyzed the relationship between the gene expression and clinicopathological parameters. The result suggests that overexpression of TSPAN1 is correlated to the prognosis of colorectal cancer patients.
A total of 88 patients with colorectal adenocarcinoma, diagnosed and treated from January 1998 to April 2000 were investigated in this study. Of the 88 cases evaluated, 46.6% (41 cases) were rectum cancers, 30.1% (27 cases) were sigmoid colon cancers, 6.8% (6 cases) were descending colon cancers, 2.3% (2 cases) were transverse colon cancers and 13.6 % (12 cases) were ascending colon cancers. The median age at the time of diagnosis was 62.2 years (range, 37-85). There were 50 male patients, 38 female patients. None of them had received chemotherapy or radiotherapy before diagnosis. After surgery, these patients with TMN stage II took oral 5-fluorouracil and patients with stage III-IV were subjected to 5-fluorouracil-based systemic chemotherapy. In order to avoid bias, each case was diagnosed by two pathologists.
The clinicopathological data were determined according to the WHO classification and TNM cancer staging[111218]. The average size of the tumor was 4 cm (range from 1.5 to 7.6 cm), 54.5% (48 cases) were cauliflower/polyp type and 45.45% (40 cases) were ulcer/sclerotic type. Adenocarcinomas were graded predominantly on the basis of the extent of glandular appearances, and divided into well (lesions exhibit glandular structures in > 95% of the tumor, grade 1, 15.9% or 14 cases), moderate (lesions have 50%-95% glands, grade 2, 44.31% or 39 cases) and poor differentiation (lesions have 5%-50% glands, grade 3, 39.77% or 35 cases). Tumor limited in submucosa (T1) and muscularis propria (T2) as stage I accounted for 32.95% (29 cases), tumor invaded through muscularis propria into subserosa or into non-peritonealized pericolic or perirectal tissues (T3) and tumor directly invades other organs or structures and/or perforates visceral peritoneum (T4) as stage II accounted for 29.54% (26 cases), and the tumor with metastasis in 1-3 regional lymph nodes (N1-3) in any T as stage III and the tumor with distant metastasis (M) in any T and N as stage IV, III and IV accounted for 37.5 % (33 cases). Vascular invasion in 26 cases (29.55%) demonstrated that vessel wall was occlusive or infiltrating damaged up to the complete destruction with a surrounding fibroinflammatory reaction[19–21]. Such clinicopathological factors as perineural invasion and desmoplasia reaction were observed and analyzed as well. The proliferation level of cancer cells was evaluated based on the expression of PCNA in tumor parenchymas.
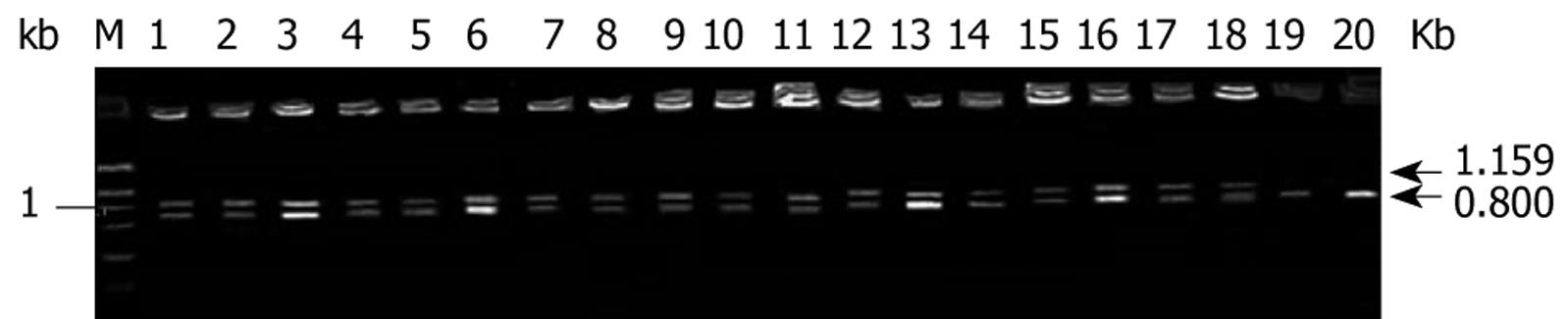
Twenty cases of fresh colorectal cancer specimens were stored in -70°C refrigerator immediately after dissection for semi-quantitative RT-PCR with co-amplification of TSPAN1 gene and an internal control β-actin. Briefly, total RNA from tumor tissues was extracted with TRLZOL reagent and the reverse transcription was performed with Rneasy Kit (Clontech, CA, USA) according to previously published protocols[14]. A 50 &mgr;L PCR reaction contains approximately 50 ng of human colorectal cancer ds-cDNA; 40 mmol/L Tricine-KOH, pH9.2; 15 mmol/L KOAc; 3.5 mmol/L Mg (OAc)2; 0.2 &mgr;mol/L 5’ TSPAN1 primer (5’-CAG-TTC-CCT-CTT-TCA-GAA-CTC-ACT-G-3’); 0.2 &mgr;mol/L 3’ TSPAN1primer (5’-ATC-CAC-CCA-GAG-GCT-CTG-CTG-ATT-TCA-CCT-3’); 0.1 &mgr;mol/L 5' β-actin primer (5’-TTA-CAC-CCT-TTC-TTG-ACA-AAA-CCT-A-3’); 0.1 &mgr;mol/L 3’β-actin primer (5’-CAA-AAG-CCT-TCA-TAC-ATC-TCA-AGT-3’); 0.2 mmol/L each of dATP, dGTP, dCTP and dTTP; and 1 &mgr;L of AdvantageTM cDNA Polymerase Mix (50X; contains KlenTaq-1 and Deep Vent polymerases). The PCR cycling was as follows: PCR tubes were preheated at 94°C for 20 s; then run 30 cycles at 96°C for 6 s (denature); 60°C for 20 s for annealing and 72°C for 1 min for extension, in a DNA thermal cycle 9600 (PE Biosystems, CA, USA). PCR products were applied to electrophoresis on 1% agarose gel analysis; the expected TSPAN1 gene was a band at 1159 bp. TSPAN1 expression was evaluated by calculating the average ratios of light density using symmetry computerized gel imaging system[14].
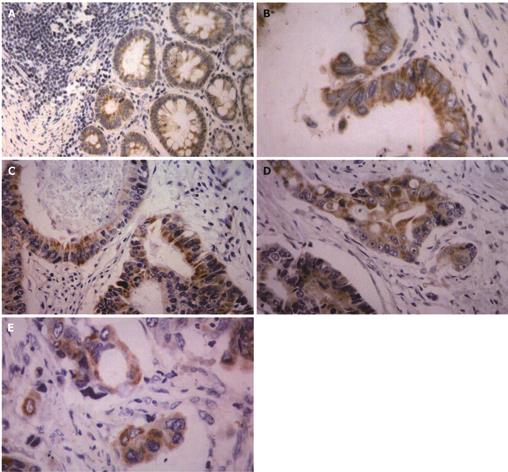
All 88 adenocarcinoma samples were routinely fixed in 40 g/L formaldehyde solution and embedded in paraffin. After slicing into 4 &mgr;m thick sections, immunohistochemistry was performed using Dako Elivision TM Plus Two-step System (PV-6000 kit, Zymed, Co., USA.). To detect the TSPAN1 and PCNA expressions in colorectal adenocarcinoma tissues, the sections were dewaxed in xylene and rinsed in alcohol and graded alcohol/water mixtures. Sections were then submitted to antigen retrieval treatment in a pressure cooker. The tissues were boiled in 0.01 mol/L, pH 6.0 citric acid buffer to retrieval antigen for 5 min. They were then treated with 0.3% hydrogen peroxide in absolute methanol to inhibit endogenous peroxidase activity for 15 min at room temperature. After blocking of background staining with diluted normal calf serum, sections were incubated overnight at 4°C with polyclonal antibodies against TSPAN1 (antibody prepared with the help of American San Francisco gene biotechnology company) and PCNA (PC10, No. 40780708, DAKO, USA), respectively. Subsequent reaction proceeded using a two step assay, immunoreaction was visualized with peroxidase-3,3’-diaminobenzidine (DAB). Finally, sections were lightly counterstained with Mayer’s haematoxylin and mounted. The negative controls were set by omitting the primary antibodies. The positive controls were the hepatocellular carcinoma with positive expressions of TSPAN1. In addition, 10 specimens from the marginal normal mucosa of tumor were used as normal controls[16].
All sections were blindly analyzed by two experienced pathologists under light microscope. Based on the estimated percentages of positive parenchyma cells and/or the immunostaining intensity, which was determined by comparing the immunoreactivity of the positive controls that were included in each experiment, staining results were divided into four categories: (-) tissues specimens: positive parenchyma cell with less than 5% of the cancer tissues and/or weakly stained; (+) tissue specimens: positive parenchyma cell with less than 25% of the cancer tissues and/or weakly stained; (++) tissues specimens: positive parenchyma cell with less than 50% of the cancer tissues and/or moderately stained, and (+++) tissue specimens: positive parenchyma cell with more than 75% of the cancer tissues and/or strongly stained[1416].
Association between TSPAN1 gene expression and other clinicopathological factors of the tumor were assessed by the Fisher’s exact test (two-sided) for categorical variables and χ2 test were used to compare ordinal variables. The grading-related data was analysed by Spearman test. Overall survival was defined as the period from the date of diagnosis to the date of death. Survival curves were determined according to the Kaplan-Meier method, and compared using Log-rank test statistical differences. Multivariate survival analysis was performed with SPSS version 11.0 Software (Chicago, IL, USA).
Total RNA was extracted from 20 cases of colorectal adenocarcinoma tissues. RT-PCR analysis of TSPAN1 mRNA expression was then performed. The positive rate of TSPAN1 mRNA expression was 90% (18/20) in the colorectal adenocarcinoma (Figure 1), and the relative amount of TSPAN1 mRNA levels in cancer tissues was assessed based on the β-actin control. The relative amounts of TSPAN1 mRNA were 0.89 ± 0.30.
TSPAN1 was mainly presented in cytoplasm and located at membrane as well. In the normal control epithelium, 3 cases presented a weakly positive staining of TSPAN1, and only 1 case presented moderately positive expression (Figure 2A). We observed TSPAN1 protein expression in 78.41% (69/88) cases of tumors, in which 17.39 % (12/69) was displayed as strong expressed (+++), 44.93% (31/69) as moderately expressed (++), and 37.68% (26/69) as weakly expressed (+). There were significant differences between colorectal adenocarcinoma and normal control epithelium (P < 0.05), (Figures 2B-E).
To investigate the role of TSPAN1 expression in colorectal cancer, we examined the correlation of TSPAN1 expression with the clinicopathological features (Table 1). We found a positive correlation with histological grade, PCNA expression, nodal metastasis and TNM stages (P = 0.001, 0.015, 0.008 and 0.002, respectively). TNM staging of colorectal cancer is more important for patient’s prognosis evaluation. The five-year survival rate of TMN stage 1 is more than 95%, while it is less 10% in patients with TNM stage III-IV. From Table 1, it can be found that the TSPAN1 expression rate and intensity in early TNM stage were lower than in late TNM stage cancer tissues. In addition, TSPAN1 expression was not associated with vascular invasion, perineural invasion and desmoplasia.
| Parameters | Cases | TSPAN1 expression intensity | P | |||
| - | + | ++ | +++ | |||
| Gender | ||||||
| Male | 50 | 9 | 14 | 21 | 6 | 0.472 |
| Female | 38 | 10 | 12 | 10 | 6 | |
| Tumor size (cm) | ||||||
| < 4.0 | 35 | 10 | 10 | 10 | 5 | 0.469 |
| > 4.0 | 53 | 9 | 16 | 21 | 7 | |
| Type | ||||||
| Cauliflower/polyp | 48 | 9 | 14 | 16 | 9 | 0.595 |
| Ulcer/infiltration | 40 | 10 | 12 | 15 | 3 | |
| Location | ||||||
| Rectum | 41 | 9 | 11 | 17 | 4 | 0.595 |
| Colon | 47 | 10 | 15 | 14 | 8 | |
| Grade | ||||||
| Well | 14 | 6 | 6 | 2 | 0 | 0.001 |
| Moderate | 39 | 9 | 14 | 14 | 2 | |
| Poor | 35 | 4 | 6 | 15 | 10 | |
| PCNA | ||||||
| + | 43 | 14 | 14 | 13 | 2 | 0.015 |
| ++/+++ | 45 | 5 | 12 | 18 | 10 | |
| Lymph node metastasis | ||||||
| No | 55 | 16 | 20 | 14 | 5 | 0.008 |
| Yes | 33 | 3 | 6 | 17 | 7 | |
| TNM stage | ||||||
| I | 29 | 11 | 9 | 7 | 2 | 0.002 |
| II | 26 | 5 | 11 | 7 | 3 | |
| III-IV | 33 | 3 | 6 | 17 | 7 | |
| Vascular invasion | ||||||
| No | 62 | 14 | 21 | 20 | 7 | 0.424 |
| Yes | 26 | 5 | 5 | 11 | 5 | |
| Perineural invasion | ||||||
| No | 67 | 16 | 22 | 22 | 7 | 0.235 |
| Yes | 21 | 3 | 4 | 9 | 5 | |
| Desmoplasia | ||||||
| No | 55 | 10 | 16 | 20 | 9 | 0.647 |
| Yes | 33 | 9 | 10 | 11 | 3 | |
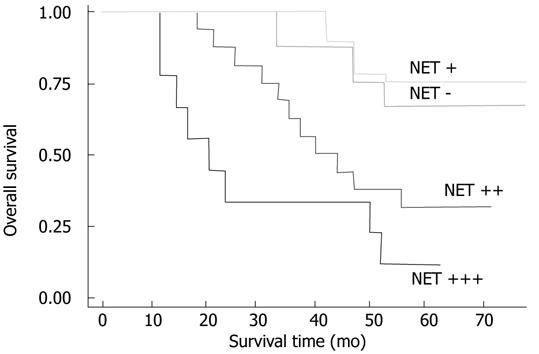
Within a period of 60 mo of the follow-up, 24 cancer-related deaths occurred, 3 of the deaths come from 9 patients with TSPAN1 negative tumors, and 21 from 33 patients in the TSPAN1 positive group. In the entire cohort, the overall survival rate of patients with TSPAN1 negative tumors were significantly higher than that of those with TSPAN1 positive tumors (63.64% vs 33.33%; log-rank test: χ2 = 15.48, P = 0.001). Kaplan-Meier estimated the overall survival rate based on cell TSPAN1 expression in the patients with a follow-up period of 60 mo (Figure 3). To compare with other clinicopathological factors, the effects of histologic grades, node status, PCNA expression, TNM stages, vascular invasion or perineural invasion on the patients’ survival were also analysed with univariate log-rank test. As shown in Table 2, the factors of cellular differentiation, node status, PCNA expression, TNM stages had a significant effect on the overall survival rate (P = 0.03, 0.001, 0.0003 and 0.002, respectively). Furthermore, univariate survival analysis was performed to investigate possible prognostic impact of TSPAN1 in colorectal cancer. As shown in Table 2, the expression of TSPAN1 correlated with a worsening of the survival probability, which was statistically significant. This was also confirmed by a multivariate survival analysis including above factors (Table 3). All of these results suggested that TSPAN1 expression in tumors was an independent prognostic factor for colorectal adenocarcinoma patients (relative risk = 0.755; 95% confidence interval: 0.302-1.208 P = 0.001).
| Parameters | 5-yr survival rate (%) | Log-rank test | |
| χ2 | P | ||
| TSPAN1 expression | |||
| - | 66.67 (6/9) | 15.48 | 0.0015 |
| + | 71.4 (5/7) | ||
| ++ | 35.3 (6/17) | ||
| +++ | 11.1 (1/9) | ||
| Grade | |||
| Well | 87.5 (7/8) | 6.91 | 0.0316 |
| Moderate | 37.5 (6/16) | ||
| Poor | 27.8 (5/18) | ||
| Node status | |||
| No | 71.6 (12/17) | 15.67 | 0.0001 |
| Yes | 24.0 (6/25) | ||
| PCNA expression | |||
| + | 63.1 (12/19) | 9.05 | 0.0026 |
| ++-+++ | 26.1 (6/23) | ||
| TNM stages | |||
| I | 83.3 (10/12) | 16.20 | 0.0030 |
| II | 62.5 (5/8) | ||
| III-IV | 13.6 (3/22) | ||
| Vascular invasion | |||
| No | 46.4 (13/28) | 1.39 | 0.2377 |
| Yes | 37.7 (5/14) | ||
| Perineural invasion | |||
| No | 44.7 (14/32) | 0.77 | 0.3795 |
| Yes | 40.0 (4/10) | ||
| Desmoplasia | |||
| No | 33.3 (3/9) | 0.02 | 0.8829 |
| Yes | 42.4 (15/33) | ||
| Variable | Multivariate analysis | Pvalue | |||
| HR | SD | Z | 95% CI | ||
| TSPAN1 expression | 0.755 | 0.231 | 3.27 | 0.302-1.208 | 0.001 |
| Grade | 0.798 | 0.318 | 2.51 | 0.175-1.421 | 0.012 |
| Node status | 1.779 | 0.509 | 3.49 | 0.781-2.778 | 0.000 |
| PCNA expression | 1.325 | 0.475 | 2.79 | 0.394-2.256 | 0.005 |
| TNM stages | 1.159 | 0.341 | 3.39 | 0.490-1.829 | 0.001 |
| Vescular invasion | 0.491 | 0.423 | 1.16 | 0.338-1.320 | 0.246 |
| Perineural invasion | 0.409 | 0.473 | 0.87 | 0.517-1.336 | 0.386 |
| Desmoplasia | 0.061 | 0.415 | 0.15 | 0.752-0.873 | 0.884 |
Many studies reported that TSPAN1 mRNA and protein were expressed in human normal tissues and carcinomas[13–17]. Serru detected TSPAN1 expression in various cell lines by RT-PCR including cervical cancer, lung cancer, squamous carcinoma, colorectal cancer and breast cancer cells[13]. Wollscheid et al[17] detected TSPAN1 mRNA level by RT-PCR and TSPAN1 protein by immunohistochemistry in cervical cancer and found that the gene was expressed in CIN III, cervical squamous cell carcinoma and adenocarcinoma, especially in all undifferentiated cervical carcinoma and adenocarcinoma. They thought TSPAN1 gene expression correlated to cell proliferation and may be used as a marker for cervical cancer prognosis. However, TSPAN1 gene expression in human colorectal cancer tissues has not been reported so far. In this study, we for the first time demonstrated that TSPAN1 mRNA and protein were extensively expressed in 90% and 78% human colorectal cancer tissues, respectively. Our results revealed that epithelial cells of the normal colon or rectum displayed a slight expression of TSPAN1 antigen (Figure 2A). There was significant difference between cancer tissues and normal control. The results are consistent with most other reported data[12–14] and suggest that the TSPAN1 expression is a specific marker for malignant transformation.
In colorectal cancer, the presence of many tumor-associated antigens and their relationship with clinical pathological parameters have been described[2223]. PCNA, a major marker for cell proliferation, is highly expressed in most tumors[24]. In this study, the finding of a significant positive correlation between TSPAN1 and PCNA expression provided further evidence to support a potential role of TSPAN1 in tumor proliferation process (Table 1). The colorectal cancer development may hence relate to the accumulation of TSPAN1 protein in tumor cells. Similarly, our previous study found that TSPAN1 expression correlated with tumor proliferation maker Ki67 expression in human gastric carcinomas[16].
Currently, the TNM stage represents the main tool for identifying prognostic differences among patients with colorectal cancer. The reported 5-year survival rate is 95% for stage I patients, 67% for stages II, and 9.4% for stage III and IV patients[25]. In our prospective 5-year follow-up study, the overall survival rate was 83.3% for stage I patients, 62.5% for stage II patients, and 13.6% for stage III and stage IV patients (Table 2). Similarly, we showed that there was a significant correlation between the overall survival rate and the disease stages. Our study revealed that there was a statistically significant association between TSPAN1 expression and the various stages of colorectal cancer, in which TSPAN1 positive staining was seen in 63.64% patients with shorter survival time (Table 3). The univariate and multivariate analyses suggested that TSPAN1 status, PCNA expression, tumor stages and nodal status were strong predictors for the final clinical outcome (Table 3). Likewise, another study in our lab also showed that TSPAN1 expression was significantly correlated with the metastasis and poor prognosis of gastric carcinoma[16]. Increasing TSPAN1 protein expression was found associated with more advanced stages of cervical carcinoma[17]. All these findings suggest that TSPAN1 over-expression status might yield unfavorable prognosis for some types of cancers. Identifying those patients with high-risk colorectal cancers by TSPAN1 expression detection would be of great benefit for improving the treatment strategies. By the way, other reports displayed that vascular invasion and perineural invasion were correlated with a poor prognosis[19–21], but in this study we found no direct effect on tumor prognosis.
Colorectal carcinoma is one of the most common cancers in western world and in China, however its molecular mechanism is still unclear. To understand the specific regulation of gene expressions between colorectal cancer and non-cancer tissues and know the genes or proteins characteristics will delineate the molecular changes and obtain useful diagnostic marker. We have demonstrated that TSPAN1 was expressed in majority of human colorectal carcinomas in the current study. TSPAN1 expression, measured by immunohistochemistry in the tumor tissues, may be a candidate gene for diagnosis and prognosis of colorectal carcinoma. The overexpression of TSPAN1 in cytoplasm is associated with higher tumor grade, metastasis, proliferation, and more advanced stages and poor prognosis in colorectal adenocarcinoma patients, suggesting a tumor-related gene role of TSPAN1 in human colorectal cancer development.
The colorectal cancer results from a combination of environmental factors, diet, lifestyle, chronic inflammation and accumulation of specific genetic alterations. TSPAN1 (GenBank Accession No. AF065388) is a new member of TM4SF located at chromosome 1 p34.1. It encodes a 241 amino acid protein. TSPAN1 was reported as a tumor-related gene recently.
TSPAN1 gene over-expression was detected in liver cancer, prostate cancer , gastric carcinoma and cervix cancer. It has been proposed that TSPAN1 plays a role in cell mitosis and/or cause cell abnormal differentiation.
In this study the authors examined fresh tumor tissues and histological sections of colorectal adenocarcinoma to determine the expression of TSPAN1 mRNA and protein, and analyzed the relationship between the gene expression and clinicopathological parameters, and found that overexpression of TSPAN1 is correlated to prognosis of colorectal cancer patients.
Testing TSPAN1 expression in tissues would be a useful tool to evaluate the prognosis of patients with colorectal cancer.
The authors examined the expression of Net-1 in colorectal tissues, a novel gene whose function has yet to be understood so far. This study is of some clinical significance.
| 1. | Parkin DM, Bray F, Ferlay J, Pisani P. Global cancer statistics, 2002. CA Cancer J Clin. 2005;55:74-108. [Cited in This Article: ] |
| 2. | Jemal A, Siegel R, Ward E, Murray T, Xu J, Smigal C, Thun MJ. Cancer statistics, 2006. CA Cancer J Clin. 2006;56:106-130. [Cited in This Article: ] |
| 3. | Ji BT, Devesa SS, Chow WH, Jin F, Gao YT. Colorectal cancer incidence trends by subsite in urban Shanghai, 1972-1994. Cancer Epidemiol Biomarkers Prev. 1998;7:661-666. [Cited in This Article: ] |
| 4. | You WC, Jin F, Devesa S, Gridley G, Schatzkin A, Yang G, Rosenberg P, Xiang YB, Hu YR, Li Q. Rapid increase in colorectal cancer rates in urban Shanghai, 1972-97, in relation to dietary changes. J Cancer Epidemiol Prev. 2002;7:143-146. [Cited in This Article: ] |
| 5. | Ogino S, Brahmandam M, Cantor M, Namgyal C, Kawasaki T, Kirkner G, Meyerhardt JA, Loda M, Fuchs CS. Distinct molecular features of colorectal carcinoma with signet ring cell component and colorectal carcinoma with mucinous component. Mod Pathol. 2006;19:59-68. [Cited in This Article: ] |
| 6. | Minoo P, Zlobec I, Baker K, Tornillo L, Terracciano L, Jass JR, Lugli A. Prognostic significance of mammalian sterile20-like kinase 1 in colorectal cancer. Mod Pathol. 2007;20:331-338. [Cited in This Article: ] |
| 7. | Givalos N, Gakiopoulou H, Skliri M, Bousboukea K, Konstantinidou AE, Korkolopoulou P, Lelouda M, Kouraklis G, Patsouris E, Karatzas G. Replication protein A is an independent prognostic indicator with potential therapeutic implications in colon cancer. Mod Pathol. 2007;20:159-166. [Cited in This Article: ] |
| 8. | Denkert C, Koch I, von Keyserlingk N, Noske A, Niesporek S, Dietel M, Weichert W. Expression of the ELAV-like protein HuR in human colon cancer: association with tumor stage and cyclooxygenase-2. Mod Pathol. 2006;19:1261-1269. [Cited in This Article: ] |
| 9. | Murata M, Iwao K, Miyoshi Y, Nagasawa Y, Ohta T, Shibata K, Oda K, Wada H, Tominaga S, Matsuda Y. Molecular and biological analysis of carcinoma of the small intestine: beta-catenin gene mutation by interstitial deletion involving exon 3 and replication error phenotype. Am J Gastroenterol. 2000;95:1576-1580. [Cited in This Article: ] |
| 10. | Debruyne PR, Bruyneel EA, Karaguni IM, Li X, Flatau G, Müller O, Zimber A, Gespach C, Mareel MM. Bile acids stimulate invasion and haptotaxis in human colorectal cancer cells through activation of multiple oncogenic signaling pathways. Oncogene. 2002;21:6740-6750. [Cited in This Article: ] |
| 11. | Chamberlain NL, Ward RL, Hawkins NJ. Clinicopathological significance of erbB-2 expression in colorectal carcinoma. Oncol Rep. 1999;6:527-531. [Cited in This Article: ] |
| 12. | Sobin LH, Wittekind Ch, editors . TNM classification of malignant Tumors. 6th edition. Wiley-Liss: New York 2002; . [Cited in This Article: ] |
| 13. | Serru V, Dessen P, Boucheix C, Rubinstein E. Sequence and expression of seven new tetraspans. Biochim Biophys Acta. 2000;1478:159-163. [Cited in This Article: ] |
| 14. | Chen L, Wang Z, Zhan X, Li DC, Zhu YY, Zhu J. Association of NET-1 gene expression with human hepatocellular carcinoma. Int J Surg Pathol. 2007;15:346-353. [Cited in This Article: ] |
| 15. | Xu J, Stolk JA, Zhang X, Silva SJ, Houghton RL, Matsumura M, Vedvick TS, Leslie KB, Badaro R, Reed SG. Identification of differentially expressed genes in human prostate cancer using subtraction and microarray. Cancer Res. 2000;60:1677-1682. [Cited in This Article: ] |
| 16. | Chen L, Li X, Wang GL, Wang Y, Zhu YY, Zhu J. Clinicopathological significance of overexpression of TSPAN1, Ki67 and CD34 in gastric carcinoma. Tumori. 2008;94:531-538. [Cited in This Article: ] |
| 17. | Wollscheid V, Kühne-Heid R, Stein I, Jansen L, Köllner S, Schneider A, Dürst M. Identification of a new proliferation-associated protein NET-1/C4.8 characteristic for a subset of high-grade cervical intraepithelial neoplasia and cervical carcinomas. Int J Cancer. 2002;99:771-775. [Cited in This Article: ] |
| 18. | Stanley RH, Lauri AA. World Health Organization Classification of Tumours: Pathology and Genetics of Tumours of the Digestive System. Lyon: IARC Press 2004; . [Cited in This Article: ] |
| 19. | Sternberg A, Amar M, Alfici R, Groisman G. Conclusions from a study of venous invasion in stage IV colorectal adenocarcinoma. J Clin Pathol. 2002;55:17-21. [Cited in This Article: ] |
| 20. | Ouchi K, Sugawara T, Ono H, Fujiya T, Kamiyama Y, Kakugawa Y, Mikuni J, Tateno H. Histologic features and clinical significance of venous invasion in colorectal carcinoma with hepatic metastasis. Cancer. 1996;78:2313-2317. [Cited in This Article: ] |
| 21. | Talbot IC, Ritchie S, Leighton M, Hughes AO, Bussey HJ, Morson BC. Invasion of veins by carcinoma of rectum: method of detection, histological features and significance. Histopathology. 1981;5:141-163. [Cited in This Article: ] |
| 22. | Seicean R, Funariu G, Seicean A. Molecular prognostic factors in rectal cancer. Rom J Gastroenterol. 2004;13:223-231. [Cited in This Article: ] |
| 23. | Graziano F, Cascinu S. Prognostic molecular markers for planning adjuvant chemotherapy trials in Dukes' B colorectal cancer patients: how much evidence is enough? Ann Oncol. 2003;14:1026-1038. [Cited in This Article: ] |
| 24. | Jaskulski D, Gatti C, Travali S, Calabretta B, Baserga R. Regulation of the proliferating cell nuclear antigen cyclin and thymidine kinase mRNA levels by growth factors. J Biol Chem. 1988;263:10175-10179. [Cited in This Article: ] |