Published online Dec 28, 2008. doi: 10.3748/wjg.14.7329
Revised: November 2, 2008
Accepted: November 9, 2008
Published online: December 28, 2008
AIM: To detect the MLH1 gene promoter germline-methylation in probands of Chinese hereditary nonpolyposis colorectal cancer (HNPCC), and to evaluate the role of methylation in MLH1 gene promoter and molecular genetics in screening for HNPCC.
METHODS: The promoter germline methylation of MLH1 gene was detected by methylation-specific PCR (MSP) in 18 probands from unrelated HNPCC families with high microsatellite-instability (MSI-H) phenotype but without germline mutations in MSH2, MLH1 and MSH6 genes. At the same time, 6 kindreds were collected with microsatellite-stability (MSS) phenotype but without germline mutations in MSH2, MLH1 and MSH6 genes as controls. The results of MSP were confirmed by clone sequencing. To ensure the reliability of the results, family H65 with nonsense germline mutation at c.2228C > A in MSH2 gene was used as the negative control and the cell line sw48 was used as the known positive control along with water as the blank control. Immunochemical staining of MLH1 protein was performed with Envision two-step method in those patients with aberrant methylation to judge whether the status of MLH1 gene methylation affects the expression of MLH1 protein.
RESULTS: Five probands with MLH1 gene promoter methylation were detected in 18 Chinese HNPCC families with MSI-H phenotype but without germline mutations in MSH2, MLH1 and MSH6 genes. Two of the five probands from families H10 and H29 displayed exhaustive-methylation, fulfilling the Japanese criteria (JC) and the Amsterdam criteria (AC), respectively. The other 3 probands presented part-methylation fulfilling the AC. Of the 13 probands with unmethylation phenotype, 8 fulfilled the JC and the Bethesda guidelines (BG), 5 fulfilled the AC. The rate of aberrant methylation in MLH1 gene in the AC group (22.2%, 4/18) was higher than that in the JC/BG groups (5.6%, 1/18) in all HNPCC families with MSI-H phenotype but without germline mutations in MSH2, MLH1 and MSH6 genes. However, no proband with methylation in MLH1 gene was found in the families with MSS phenotype and without germline mutations in MSH2, MLH1 and MSH6 genes. No expression of MLH1 protein was found in tumor tissues from two patients with exhaustive-methylation phenotype, whereas positive expression of MLH1 protein was observed in tumor tissues from patients with partial methylation phenotype (excluding family H42 without tumor tissue), indicating that exhaustive-methylation of MLH1 gene can cause defective expression of MLH1 protein.
CONCLUSION: Methylation phenotype of MLH1 gene is correlated with microsatellite phenotype of MMR genes, especially with MSI-H. Exhaustive-methylation of MLH1 gene can silence the expression of MLH1 protein. MLH1 promoter methylation analysis is a promising tool for molecular genetics screening for HNPCC.
-
Citation: Zhou HH, Yan SY, Zhou XY, Du X, Zhang TM, Cai X, Lu YM, Cai SJ, Shi DR.
MLH1 promoter germline-methylation in selected probands of Chinese hereditary non-polyposis colorectal cancer families. World J Gastroenterol 2008; 14(48): 7329-7334 - URL: https://www.wjgnet.com/1007-9327/full/v14/i48/7329.htm
- DOI: https://dx.doi.org/10.3748/wjg.14.7329
| Case | Gender | Age (yr) | Criteria | MSI | MLH1/MSH2/MSH6 mutation study |
| H21 | M | 38 | AC | MSI-H | NM |
| H22 | M | 46 | AC | MSI-H | NM |
| H28 | F | 30 | AC | MSI-H | NM |
| H29 | F | 37 | AC | MSI-H | NM |
| H32 | M | 51 | AC | MSI-H | NM |
| H42 | M | 65 | AC | MSI-H | NM |
| H46 | M | 48 | AC | MSI-H | NM |
| H57 | F | 47 | ACII | MSI-H | NM |
| H63 | F | 47 | AC | MSI-H | NM |
| H10 | M | 41 | JC | MSI-H | NM |
| H12 | F | 50 | JC | MSI-H | NM |
| H41 | M | 46 | JC | MSI-H | NM |
| H55 | M | 49 | JC | MSI-H | NM |
| H7 | M | 38 | BG | MSI-H | NM |
| H8 | M | 43 | BG | MSI-H | NM |
| H30 | M | 48 | BG | MSI-H | NM |
| H35 | F | 38 | BG | MSI-H | NM |
| H51 | F | 27 | BG | MSI-H | NM |
| Case | Gender | Age (yr) | Criteria | MSI | MLH1/MSH2/MSH6 mutation study |
| H16 | F | 44 | JC | MSS | NM |
| H20 | F | 54 | BG | MSS | NM |
| H44 | F | 39 | BG | MSS | NM |
| H48 | M | 28 | BG | MSS | NM |
| H50 | M | 55 | BG | MSS | NM |
| H54 | M | 43 | BG | MSS | NM |
Hereditary non-polyposis colorectal cancer (HNPCC), also known as Lynch syndrome, is characterized by an autosomal dominant inheritance of early-onset microsatellite instability (MSI)-positive colorectal cancer and an increased risk of other cancers, including cancers of the endometrium, stomach, ovary, urinary tract, pancreas, and small bowel. HNPCC accounts for 5%-10% of all colorectal cancers and is caused by a mutation in one of the following DNA mismatch repair (MMR) genes: MLH1, MSH2, MSH6, PMS1 and PMS2[1-3]. Germline mutations in MLH1 and MSH2 account for > 90% of all known MMR mutations in HNPCC[4], and germline mutations in MSH6 account for 5%-10%, whereas mutations in other genes are rare[3,5]. MSI has been observed in approximately 13% of sporadic colorectal cancers (CRC) and in virtually all CRC arising in patients with HNPCC. Germline mutations in MMR genes, high-frequency microsatellite instability (MSI-H) and loss of MMR protein expression are the hallmarks of HNPCC. Epigenetic silencing is usually considered a kind of somatic phenomenon and somatic MLH1 promoter hypermethylation is generally accepted in the tumorigenesis of sporadic tumours. However, little is known about the maintenance of epigenetic state in the germline[6] and abnormal MLH1 gene promoter methylation in normal body cells is controversially discussed as a mechanism predisposing patients to HNPCC. Recently, aberrant methylation in MMR genes, MLH1 or MSH2, has been supposed as a basic factor for cancer[7]. Promoter hypermethylation in MLH1 gene of colorectal tumors correlates well with loss of MLH1 protein in sporadic MSI-positive cases[8,9]. This study was to investigate the MLH1 gene germline epimutation by methylation-specific PCR (MSP) in 18 Chinese HNPCC kindreds with MSI-H but without germline mutations in MSH2, MLH1, or MSH gene, in order to identify HNPCC families and provide experimental information for HNPCC database.
From January 1998 to October 2005, 24 Chinese HNPCC families fulfilling different clinical criteria were registered at the Department of Abdominal Surgery in Shanghai Cancer Hospital/Institution. Germline mutations in MLH1, MSH2 and MSH6 genes were excluded by DNA-PCR-based sequencing in the probands of all Chinese HNPCC families[10-12]. Of them, 18 unrelated HNPCC probands were selected for the study objects with the phenotype of MSI-H, and the remaining 6 were for the control group with the phenotype of microsatellite stability (MSS). Each participant was asked to give 10 microliters of peripheral blood and consented for access to archival tumor tissue. The characteristics of the selected cases are listed in Tables 1 and 2. To ensure the reliability of the results, family H65 with nonsense germline mutation at c.2228C > A in MSH2 gene was used as the negative control and the cell line sw48 was used as the known positive control for the methylation in MLH1 gene as well as water as the blank control. This study was proved by the Medical Ethical Committee of Cancer Hospital, Fudan University. The procedures of the study were in accordance with the international rules and regulations.
Genomic DNA from peripheral blood and the cell line sw48 was extracted with the QIAGEN (Hilden, Germany) DNA extraction kit following its manufacturer’s introductions. Concentration of the genomic DNA was determined with an ultraviolet spectrophotometer (Beckman, DU640 type).
MSP exploits the effect of sodium bisulfite on DNA, which efficiently converts unmethylated cytosine to uracil with methylated cytosine unchanged. Consequently, after treatment, methylated and unmethylated alleles have different sequences that can be used to design allele-specific primers.
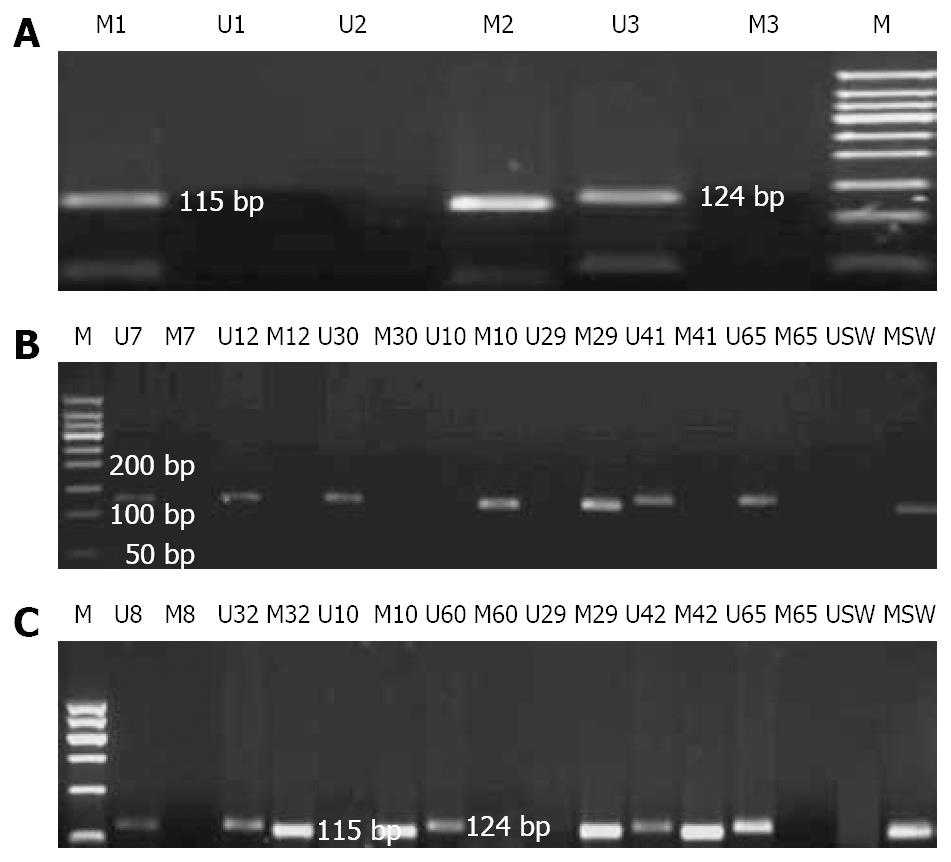
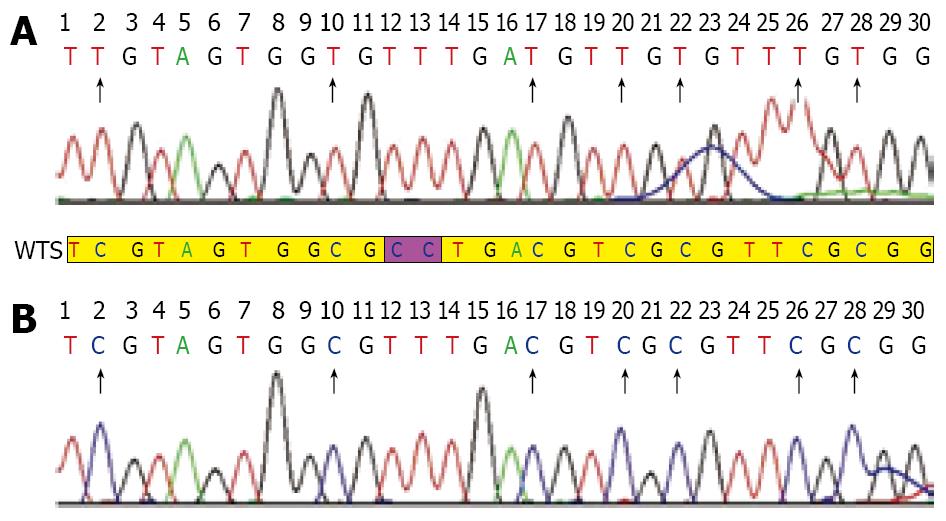
Genomic DNA was modified with sodium bisulfite as described previously[13,14]. The modified DNA was then subjected to MSP using primer pairs engineered to amplify either methylated or unmethylated DNA. Methylated and unmethylated primer pair sequences were also designed as previously described[15] and synthesized (Sangon, shanghai). PCR was carried out with HotstarTaq DNA polymerase (Cat. No. 203203): preheating at 94°C for 10 min, followed by 40 cycles of denaturation at 94°C for 45 s, annealing at 58°C for 45 s and extension at 72°C for 45 s, and a final elongation at 72°C for 7 min. PCR products were subjected to 2% agarose gel electrophoresis. The products of exhaustive-methylation only indicated a methylated band of 124 bp and the unmethylated products only indicated an unmethylated band of 115 bp, while the partially methylated products indicated both of them. After observation of clear and expected bands, the products were purified using the QIAquick gel extraction kit (Qiagen) and sequenced on a 3700 DNA sequence system (ABI, USA) in order to check the correct bisulfite modification. Appropriate positive and negative reference samples were included. Each result of sequencing was analyzed by DNA Star 5.08 bioanalysis software.
A monoclonal antibody against MLH1 (Pharmingen, San Diego, CA, USA) was prepared at a 1:40 dilution and detected by the Envision two-step method to judge whether the status of methylation in MLH1 gene would affect the expression of MLH1 protein. The expression of MLH1 was diminished in cancer tissues in the absence of detectable nuclear staining of neoplastic cells. Infiltrating lymphocytes and normal colonic crypt epithelium next to the tumor area served as internal positive controls.
Five probands with MLH1 gene methylation were found in 18 unrelated Chinese HNPCC families with MSI-H phenotype but without germline mutations in MSH2, MLH1 and MSH6 genes. The rate of abnormal methylation in MLH1 gene was approximately 27.8% (5/18). Among the 18 patients, 2 displayed exhaustively methylated phenotype and the other 3 presented partially-methylated phenotype. The exhaustive methylation accounted for 11.1% (2/18) in the HNPCC families with MSI-H but without germline mutations in MSH2, MLH1 and MSH6 genes. Perhaps, the changes might be much lower in all unselected HNPCC families. Among the 13 probands with unmethylation phenotype, 8 fulfilled the Japanese criteria (JC)/Bethesda guidelines (BG), 5 fulfilled the Amsterdam criteria (AC). All probands with partially-methylated phenotype fulfilled the AC, whereas probands of families H10 and H29 displaying exhaustively-methylated phenotype fulfilled the JC and AC, respectively. The rate of aberrant methylation in MLH1 gene in the AC group (22.2%, 4/18) was higher than that in the JC/BG groups (5.6%, 1/18) in all HNPCC families with MSI-H phenotype and without germline mutations in MSH2, MLH1 and MSH6 genes. However, no proband with methylation in MLH1 gene was found in HNPCC families with MSS phenotype but without germline mutations in MSH2, MLH1 and MSH6 genes. In our study, the expected size of bands was clear and specific. The study was repeated in triplicate to make sure all results credible (Figure 1A-C). Moreover, all exhaustively and partially methylated PCR products were purified and clone-sequenced in order to further substantiate the results of MSP (Figure 2). We believed that the methylation in MLH1 gene might be related with microsatellite phenotype. No expression of MLH1 protein was observed in tumor tissues from two patients with exhaustively methylated phenotype, while positive expression of MLH1 protein was found in tumor tissues from patients with partially methylated phenotype (excluding family H42 without tumor tissue), suggesting that exhaustive-methylation in MLH1 gene can cause defective expression of MLH1 protein and influence its function while the partial methylation in MLH1 gene may have no impact on the expression of MLH1 protein.
HNPCC syndrome is the most common form of hereditary colorectal cancer. Predisposed individuals have a higher risk of developing cancer. The syndrome is primarily due to heterozygous germline mutations in MLH1, MSH2, MSH6 and PMS2 genes. The resulting mismatch repair deficiency leads to MSI which is the hallmark of tumors arising within this syndrome, as well as a variable proportion of sporadic tumors. Diagnostic guidelines and criteria for molecular testing of suspected families have been proposed and continuously updated. However, not all families fulfilling these criteria show mutations in MMR genes and/or MSI implicating other unknown carcinogenic mechanisms and predisposition genes. This subset of tumors is the focus of current clinical and molecular research.
Germline mutations in the coding regions of MSH2 and MLH1 genes are known to be responsible for up to 45%-64% of all HNPCC families[16], and those of MSH6 account for 10% of HNPCC kindreds[17]. We have previously detected germline mutations in the entire coding regions of MSH2, MLH1 and MSH6 genes in 24 probands meeting the AC, 15 probands fulfilling the JC and 19 probands meeting the BG by PCR-gene-sequencing with 20 germline mutations detected including two mutations occurring in a same patient and three novel mutations[10,11]. Subsequently, 3 new mutations are found by mRNA-based PCR sequencing[12]. It was speculated that the remaining probands without mutations in MSH2, MLH1, and MSH6 genes might be associated with other aberrant types of genes. It was reported that DNA methylation associated with transcriptional silencing of MLH1 is the underlying cause of MMR defects in most sporadic colorectal cancers with a MSI+ phenotype[9,18]. Moreover, reversal of methylation with 5-aza-deoxycytidine not only results in reexpression of MLH1 protein, but also restoration of the MMR capacity in MMR-deficient cell lines[9]. These findings further substantiate that the promoter methylation in MLH1 gene is another deficient mechanism of MLH1 gene.
Hypermethylation of CpG island in the promoter sequence has been proved to be an important mechanism of gene silencing and is particularly associated with transcriptional silencing of tumor suppressor genes in sporadic cancers[19,20]. Germline mutations might occur in individuals with a well-characterized genetic disease but lack an identifiable mutation in known disease genes[21]. It was recently reported that monoallelic promoter hypermethylation in MLH1 gene is observed in peripheral blood from a number of patients with early-onset colorectal cancer[7,22-24]. The above results indicate that MLH1 promoter-germline mutation might be related to HNPCC.
Our study demonstrated 5 probands with MLH1 gene methylation (including 2 exhaustive-methylations which fulfill the JC and the AC, respectively, and 3 part-methylations fulfilling the AC) in 18 unrelated Chinese HNPCC families with MSI-H phenotype but without germline mutations in MSH2, MLH1 and MSH6 genes. The rate of aberrant methylation in MLH1 gene (22.2%, 4/18) was higher in probands fulfilling the AC than that (5.6%, 1/18) in those meeting the JC and BG. Of the 13 probands with unmethylated phenotype, 8 fulfilled the JC and BG (61.5%, 8/13), 5 fulfilled the AC (38.5%, 5/13). However, no proband was detected with the aberrant methylation in MLH1 gene in the 6 suspected HNPCC families with MSS phenotype and without germline mutations in MSH2, MLH1 and MSH6 genes. These findings illuminate that the promoter methylation in MLH1 gene is likely another underlying cause of MMR defect in HNPCC fulfilling the AC. In order to ravel whether the aberrant methylation in MLH1 gene influences the expression of MLH1 protein, immunostaining of MLH1 protein was carried out in 5 probands with MLH1 aberrant methylation in our study. No expression of MLH1 protein was found in 2 probands with exhaustively methylated phenotype, whereas positive expression of MLH1 protein was observed in 2 probands with partially methylated phenotype (excluding family H42 without tumor tissue) suggesting that exhaustive methylation in MLH1 gene can cause defective expression of MLH1 protein and influence its function while partial methylation of MLH1 gene may have no impact on the expression of MLH1 gene, revealing that methylation in MLH1 gene may be related with the microsatellite phenotype and influence the expression of MLH1 protein and its function, which is consistent with the reported findings in other studies[8,9].
In neoplastic cells, stable allele-specific loss of transcription due to aberrant methylation in an unmutated promoter region can identify hypermethylation as a direct mechanism of tumor suppressor gene inactivation[25]. Moreover, the promoter methylation can be passed in somatic mitosis, which is reversible. Persons with hypermethylation in MLH1 alleles of somatic cells can predispose to the development of cancer in patterns with hereditary nonpolyposis colorectal cancer. It was reported that epimutation can be transmitted from a mother to her son[26], which is consistent with transgenerational epigenetic inheritance.
In the present study, the rate of aberrant methylation in MLH1 gene was only 27.8% (5/18) in selected HNPCC with MSI-H phenotype but without germline mutations in MLH1, MSH2 and MSH6 genes. Among the probands with aberrant methylation, the rate of methylation in those fulfilling the AC accounted for 80% (4/5), which was significantly higher than that [20% (1/5)] in those meeting the JC and BG. Methylation analysis of the MLH1 promoter should be performed for all early-onset or multiple colorectal cancer patients with MSI-H tumors or loss of MLH1 protein expression due to unknown causes in HNPCC probands fulfilling the AC.
There is evidence that aberrant methylation in the promoter region of MLH1 alleles is functionally equivalent to a pathogenic MLH1 germline mutation and mimics the clinical phenotype of Lynch syndrome. ‘Sporadic’ HNPCC-patients need to be treated Lynch syndrome patients. Individuals carrying MLH1 germline epimutations are at a high risk of developing colorectal and other tumors and should be considered carriers of germline mutations. Inheritance should be discarded in each case, until more conclusive data are obtained. MLH1 promoter methylation analysis should be performed at least for the first degree relatives with positive methylation to exclude the inheritance of a familial epimutation[27]. Identification of hypermethylation as an epigenetic defect has important implications for surveillance reco-mmendations, since these patients should be treated like Lynch syndrome patients.The heritability of methylation needs to be further investiged.
Germline mutations in mismatched repair genes, such as MLH1, MSH2 and MSH6, lead to hereditary nonpolyposis colorectal cancer (HNPCC) and not all families fulfilling these criteria show mutations in mismatched repair genes. It is well known that MLH1 promoter methylation is related with sporadic colorectal cancer. However, MLH1 promoter germline-methylation in Chinese HNPCC patients has not yet been reported.
Germline mutations in MMR genes, such as MSH2, MLH1 and MSH6 contribute to the early diagnosis of HNPCC and screening of HNPCC families. Few studies on MLH1 promoter germline-methylation are available.
Five patients with MLH1 gene methylation were found in this study by methylation-specific PCR in 18 unrelated Chinese HNPCC probands with high microsatellite-instability phenotype but without germline mutations in MSH2, MLH1 and MSH6 gene. The rate of abnormal methylation in MLH1 gene was approximately 27.8% (5/18) and the rate (22.2%, 4/18) in probrands fulfilling the Amsterdam criteria, which was higher than that (5.6%, 1/18) in those meeting the Japanese criteria/Bethesda guidelines.
MLH1 promoter methylation analysis can be used for the microsatellite phenotype of mismatched repair genes and is a promising tool for molecular genetics screening of HNPCC.
HNPCC is an abbreviation of hereditary nonpolyposis colorectal cancer; MSP is an abbreviation of methylation-specific PCR.
In this study, MLH1 promoter germline-methylation was detected in 18 unrelated Chinese HNPCC probands with high microsatellite-instability phenotype but without germline mutations in MSH2, MLH1 and MSH6 gene. The rate of aberrant methylation in probands meeting theAmsterdam criteria was higher than that in those fulfilling the Japanese criteria/Bethesda guidelines. However, the function of MLH1 promoter germline-methylation should be further studied with a large of samples.
Peer reviewer: Jose JG Marin, Professor, Head of the Departamento Physiology and Pharmacology, University of Salamanca, CIBERehd, Campus Miguel de Unamuno, ED-S09, Salamanca 37007, Spain
S- Editor Li LF L- Editor Wang XL E- Editor Ma WH
| 1. | Miyaki M, Konishi M, Tanaka K, Kikuchi-Yanoshita R, Muraoka M, Yasuno M, Igari T, Koike M, Chiba M, Mori T. Germline mutation of MSH6 as the cause of hereditary nonpolyposis colorectal cancer. Nat Genet. 1997;17:271-272. |
| 2. | Muller A, Fishel R. Mismatch repair and the hereditary non-polyposis colorectal cancer syndrome (HNPCC). Cancer Invest. 2002;20:102-109. |
| 3. | Peltomaki P, Vasen HF. Mutations predisposing to hereditary nonpolyposis colorectal cancer: database and results of a collaborative study. The International Collaborative Group on Hereditary Nonpolyposis Colorectal Cancer. Gastroenterology. 1997;113:1146-1158. |
| 4. | Peltomaki P. Deficient DNA mismatch repair: a common etiologic factor for colon cancer. Hum Mol Genet. 2001;10:735-740. |
| 5. | Liu B, Parsons R, Papadopoulos N, Nicolaides NC, Lynch HT, Watson P, Jass JR, Dunlop M, Wyllie A, Peltomaki P. Analysis of mismatch repair genes in hereditary non-polyposis colorectal cancer patients. Nat Med. 1996;2:169-174. |
| 6. | Reik W, Dean W, Walter J. Epigenetic reprogramming in mammalian development. Science. 2001;293:1089-1093. |
| 7. | Gazzoli I, Loda M, Garber J, Syngal S, Kolodner RD. A hereditary nonpolyposis colorectal carcinoma case associated with hypermethylation of the MLH1 gene in normal tissue and loss of heterozygosity of the unmethylated allele in the resulting microsatellite instability-high tumor. Cancer Res. 2002;62:3925-3928. |
| 8. | Miyakura Y, Sugano K, Konishi F, Ichikawa A, Maekawa M, Shitoh K, Igarashi S, Kotake K, Koyama Y, Nagai H. Extensive methylation of hMLH1 promoter region predominates in proximal colon cancer with microsatellite instability. Gastroenterology. 2001;121:1300-1309. |
| 9. | Herman JG, Umar A, Polyak K, Graff JR, Ahuja N, Issa JP, Markowitz S, Willson JK, Hamilton SR, Kinzler KW. Incidence and functional consequences of hMLH1 promoter hypermethylation in colorectal carcinoma. Proc Natl Acad Sci USA. 1998;95:6870-6875. |
| 10. | Cai Q, Sun MH, Fu G, Ding CW, Mo SJ, Cai SJ, Ren SX, Min DL, Xu XL, Zhu WP. [Mutation analysis of hMSH2 and hMLH1 genes in Chinese hereditary nonpolyposis colorectal cancer families]. Zhonghua Bing Li Xue Za Zhi. 2003;32:323-328. |
| 11. | Yan SY, Zhou XY, Du X, Zhang TM, Lu YM, Cai SJ, Xu XL, Yu BH, Zhou HH, Shi DR. Three novel missense germline mutations in different exons of MSH6 gene in Chinese hereditary non-polyposis colorectal cancer families. World J Gastroenterol. 2007;13:5021-5024. |
| 12. | Wang CF, Zhou XY, Zhang TM, Sun MH, Xu Y, Shi DR. [The analysis for mRNA mutation of MLH1, MSH2 genes and the gene diagnosis for hereditary nonpolyposis colorectal cancer]. Zhonghua Yixue Yichuanxue Zazhi. 2006;23:32-36. |
| 13. | Herman JG, Graff JR, Myohanen S, Nelkin BD, Baylin SB. Methylation-specific PCR: a novel PCR assay for methylation status of CpG islands. Proc Natl Acad Sci USA. 1996;93:9821-9826. |
| 14. | Grady WM, Rajput A, Lutterbaugh JD, Markowitz SD. Detection of aberrantly methylated hMLH1 promoter DNA in the serum of patients with microsatellite unstable colon cancer. Cancer Res. 2001;61:900-902. |
| 15. | Chung WB, Hong SH, Kim JA, Sohn YK, Kim BW, Kim JW. Hypermethylation of tumor-related genes in genitourinary cancer cell lines. J Korean Med Sci. 2001;16:756-761. |
| 16. | Shin KH, Shin JH, Kim JH, Park JG. Mutational analysis of promoters of mismatch repair genes hMSH2 and hMLH1 in hereditary nonpolyposis colorectal cancer and early onset colorectal cancer patients: identification of three novel germ-line mutations in promoter of the hMSH2 gene. Cancer Res. 2002;62:38-42. |
| 17. | Kariola R, Raevaara TE, Lonnqvist KE, Nystrom-Lahti M. Functional analysis of MSH6 mutations linked to kindreds with putative hereditary non-polyposis colorectal cancer syndrome. Hum Mol Genet. 2002;11:1303-1310. |
| 18. | Cunningham JM, Christensen ER, Tester DJ, Kim CY, Roche PC, Burgart LJ, Thibodeau SN. Hypermethylation of the hMLH1 promoter in colon cancer with microsatellite instability. Cancer Res. 1998;58:3455-3460. |
| 20. | Rountree MR, Bachman KE, Herman JG, Baylin SB. DNA methylation, chromatin inheritance, and cancer. Oncogene. 2001;20:3156-3165. |
| 21. | Martin DI, Ward R, Suter CM. Germline epimutation: A basis for epigenetic disease in humans. Ann N Y Acad Sci. 2005;1054:68-77. |
| 22. | Miyakura Y, Sugano K, Akasu T, Yoshida T, Maekawa M, Saitoh S, Sasaki H, Nomizu T, Konishi F, Fujita S. Extensive but hemiallelic methylation of the hMLH1 promoter region in early-onset sporadic colon cancers with microsatellite instability. Clin Gastroenterol Hepatol. 2004;2:147-156. |
| 23. | Suter CM, Martin DI, Ward RL. Germline epimutation of MLH1 in individuals with multiple cancers. Nat Genet. 2004;36:497-501. |
| 24. | Hitchins M, Williams R, Cheong K, Halani N, Lin VA, Packham D, Ku S, Buckle A, Hawkins N, Burn J. MLH1 germline epimutations as a factor in hereditary nonpolyposis colorectal cancer. Gastroenterology. 2005;129:1392-1399. |
| 25. | Myohanen SK, Baylin SB, Herman JG. Hypermethylation can selectively silence individual p16ink4A alleles in neoplasia. Cancer Res. 1998;58:591-593. |
| 26. | Hitchins MP, Wong JJ, Suthers G, Suter CM, Martin DI, Hawkins NJ, Ward RL. Inheritance of a cancer-associated MLH1 germ-line epimutation. N Engl J Med. 2007;356:697-705. |
| 27. | Morak M, Schackert HK, Rahner N, Betz B, Ebert M, Walldorf C, Royer-Pokora B, Schulmann K, von Knebel-Doeberitz M, Dietmaier W. Further evidence for heritability of an epimutation in one of 12 cases with MLH1 promoter methylation in blood cells clinically displaying HNPCC. Eur J Hum Genet. 2008;16:804-811. |