Published online May 7, 2007. doi: 10.3748/wjg.v13.i17.2446
Revised: December 28, 2006
Accepted: March 21, 2007
Published online: May 7, 2007
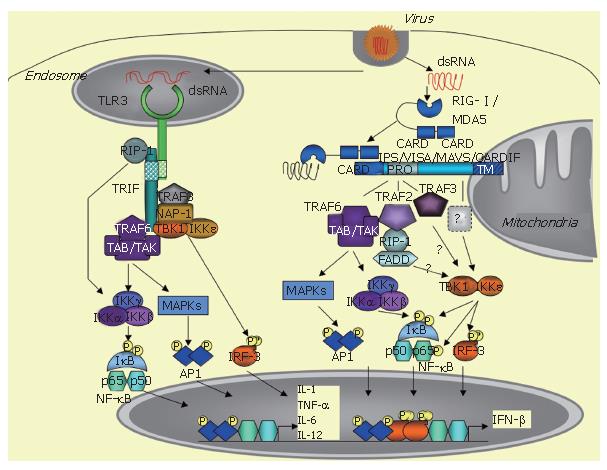
The innate immune response is triggered by a variety of pathogens, including viruses, and requires rapid induction of type I interferons (IFN), such as IFNβ and IFNα. IFN induction occurs when specific pathogen motifs bind to specific cellular receptors. In non-professional immune, virally-infected cells, IFN induction is essentially initiated after the binding of dsRNA structures to TLR3 receptors or to intracytosolic RNA helicases, such as RIG-I /MDA5. This leads to the recruitment of specific adaptors, such as TRIF for TLR3 and the mitochondrial-associated IPS-1/VISA/MAVS/CARDIF adapter protein for the RNA helicases, and the ultimate recruitment of kinases, such as MAPKs, the canonical IKK complex and the TBK1/IKKε kinases, which activate the transcription factors ATF-2/c-jun, NF-κB and IRF3, respectively. The coordinated action of these transcription factors leads to induction of IFN and of pro-inflammatory cytokines and to the establishment of the innate immune response. HCV can cleave both the adapters TRIF and IPS-1/VISA/MAVS/CARDIF through the action of its NS3/4A protease. This provokes abrogation of the induction of the IFN and cytokine pathways and favours viral propagation and presumably HCV chronic infection.
- Citation: Meurs EF, Breiman A. The interferon inducing pathways and the hepatitis C virus. World J Gastroenterol 2007; 13(17): 2446-2454
- URL: https://www.wjgnet.com/1007-9327/full/v13/i17/2446.htm
- DOI: https://dx.doi.org/10.3748/wjg.v13.i17.2446
The innate immune response is triggered in response to a variety of pathogens, such as bacteria and viruses, and is essential for a rapid limitation in the spread or action of these pathogens. The type I interferons, represented by IFNβ and by different subtypes of IFNα, play an important role in this process as they can mount an immediate antiviral response and stimulate adaptative immunity[1] Type I interferons are secreted proteins that exert their function after binding to specific IFNAR receptors and after activation of the JAK/STAT signalling pathway[2]. They are potent and can induce over 300 genes, collectively referred to as interferon stimulated genes (ISGs)[3]. Because of their antiviral, antiproliferative and immunomodulatory activities, IFNs are used for the treatment of different tumors and viral infections, such as HBV and HCV. Under normal physiological and health conditions, however, IFNs are expressed at a minimum level and their induction in response to a pathogen involves a complex and well orchestrated cellular machinery.
Microbial agents are recognized through some of their motifs or pathogen-associated molecular patterns (PAMPS) by specific cellular receptors, referred to as PRRs (pathogen-recognition receptors). The PAMPs responsible for IFN induction can be bacterial external compounds, such as lipopolysacharides (LPS), viral envelopes and nucleic acids including dsRNA, ssRNA and DNA. The PRRs are members of the toll-like receptors (TLRs) family, located either at cellular (TLR4 for LPS and viral envelops) or endosomal membranes (TLR3 for dsRNA, TLR7/8 for ssRNA and TLR9 for DNA)[4-7]. In addition to this, another route of induction takes place in the cytosol through activation of specific RNA helicases, such as RIG-I and MDA5[8,9]. The cellular type plays an important role in the specificity of induction, since IFN is induced in the immune plasmacytoid cells through TLR7/8 (ssRNA) and TLR9 (DNA), while its induction in the non-professional immune cells, including dendritic cells (DCs), requires the endosomal TLR3 or the intracytoplasmic RNA helicase[10].
At the transcriptional level, IFN induction requires the conjugated action of the three transcription factors: ATF2/c-jun, NF-κB and IRF3. ATF2/c-jun and NF-κB are activated in response to various stimuli, such as growth factors and pro-inflammatory molecules, by phosphorylation through the MAPK cascade and the canonical IKK α/β/γ complex, respectively. IRF3 phosphorylation is triggered by a viral infection, after incubation of cells with dsRNA or after introduction of dsRNA by transfection[11]. The kinases responsible for IRF3 phosphorylation, which were referred to as VAK for virus-activated kinases for some years[11], were identified in 2003 as the two non-canonical IκB kinases: TBK1 (Tank binding kinase 1, also known as NAK for NF-κB activating kinase) and IKKε[12,13].
After binding to their respective PAMPs, the TLRs recruit adaptor proteins through homotypic interactions with their cyoplasmic TIR (Toll/IL-1 receptor) domain. All TLRs, except TLR3, recruit MyD88, which links them to the NF-κB and MAPK pathways through activation of the IRAKs (IL-1 receptor associated kinases) and TRAF6 (TNF receptor associated factor 6). In contrast, TLR3, upon dsRNA binding, recruits a different adapter, named TRIF (TIR domain-containing adapter inducing IFN-β) or TICAM (TIR-containing adaptor molecule-1)[14,15]. TLR4 can also induce IFN through TRIF, but, in that case, it recruits TRIF indirectly through interaction with another adaptor called TRAM (TRIF related adaptor molecule) or TICAM-2[16]. TRIF interacts with a number of signaling molecules, such as TRAF6, which in turn activate the NF-κB and MAPK pathways[17,18]. In addition, it can also activate NF-κB through interaction with the kinase RIP-1 (receptor interacting protein-1)[19]. Importantly, TRIF is involved in IRF3 activation by recruiting TBK1 and, presumably, also IKKε, although this remains controversial[13,17]. TRIF can also recruit TRAF6, which activates the NF-κB and the MAPK pathways[17,18], NAP-1 (NAK associated protein-1) and TRAF3, which are both involved in IFN induction, although their exact role still needs to be clarified[20,22]. Through all these different interactions, TRIF triggers induction of IFN and pro-inflammatory cytokines (Figure 1).
In accord with a role for TLR3 in IFN induction in response to dsRNA, TLR3 deficient mice present a strong reduction in their ability to induce IFN and proinflammatory cytokines when injected with synthetic or viral dsRNA. These mice are also more susceptible to infection by MCMV[5]. TLR3 is expressed in endosomal compartments and is abundant in conventional dendritic cells, therefore allowing immediate activation of the immune response. TLR3 has also been shown to be important in crosspriming, therefore allowing CD8+ T-cell response at the site of virally-infected tissue cells[23]. However, the role of TLR3 in the antiviral response is probably more complex. For instance, infection of TLR3-deficient mice with West Nile virus (WNV) resulted in a better survival of these mice to the infection than the TLR3-wt mice, in which the infection provoked a TLR3-dependent inflammatory response, associated with brain penetration of the virus and neuronal injury[24]. Similarly, TLR3-deficient mice had an unexpected survival advantage to Influenza A virus (IAV), which is a highly contagious acute respiratory disease, despite a higher viral production in the lungs, because these animals displayed significantly reduced inflammatory mediators than the wt animals in the bronchoalveolar airspace[25].
Like TLR3, the TLR 7, 8 and 9 are expressed in en-dosomal compartments but their expression is restricted to a subtype of dendritic cells, which are the plasmacytoid dendritic cells (pDCs). These latter cells represent less than 1% of the circulating PBMCs but can release massive amounts of IFN-α in the blood. They were previously also named NIPC for Natural Interferon Producing Cells[1]. Induction of IFN by these TLRs, in response to ssRNA (TLR7.8) or DNA (TLR9) is exclusively MyD88-dependent and does not require TRIF. In this situation, the signaling events leading to IFN induction involve recruitment of TRAF6, IRAK4 and the transcription factor IRF7. This provokes IRF-7 phosphorylation and direct induction of IFNα. This signaling process is possible because of the constitutive presence of IRF7 in the pDCs[26,28].
TLR3- and TRIF-deficient mice are impaired in their ability to induce IFN in response to dsRNA and they may become sensitive to viral infections[5,29]. However, their response to dsRNA is not totally abolished and they are still resistant to some viral infections, such as VSV (vesicular stomatitis virus) or SeV (Sendai virus)[30]. This suggested the existence of novel IFN-inducing pathways, which were independent of the TLR3/TRIF axis. A role for the dsRNA-dependent protein kinase PKR was unlikely since earlier studies showed that this kinase was not required for IRF3 or IRF7 phosphorylation[31]. Furthermore, the induction of T cell stimulatory molecules was normal and induction of IFN-β was only slightly suppressed in DC from PKR-deficient mice[32].
Human K562 cells lack the entire IFN-encoding locus and do not activate IRF3 after virus infection, unless they are treated with IFNβ. An expression cDNA library generated from IFNβ-treated K562 cells was therefore generated and screened for stimulation of the transcription of an IRF-dependent reporter in the presence of the synthetic dsRNA poly(I)-poly(C), after transfection in murine L929 cells. This led to the isolation of one clone encoding for the caspase activation recruitment domain (CARD)-containing the N terminal domain of the DexD/H box helicase RIG-I [8]. The presence of CARD allows recruitment of proteins through homotypic interactions. RIG-I was initially described as being induced by Retinoic Acid (RIG = retinoic acid inducible gene)[33]. It was also shown to be induced by IFNγ[34], TNF-α and IFNα[35]. RIG-I belongs to a family of RNA helicases that also contains MDA-5 (melanoma differentiation associated gene-5)[36] and LGP2[37]. MDA-5 is highly homologous to RIG-I (23% identity in the CARD and 35% identity in the helicase domain). Both proteins bind dsRNA and transmit signaling through their RNA helicase/ATPase domain, probably by a conformational change, which enables their N-terminal CARD domain to initiate the downstream signaling events leading to ATF2/C-jun, NF-κB and IRF3 activation. In contrast, LGP2, which shows 31% and 41% identity with the RNA helicase domains of RIG-I and MDA5, respectively, lacks the CARD domain and thus probably has a negative regulatory role on the RIG-I /MDA5 pathway[38]. It is interesting to note that all three RNA helicases, i.e., RIG-I , MDA5 and LGP2, are IFN-inducible proteins. Yet, they are directly involved in the very first step of IFN induction, at least for RIG-I and MD-5. This indicates that the early steps of IFN induction already require significant cytosolic expression of these RNA helicases. Accordingly, RIG-I can be independently induced in response to different stimuli, such as TNF-α[35]. It is therefore possible that virally-induced inflammatory processes are important to generate the required amounts of RIG-I necessary for triggering IFN induction. Although RIG-I and MDA5 present strong similarities, they apparently do not have similar functions in cells. For instance, deletion of the RIG-I gene is lethal with most of the embryos dying between 12 and 14 d and with mice born alive dying after 3 wk. The developmental defect of the RIG-I -/- embryos was linked to massive liver degeneration[10]. In contrast, MDA-5 deficient mice are healthy[39]. Most cell types derived from the RIG-I deficient embryo are unable to produce type I IFN and inflammatory cytokines, such as IL-6, upon SeV infection[10]. Interestingly, systematic comparison of RIGI and MDA5 deficient MEFs showed that the two RNA helicases differ in their specificity for IFN induction. RIG-I was required for IFN induction by in vitro transcribed RNAs and by the following viruses: SeV, NDV, influenza virus, VSV or JEV, while MDA5 was required for IFN induction by poly(I)-poly(C) and the picornaviruses EMCV, Theiler and Mengo[39,40]. The reason for this discrepancy was recently solved independently by two groups who showed that the ligand for RIG-I is an uncapped 5' triphosphate RNA, which is a situation found in viruses of the Flaviviridae family, including HCV, and non segmented viruses, such as Paramyxoviruses and rhabdoviruses. In contrast, MDA5 recognize viruses with protected 5' RNA ends, such as in the case of picornaviruses[41,42].
In 2005, four groups independently reported the identification of an adapter protein that links the RNA helicase to the downstream MAPK, NF-κB and IRF3 signaling pathways. This protein is referred to as IPS-1 (Interferon-β Promoter Stimulator 1[43]), VISA (Virus-Induced Signaling Adaptor[44]), MAVS (Mitochondrial AntiViral Signaling[45]) and CARDIF (CARD adapter inducing IFNβ[46]). This protein was previously identified in a cDNA library screen as a NF-κB activating molecule[47]. In the absence of a consensus name for this protein, it will be referred to here as CARDIF, in acknowledgment of the group that first presented the sensitivity of this protein to cleavage by the HCV NS3/4A protease (46; see below). The particularity of this adapter protein is localization to the mitochondrial membrane through a specific transmembrane domain located at its C terminus[45]. The 540 residue CARDIF protein associates with the tandem CARD domain of RIG-I through its own N-terminal 1-77 CARD-like domain. Coprecipitation assays showed that CARDIF associates strongly with RIG-I and weakly with MDA5[46]. Another important feature of CARDIF is a proline rich region (103-173 residues) near the N terminus, through which it interacts with several signaling components including TRAF6, TRAF2[44], RIP1, FADD[43] and more recently, TRAF3[48] (Figure 1). It is still unclear how TBK1 and IKKε associate with CARDIF. In one report, TBK1 was found to associate with CARDIF and IKKε was not examined[44]. In another study, neither of these kinases was found to associate with CARDIF but the data were presented only for TBK1[43]. In contrast, a strong association of CARDIF with IKKε and no interaction with TBK1 was presented[46]. In accord with the latter, recent confocal microscopy analysis demonstrated a tight colocalization for IKKε with the mitochondrial protein CARDIF, whereas TBK1 was associated with other vesicles[49]. Using coprecipitation techniques, our group also recently confirmed association of CARDIF with IKKε but not with TBKI (Vitour et al, unpublished observations). The CARDIF deficient mice are viable and fertile. Upon viral infection, such as VSV, they can produce IFNα and IFNβ in their sera, as measured by ELISA, presumably through TLR activation, for instance in pDCs. However, they failed to produce IFNα, IFNβ and IL-6 after poly(I)-poly(C) injection, which is reminiscent of the defect in MDA5 deficient mice to respond to poly(I)-poly(C)[39]. Infection of the CARDIF -/- and -/+ mice with different concentrations of VSV showed a VSV-induced mortality that was both dependent on the CARDIF gene dosage and viral titer. Since VSV was shown to induce IFN through RIG-I , these in vivo experiments firmly demonstrate that CARDIF is involved in both the IFN inducing pathways mediated by RIG-I and MDA5[50].
The TBK1 and IKKε kinases play an essential role in the induction of IFN and inflammatory cytokines through their ability to phosphorylate serine residues at the C terminus of both IRF3 and IRF7. This provokes a change in the conformation of these transcription factors, promoting their dimerization and then their binding to their DNA consensus binding sites[12,13,51]. The two kinases are enzymatically similar with strong sequence identity. Accordingly, they behave similarly in their ability to activate IRF3 and IRF7 and in their ability to phosphorylate the IκBα inhibitor of NF-κB at its Ser 36 residue, whereas the two structurally related IKKα and IKKβ kinases phosphorylate IκBα at residues Ser32 and Ser36[52]. Although very similar, TBK1 and IKKε present some differences that may be of importance. For instance, deletion of the TBK1 gene leads to embryonic lethality at d 15[53], wheras IKKε deficient mice are viable[54]. Another difference is the fact that IKKε is more closely associated with CARDIF than TBK1[46,49]. Finally, both IKKε and TBK1 were shown to sustain the NF-κB transcriptional activity through the phosphorylation of specific serine residues at the C terminal transactivation domain of the cRel[55] or RelA p65 subunit[56,57]. Interestingly, however, IKKε was found to play a more critical role than TBK1 in controlling the basal/constitutive p65 phosphorylation[57]. This new finding coupled to the fact that IKKε can sustain its own expression via NF-κB and c/EBPδ transcription factors[58,59], whereas expression of TBK1 is constitutive, provides a link to suspect a role for IKKε in controlling the proliferation of certain cancer cells. In contrast, TBK1 could play a major role in IFNβ induction. Indeed, studies with TBK1 and IKKε murine deficient cells pointed out a more important role for TBK1 than for IKKε in IFN induction in response to LPS, dsRNA (delivered intracytoplasmically) and to virus infection. However, use of the IKKε/TBK1 doubly deficient cells revealed a complete abolition of IFNβ induction[54].
The current treatment against HCV, a combination of pegylated IFN and ribavirin, leads to viral clearance in 50% to 80% of cases. The efficacy of treatment depends on several factors, such as age and sex of the patients, viral parameters, such as genotypes and viral load, and host immune parameters[60]. HCV can interfere with the cellular response to IFN through some of its proteins, which can target the JAK/STAT signaling pathway that is activated in response to the binding of IFN to its receptor[61-63], or through other interactions leading to inhibition of the induction or the function of some ISGs[64]. In 2003, the HCV NS3/4A protease was shown to interfere with IFN induction by preventing IRF3 phosphorylation[65]. This important finding was achieved at the same time as the identification of the two kinases leading to IRF3-phosphorylating kinases, TBK1 and IKKε[12,13].
The HCV 70 Kda NS3 protein presents a serine proteinase domain at its N terminus (aa 1-180) and an RNA helicase domain at its C-terminus (aa 181-631) (reviewed in[66]). Its helicase activity is coordinated by ATP and allows NS3 to move along the RNA like an inchworm to catalyse RNA unwinding[67]. This activity is important for HCV replication. The protease activity catalyses the following cleavages of the viral polyprotein: NS3-NS4A, NS4A-NS4B, NS4B-NS5A and NS5A-NS5B with the following efficiency: NS5A/5B > NS4A/4B > NS4B/5A. Cleavage between NS3 and NS4A is an intramolecular reaction and the rest of the cleavages are mediated in trans[68]. NS4A is a small protein of 54 aa that acts as a cofactor to enhance the NS3 protease activity. For this reason, and also because the NS3 requires a noncatalytic structural zinc ion for its protease activity, this enzyme is unique among the other members of the trypsin superfamily to which it belongs[66]. The catalytic domain of NS3 is formed by a triad of three important residues, S139, H57 and D81. After cleavage of the NS3/4A junction, the C-terminus of NS3 forms a β-strand that occupies the proteinase active site and thus protects it. In contrast with other proteinases, the substrate binding site of NS3 is shallow and solvent-exposed and its selective recognition of substrates requires extended contact surface. The active site of NS3 is well conserved among the different genotypes and the HCV sequences that are cleaved by NS3/4A have the consensus sequence D/E-XXXX-C/T↓S/A-XX-L/W/Y (Table 1). The cleavage sequence and protease specificity of NS3/4A protease have been well characterized since 1993 and it was conceivable that, due to its ability to cleave in trans, the NS3/4A protease may be able to cleave cellular proteins in addition to the processing of the viral proteins.
| Name | Genotype (GI) | NS3/4A | NS4A/4B | NS4B/5A | NS5A/5B | Reference |
| H77 | 1a (GI: 2316097 ) | MSADLEVVT STWVLVGG | QEFDEMEEC SQHLPYIE | ISSECTTPC SGSWLRDI | ADTEDVVCC SSYSWTGA | Kolykhalov et al, 1997 |
| HCV-N | 1b (GI: 23957856) | MSADLEVVT STWVLVGG | REFDEMEEC ASHLPYIE | INEDCSTPC SGSWLRDV | EAGESVVCC SMSYTWTG | Beard et al, 1999 |
| JFH1 | 2a (GI: 13122261) | MQADLEVMT STWVLAGG | EAFDEMEEC ASRAALIE | ITEDCPIPC SGSWLRDV | EEDDTTVCC SMSYSWTG | Kato et al, 2001 |
| NZL1 | 3a (GI: 514395) | MSADLEVTT STWVLLGG | QQYDEMEEC SQAAPYIE | INEDYPSPC SDDWLRTI | SEEQSVVCC SMSYSWTG | Sakamoto et al, 1994 |
| ED43 | 4a (GI: 2252489) | MSADLEVVT STWVLVGG | QQFDEMEEC SKHLPLVE | INEDCSTPC STPCAESW | SGSEDVVCC SMSYSWTG | Chamberlain et al, 1997 |
| SA13 | 5a (GI: 3660725) | MSADLEVIT STWVLVGG | QQFDEMEEC SASLPYMD | IGEDYSTPC DGTWLRAI | SDEDSVVCC SMSYSWTG | Bukh et al, 1998 |
| 6a33 | 6a (GI: 57791993) | MSADLEVIT STWVLVGG | QQFDEMEEC SRHIPYLAE | VNEDTATPC ATSWLRDV | SDQDDVVCC SMSYSWTG | Zhou et al, 2004 |
Analysis of the mechanism(s) by which NS3/4A was inhibiting the IRF3 phosphorylation allowed determine that this protease was acting upstream of the two TBK1/IKKε kinases and was affecting both the TLR3/TRIF pathway and the RIG-I helicase pathway[69,70]. Disruption of IFN induction through TLR3/TRIF was shown to be due to the cleavage of the TLR3 adapter TRIF by NS3/4A[71], while disruption of IFN induction through the RIG-I pathway was due to the cleavage of the mitochondrial adapter protein IPS-1/MAVS/VISA/CARDIF[46,49,72]. Cleavage of TRIF occurs between its Cys372 and Ser373 residues, which separates its TIR domain from the TBK1-interacting N terminus domain. It is interesting to note that the TRIF cleavage site (PSSTPC↓SAHLT) differs from the HCV consensus cleavage site by a proline at the P6 position instead of an acidic residue. Strikingly, this proline is preceded by a stretch of 7 prolines and it is thought that this particular sequence may enhance the affinity of TRIF for the NS3 protease[73]. This stretch of proline, which can form a left-handed polyproline II helix may compensate for the absence of acidic residue at the P6 position, which normally is contributing to enhance the Km values in the viral natural substrates. A helix composed of hydrophobic residues was identified in the NS3 protease domain, not far from the protease active site and may represent a possible site to anchor TRIF near the active site of NS3[73]. The CARDIF cleavage site at the 508 residue, EREVPC↓HRPS, presents more similarity with the HCV consensus cleavage site, except that there is a histidine residue at position P'1 instead of a serine or alanine[46]. Because of this, it is possible to conceive that several cellular substrates for NS3/4A exist. However, they may be difficult to depict, based solely on sequence examination, if, similarly to TRIF and CARDIF, they diverge from the consensus NS3/4A cleavage sequence. The highly specific product-based macrocyclic NS3 protease inhibitor BILN 2061[74] and the less toxic new generation of another class of NS3 inhibitors, referred to as electrophilic or serine-trap inhibitors, such as VX-950[75] and SCH6[76], probably exert their high inhibitory effect on HCV infection, not only by preventing HCV expression[77] but also by preventing NS3/4A to interfere with IFN induction.
The ability of HCV to inhibit the early events of IFN induction emphasizes the importance of the IFN signalling pathways and may therefore represent one of the mechanisms by which this virus compromises the host immune response and favours its propagation. Indeed, the in vitro propagation of infectious particles of HCV genotype 2a[77-79] and HCV of genotype 1a[80] is now highly promoted by infecting a Huh7 cellular clone that was previously isolated for its high susceptibility to HCV replicons[81]. The particularity of this clone, known as Huh7.5, is to contain a mutation in the first CARD domain of RIG-I . This mutation does not prevent the binding of RIG-I to dsRNA but abolishes its ability to activate downstream elements and is likely the one responsible for the inability of this clone to induce IFNβ and early ISGs in response to a viral or dsRNA stimulus. Indeed, complementation of Huh7.5 cells with a plasmid expressing RIG-I restores ISG56 induction in response to SeV or with in vitro transcribed HCV dsRNA-containing structures, such as its NTRs (non translated regions)[82]. The 5' and 3' HCV NTRs were proposed as the HCV elements required for RIG-I activation immediately after internalisation of the viral genome into the cytosol[82]. This was confirmed recently with a study showing the importance of 5' triphosphate in blunt-end dsRNA to signals to IFN induction and allowing the cell to discriminate between self and non self[83] and with the identification of 5' triphosphate RNAs as being the ligand for RIG-I [41,42].
The possibility of using the JFH1 recombinant virus of genotype 2a to infect cell cultures in vitro now gives the possibility to analyse the interaction of HCV with IFN induction pathways in more natural conditions of infection. Indeed, specific cleavage of CARDIF, but not of CARDIF C508A in which the NS3/4A cleavage site has been abolished, could be demonstrated after transient transfection in JFH1-infected Huh7.5 cells[46]. In support of cleavage of CARDIF by NS3/4A, subcellular redistribution of endogenous CARDIF from the mitochondria to the cytosol could be demonstrated in COS cells after transient transfection with NS3/4A-encoding plasmids, in HCV replicon cells[84], in JFH1-infected Huh7 cells and in a liver biopsy from a patient with chronic HCV infection[84]. Much work is needed yet to fully understand the exact relationship between the RNA helicase/CARDIF/kinases pathway and the ability of HCV to escape the cellular defense. For instance, the NS3/4A inhibitor BILN2061 was shown to restore the IFN induction in response to ectopically added CARDIF in JFH1-infected cells[85] but definite proof that the NS3/4A inhibitor BILN2061 can restore IFN induction in HCV-infected cells remains to be established. JFH1 was reported to infect Huh7 cells with a viral progeny reaching similar levels to those obtained from Huh7.5 cells, after a lag of 7 d. This delay was first explained by the ability of JFH1 to induce an antiviral response in the Huh7 cells, but not in the Huh7.5 cells where the RIG-I pathway is defective and cannot recruit the downstream CARDIF adapter[79]. In a follow-up study, however, it was shown that JFH1 fails to induce IFNβ in the Huh7 cells, as well as early ISGs, such as ISG15 or ISG56, from the onset of infection. Interestingly, these authors could demonstrate that JFH1 was blocking IFNβ induction upstream of the TBK1/IKKε kinases, presumably through CARDIF cleavage. However, overexpression of CARDIF in those infected-cells could not restore dsRNA-induced IFN-β promoter activity and RIG-I overexpression could only partially restore it. A current hypothesis is that, in addition to cleaving CARDIF through its NS34A protease activity, HCV infection also provokes RIG-I inactivation through a process independent of NS34A. This suggests the existence of a RIG-I dependent signaling pathway that could by-pass CARDIF to trigger IFNβ expression, and thus represents an additional threat for the virus[85]. In support of this, in a recent study, we showed that expression levels of RIG-I (and of the other RNA helicases MDA5 and LGP2) were down-regulated in liver biopsies from HCV chronically-infected patients. In these biopsies, the expression levels of IKKε-, but not those of TBK1, were also down-regulated[86]. Interestingly, IKKε, when overexpressed, can provoke inhibition of HCV expression in a replicon system and we demonstrated that its antiviral action can occur rapidly, in the absence of IFN induction, through the action of one or several genes induced through activation of IRF3, NF-κB and c/EBPδ[70,86]. RIG-I belongs to the genes induced by IKKε and it is possible to hypothesize that HCV chronic infection thrives in an environment with low RIG-I and IKKε expresssion and/or activity. In line with the down-regulation of IKKε, decreased expression of the NF-κB RelA subunit, one of IKKε susbtrates, was found to be associated with enhanced fibrosis progression in the liver of patients with chronic hepatitis C[87].
The recent identification of different partners from the TLR- and RNA helicase-IFN inducing pathways, coupled with the possibility of using cell culture systems infected with recombinant HCV, now allows rapid progress in the comprehension of the relative importance of these pathways in cellular defence and in their ability to interfere with HCV propagation.
S- Editor Liu Y L- Editor Lutze M E- Editor Chen GJ
| 1. | Le Bon A, Tough DF. Links between innate and adaptive immunity via type I interferon. Curr Opin Immunol. 2002;14:432-436. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 448] [Cited by in F6Publishing: 453] [Article Influence: 20.6] [Reference Citation Analysis (0)] |
| 2. | Brierley MM, Fish EN. Stats: multifaceted regulators of transcription. J Interferon Cytokine Res. 2005;25:733-744. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 145] [Cited by in F6Publishing: 147] [Article Influence: 8.2] [Reference Citation Analysis (0)] |
| 3. | de Veer MJ, Holko M, Frevel M, Walker E, Der S, Paranjape JM, Silverman RH, Williams BR. Functional classification of interferon-stimulated genes identified using microarrays. J Leukoc Biol. 2001;69:912-920. [PubMed] [Cited in This Article: ] |
| 4. | Hemmi H, Takeuchi O, Kawai T, Kaisho T, Sato S, Sanjo H, Matsumoto M, Hoshino K, Wagner H, Takeda K. A Toll-like receptor recognizes bacterial DNA. Nature. 2000;408:740-745. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 4803] [Cited by in F6Publishing: 4695] [Article Influence: 195.6] [Reference Citation Analysis (0)] |
| 5. | Alexopoulou L, Holt AC, Medzhitov R, Flavell RA. Recognition of double-stranded RNA and activation of NF-kappaB by Toll-like receptor 3. Nature. 2001;413:732-738. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 4510] [Cited by in F6Publishing: 4482] [Article Influence: 194.9] [Reference Citation Analysis (0)] |
| 6. | Diebold SS, Kaisho T, Hemmi H, Akira S, Reis e Sousa C. Innate antiviral responses by means of TLR7-mediated recognition of single-stranded RNA. Science. 2004;303:1529-1531. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 2469] [Cited by in F6Publishing: 2471] [Article Influence: 123.6] [Reference Citation Analysis (0)] |
| 7. | Heil F, Hemmi H, Hochrein H, Ampenberger F, Kirschning C, Akira S, Lipford G, Wagner H, Bauer S. Species-specific recognition of single-stranded RNA via toll-like receptor 7 and 8. Science. 2004;303:1526-1529. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 2734] [Cited by in F6Publishing: 2751] [Article Influence: 137.6] [Reference Citation Analysis (0)] |
| 8. | Yoneyama M, Kikuchi M, Natsukawa T, Shinobu N, Imaizumi T, Miyagishi M, Taira K, Akira S, Fujita T. The RNA helicase RIG-I has an essential function in double-stranded RNA-induced innate antiviral responses. Nat Immunol. 2004;5:730-737. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 2934] [Cited by in F6Publishing: 2984] [Article Influence: 149.2] [Reference Citation Analysis (0)] |
| 9. | Andrejeva J, Childs KS, Young DF, Carlos TS, Stock N, Goodbourn S, Randall RE. The V proteins of paramyxoviruses bind the IFN-inducible RNA helicase, mda-5, and inhibit its activation of the IFN-beta promoter. Proc Natl Acad Sci USA. 2004;101:17264-17269. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 751] [Cited by in F6Publishing: 779] [Article Influence: 39.0] [Reference Citation Analysis (0)] |
| 10. | Kato H, Sato S, Yoneyama M, Yamamoto M, Uematsu S, Matsui K, Tsujimura T, Takeda K, Fujita T, Takeuchi O. Cell type-specific involvement of RIG-I in antiviral response. Immunity. 2005;23:19-28. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1024] [Cited by in F6Publishing: 1022] [Article Influence: 53.8] [Reference Citation Analysis (0)] |
| 11. | Servant MJ, Grandvaux N, Hiscott J. Multiple signaling pathways leading to the activation of interferon regulatory factor 3. Biochem Pharmacol. 2002;64:985-992. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 128] [Cited by in F6Publishing: 132] [Article Influence: 6.0] [Reference Citation Analysis (0)] |
| 12. | Sharma S, tenOever BR, Grandvaux N, Zhou GP, Lin R, Hiscott J. Triggering the interferon antiviral response through an IKK-related pathway. Science. 2003;300:1148-1151. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1317] [Cited by in F6Publishing: 1288] [Article Influence: 61.3] [Reference Citation Analysis (0)] |
| 13. | Fitzgerald KA, McWhirter SM, Faia KL, Rowe DC, Latz E, Golenbock DT, Coyle AJ, Liao SM, Maniatis T. IKKepsilon and TBK1 are essential components of the IRF3 signaling pathway. Nat Immunol. 2003;4:491-496. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 2013] [Cited by in F6Publishing: 2041] [Article Influence: 97.2] [Reference Citation Analysis (0)] |
| 14. | Yamamoto M, Sato S, Mori K, Hoshino K, Takeuchi O, Takeda K, Akira S. Cutting edge: a novel Toll/IL-1 receptor domain-containing adapter that preferentially activates the IFN-beta promoter in the Toll-like receptor signaling. J Immunol. 2002;169:6668-6672. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 940] [Cited by in F6Publishing: 890] [Article Influence: 40.5] [Reference Citation Analysis (0)] |
| 15. | Oshiumi H, Matsumoto M, Funami K, Akazawa T, Seya T. TICAM-1, an adaptor molecule that participates in Toll-like receptor 3-mediated interferon-beta induction. Nat Immunol. 2003;4:161-167. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 931] [Cited by in F6Publishing: 903] [Article Influence: 43.0] [Reference Citation Analysis (0)] |
| 16. | Oshiumi H, Sasai M, Shida K, Fujita T, Matsumoto M, Seya T. TIR-containing adapter molecule (TICAM)-2, a bridging adapter recruiting to toll-like receptor 4 TICAM-1 that induces interferon-beta. J Biol Chem. 2003;278:49751-49762. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 288] [Cited by in F6Publishing: 296] [Article Influence: 14.1] [Reference Citation Analysis (0)] |
| 17. | Sato S, Sugiyama M, Yamamoto M, Watanabe Y, Kawai T, Takeda K, Akira S. Toll/IL-1 receptor domain-containing adaptor inducing IFN-beta (TRIF) associates with TNF receptor-associated factor 6 and TANK-binding kinase 1, and activates two distinct transcription factors, NF-kappa B and IFN-regulatory factor-3, in the Toll-like receptor signaling. J Immunol. 2003;171:4304-4310. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 521] [Cited by in F6Publishing: 523] [Article Influence: 26.2] [Reference Citation Analysis (0)] |
| 18. | Jiang Z, Mak TW, Sen G, Li X. Toll-like receptor 3-mediated activation of NF-kappaB and IRF3 diverges at Toll-IL-1 receptor domain-containing adapter inducing IFN-beta. Proc Natl Acad Sci USA. 2004;101:3533-3538. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 275] [Cited by in F6Publishing: 290] [Article Influence: 14.5] [Reference Citation Analysis (0)] |
| 19. | Meylan E, Burns K, Hofmann K, Blancheteau V, Martinon F, Kelliher M, Tschopp J. RIP1 is an essential mediator of Toll-like receptor 3-induced NF-kappa B activation. Nat Immunol. 2004;5:503-507. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 625] [Cited by in F6Publishing: 608] [Article Influence: 30.4] [Reference Citation Analysis (0)] |
| 20. | Sasai M, Oshiumi H, Matsumoto M, Inoue N, Fujita F, Nakanishi M, Seya T. Cutting Edge: NF-kappaB-activating kinase-associated protein 1 participates in TLR3/Toll-IL-1 homology domain-containing adapter molecule-1-mediated IFN regulatory factor 3 activation. J Immunol. 2005;174:27-30. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 102] [Cited by in F6Publishing: 97] [Article Influence: 5.1] [Reference Citation Analysis (0)] |
| 21. | Häcker H, Redecke V, Blagoev B, Kratchmarova I, Hsu LC, Wang GG, Kamps MP, Raz E, Wagner H, Häcker G. Specificity in Toll-like receptor signalling through distinct effector functions of TRAF3 and TRAF6. Nature. 2006;439:204-207. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 692] [Cited by in F6Publishing: 700] [Article Influence: 36.8] [Reference Citation Analysis (0)] |
| 22. | Oganesyan G, Saha SK, Guo B, He JQ, Shahangian A, Zarnegar B, Perry A, Cheng G. Critical role of TRAF3 in the Toll-like receptor-dependent and -independent antiviral response. Nature. 2006;439:208-211. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 664] [Cited by in F6Publishing: 654] [Article Influence: 34.4] [Reference Citation Analysis (0)] |
| 23. | Schulz O, Diebold SS, Chen M, Näslund TI, Nolte MA, Alexopoulou L, Azuma YT, Flavell RA, Liljeström P, Reis e Sousa C. Toll-like receptor 3 promotes cross-priming to virus-infected cells. Nature. 2005;433:887-892. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 686] [Cited by in F6Publishing: 671] [Article Influence: 35.3] [Reference Citation Analysis (0)] |
| 24. | Wang T, Town T, Alexopoulou L, Anderson JF, Fikrig E, Flavell RA. Toll-like receptor 3 mediates West Nile virus entry into the brain causing lethal encephalitis. Nat Med. 2004;10:1366-1373. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 819] [Cited by in F6Publishing: 822] [Article Influence: 41.1] [Reference Citation Analysis (0)] |
| 25. | Le Goffic R, Balloy V, Lagranderie M, Alexopoulou L, Escriou N, Flavell R, Chignard M, Si-Tahar M. Detrimental contribution of the Toll-like receptor (TLR)3 to influenza A virus-induced acute pneumonia. PLoS Pathog. 2006;2:e53. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 391] [Cited by in F6Publishing: 399] [Article Influence: 22.2] [Reference Citation Analysis (0)] |
| 26. | Kawai T, Sato S, Ishii KJ, Coban C, Hemmi H, Yamamoto M, Terai K, Matsuda M, Inoue J, Uematsu S. Interferon-alpha induction through Toll-like receptors involves a direct interaction of IRF7 with MyD88 and TRAF6. Nat Immunol. 2004;5:1061-1068. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 745] [Cited by in F6Publishing: 750] [Article Influence: 37.5] [Reference Citation Analysis (0)] |
| 27. | Honda K, Yanai H, Mizutani T, Negishi H, Shimada N, Suzuki N, Ohba Y, Takaoka A, Yeh WC, Taniguchi T. Role of a transductional-transcriptional processor complex involving MyD88 and IRF-7 in Toll-like receptor signaling. Proc Natl Acad Sci USA. 2004;101:15416-15421. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 381] [Cited by in F6Publishing: 376] [Article Influence: 18.8] [Reference Citation Analysis (0)] |
| 28. | Uematsu S, Sato S, Yamamoto M, Hirotani T, Kato H, Takeshita F, Matsuda M, Coban C, Ishii KJ, Kawai T. Interleukin-1 receptor-associated kinase-1 plays an essential role for Toll-like receptor (TLR)7- and TLR9-mediated interferon-{alpha} induction. J Exp Med. 2005;201:915-923. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 379] [Cited by in F6Publishing: 372] [Article Influence: 19.6] [Reference Citation Analysis (0)] |
| 29. | Yamamoto M, Sato S, Hemmi H, Hoshino K, Kaisho T, Sanjo H, Takeuchi O, Sugiyama M, Okabe M, Takeda K. Role of adaptor TRIF in the MyD88-independent toll-like receptor signaling pathway. Science. 2003;301:640-643. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 2377] [Cited by in F6Publishing: 2376] [Article Influence: 113.1] [Reference Citation Analysis (0)] |
| 30. | Edelmann KH, Richardson-Burns S, Alexopoulou L, Tyler KL, Flavell RA, Oldstone MB. Does Toll-like receptor 3 play a biological role in virus infections? Virology. 2004;322:231-238. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 275] [Cited by in F6Publishing: 269] [Article Influence: 13.5] [Reference Citation Analysis (0)] |
| 31. | Smith EJ, Marié I, Prakash A, García-Sastre A, Levy DE. IRF3 and IRF7 phosphorylation in virus-infected cells does not require double-stranded RNA-dependent protein kinase R or Ikappa B kinase but is blocked by Vaccinia virus E3L protein. J Biol Chem. 2001;276:8951-8957. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 190] [Cited by in F6Publishing: 198] [Article Influence: 8.6] [Reference Citation Analysis (0)] |
| 32. | Honda K, Sakaguchi S, Nakajima C, Watanabe A, Yanai H, Matsumoto M, Ohteki T, Kaisho T, Takaoka A, Akira S. Selective contribution of IFN-alpha/beta signaling to the maturation of dendritic cells induced by double-stranded RNA or viral infection. Proc Natl Acad Sci USA. 2003;100:10872-10877. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 309] [Cited by in F6Publishing: 297] [Article Influence: 14.1] [Reference Citation Analysis (0)] |
| 33. | Sun YW. RIG-I, a human homolog gene of RNA helicase is induced by Retinoic Acid during the differentiation of acute promyelocytic leukemia cell.Thesis Shangai second Medical Univ.1997.. . [Cited in This Article: ] |
| 34. | Cui XF, Imaizumi T, Yoshida H, Borden EC, Satoh K. Retinoic acid-inducible gene-I is induced by interferon-gamma and regulates the expression of interferon-gamma stimulated gene 15 in MCF-7 cells. Biochem Cell Biol. 2004;82:401-405. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 50] [Cited by in F6Publishing: 54] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 35. | Matikainen S, Sirén J, Tissari J, Veckman V, Pirhonen J, Severa M, Sun Q, Lin R, Meri S, Uzé G. Tumor necrosis factor alpha enhances influenza A virus-induced expression of antiviral cytokines by activating RIG-I gene expression. J Virol. 2006;80:3515-3522. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 104] [Cited by in F6Publishing: 116] [Article Influence: 6.4] [Reference Citation Analysis (0)] |
| 36. | Kang DC, Gopalkrishnan RV, Wu Q, Jankowsky E, Pyle AM, Fisher PB. mda-5: An interferon-inducible putative RNA helicase with double-stranded RNA-dependent ATPase activity and melanoma growth-suppressive properties. Proc Natl Acad Sci USA. 2002;99:637-642. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 494] [Cited by in F6Publishing: 492] [Article Influence: 22.4] [Reference Citation Analysis (0)] |
| 37. | Rothenfusser S, Goutagny N, DiPerna G, Gong M, Monks BG, Schoenemeyer A, Yamamoto M, Akira S, Fitzgerald KA. The RNA helicase Lgp2 inhibits TLR-independent sensing of viral replication by retinoic acid-inducible gene-I. J Immunol. 2005;175:5260-5268. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 438] [Cited by in F6Publishing: 454] [Article Influence: 23.9] [Reference Citation Analysis (0)] |
| 38. | Yoneyama M, Kikuchi M, Matsumoto K, Imaizumi T, Miyagishi M, Taira K, Foy E, Loo YM, Gale M, Akira S. Shared and unique functions of the DExD/H-box helicases RIG-I, MDA5, and LGP2 in antiviral innate immunity. J Immunol. 2005;175:2851-2858. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1264] [Cited by in F6Publishing: 1221] [Article Influence: 64.3] [Reference Citation Analysis (0)] |
| 39. | Kato H, Takeuchi O, Sato S, Yoneyama M, Yamamoto M, Matsui K, Uematsu S, Jung A, Kawai T, Ishii KJ. Differential roles of MDA5 and RIG-I helicases in the recognition of RNA viruses. Nature. 2006;441:101-105. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 2766] [Cited by in F6Publishing: 2803] [Article Influence: 155.7] [Reference Citation Analysis (0)] |
| 40. | Gitlin L, Barchet W, Gilfillan S, Cella M, Beutler B, Flavell RA, Diamond MS, Colonna M. Essential role of mda-5 in type I IFN responses to polyriboinosinic: polyribocytidylic acid and encephalomyocarditis picornavirus. Proc Natl Acad Sci USA. 2006;103:8459-8464. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 869] [Cited by in F6Publishing: 865] [Article Influence: 48.1] [Reference Citation Analysis (0)] |
| 41. | Hornung V, Ellegast J, Kim S, Brzózka K, Jung A, Kato H, Poeck H, Akira S, Conzelmann KK, Schlee M. 5'-Triphosphate RNA is the ligand for RIG-I. Science. 2006;314:994-997. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1879] [Cited by in F6Publishing: 1804] [Article Influence: 100.2] [Reference Citation Analysis (0)] |
| 42. | Pichlmair A, Schulz O, Tan CP, Näslund TI, Liljeström P, Weber F, Reis e Sousa C. RIG-I-mediated antiviral responses to single-stranded RNA bearing 5'-phosphates. Science. 2006;314:997-1001. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1735] [Cited by in F6Publishing: 1670] [Article Influence: 92.8] [Reference Citation Analysis (0)] |
| 43. | Kawai T, Takahashi K, Sato S, Coban C, Kumar H, Kato H, Ishii KJ, Takeuchi O, Akira S. IPS-1, an adaptor triggering RIG-I- and Mda5-mediated type I interferon induction. Nat Immunol. 2005;6:981-988. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 2018] [Cited by in F6Publishing: 1966] [Article Influence: 103.5] [Reference Citation Analysis (0)] |
| 44. | Xu LG, Wang YY, Han KJ, Li LY, Zhai Z, Shu HB. VISA is an adapter protein required for virus-triggered IFN-beta signaling. Mol Cell. 2005;19:727-740. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1529] [Cited by in F6Publishing: 1492] [Article Influence: 78.5] [Reference Citation Analysis (0)] |
| 45. | Seth RB, Sun L, Ea CK, Chen ZJ. Identification and characterization of MAVS, a mitochondrial antiviral signaling protein that activates NF-kappaB and IRF 3. Cell. 2005;122:669-682. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 2373] [Cited by in F6Publishing: 2496] [Article Influence: 131.4] [Reference Citation Analysis (0)] |
| 46. | Meylan E, Curran J, Hofmann K, Moradpour D, Binder M, Bartenschlager R, Tschopp J. Cardif is an adaptor protein in the RIG-I antiviral pathway and is targeted by hepatitis C virus. Nature. 2005;437:1167-1172. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1933] [Cited by in F6Publishing: 1854] [Article Influence: 97.6] [Reference Citation Analysis (0)] |
| 47. | Matsuda A, Suzuki Y, Honda G, Muramatsu S, Matsuzaki O, Nagano Y, Doi T, Shimotohno K, Harada T, Nishida E. Large-scale identification and characterization of human genes that activate NF-kappaB and MAPK signaling pathways. Oncogene. 2003;22:3307-3318. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 310] [Cited by in F6Publishing: 327] [Article Influence: 15.6] [Reference Citation Analysis (0)] |
| 48. | Saha SK, Pietras EM, He JQ, Kang JR, Liu SY, Oganesyan G, Shahangian A, Zarnegar B, Shiba TL, Wang Y. Regulation of antiviral responses by a direct and specific interaction between TRAF3 and Cardif. EMBO J. 2006;25:3257-3263. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 324] [Cited by in F6Publishing: 322] [Article Influence: 17.9] [Reference Citation Analysis (0)] |
| 49. | Lin R, Lacoste J, Nakhaei P, Sun Q, Yang L, Paz S, Wilkinson P, Julkunen I, Vitour D, Meurs E. Dissociation of a MAVS/IPS-1/VISA/Cardif-IKKepsilon molecular complex from the mitochondrial outer membrane by hepatitis C virus NS3-4A proteolytic cleavage. J Virol. 2006;80:6072-6083. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 177] [Cited by in F6Publishing: 183] [Article Influence: 10.2] [Reference Citation Analysis (0)] |
| 50. | Sun Q, Sun L, Liu HH, Chen X, Seth RB, Forman J, Chen ZJ. The specific and essential role of MAVS in antiviral innate immune responses. Immunity. 2006;24:633-642. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 464] [Cited by in F6Publishing: 464] [Article Influence: 25.8] [Reference Citation Analysis (0)] |
| 51. | Servant MJ, Grandvaux N, tenOever BR, Duguay D, Lin R, Hiscott J. Identification of the minimal phosphoacceptor site required for in vivo activation of interferon regulatory factor 3 in response to virus and double-stranded RNA. J Biol Chem. 2003;278:9441-9447. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 190] [Cited by in F6Publishing: 193] [Article Influence: 9.2] [Reference Citation Analysis (0)] |
| 52. | Peters RT, Liao SM, Maniatis T. IKKepsilon is part of a novel PMA-inducible IkappaB kinase complex. Mol Cell. 2000;5:513-522. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 268] [Cited by in F6Publishing: 278] [Article Influence: 11.6] [Reference Citation Analysis (0)] |
| 53. | Bonnard M, Mirtsos C, Suzuki S, Graham K, Huang J, Ng M, Itié A, Wakeham A, Shahinian A, Henzel WJ. Deficiency of T2K leads to apoptotic liver degeneration and impaired NF-kappaB-dependent gene transcription. EMBO J. 2000;19:4976-4985. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 302] [Cited by in F6Publishing: 294] [Article Influence: 12.3] [Reference Citation Analysis (0)] |
| 54. | Hemmi H, Takeuchi O, Sato S, Yamamoto M, Kaisho T, Sanjo H, Kawai T, Hoshino K, Takeda K, Akira S. The roles of two IkappaB kinase-related kinases in lipopolysaccharide and double stranded RNA signaling and viral infection. J Exp Med. 2004;199:1641-1650. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 471] [Cited by in F6Publishing: 443] [Article Influence: 22.2] [Reference Citation Analysis (0)] |
| 55. | Harris J, Olière S, Sharma S, Sun Q, Lin R, Hiscott J, Grandvaux N. Nuclear accumulation of cRel following C-terminal phosphorylation by TBK1/IKK epsilon. J Immunol. 2006;177:2527-2535. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 84] [Cited by in F6Publishing: 90] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 56. | Mattioli I, Geng H, Sebald A, Hodel M, Bucher C, Kracht M, Schmitz ML. Inducible phosphorylation of NF-kappa B p65 at serine 468 by T cell costimulation is mediated by IKK epsilon. J Biol Chem. 2006;281:6175-6183. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 93] [Cited by in F6Publishing: 100] [Article Influence: 5.6] [Reference Citation Analysis (0)] |
| 57. | Adli M, Baldwin AS. IKK-i/IKKepsilon controls constitutive, cancer cell-associated NF-kappaB activity via regulation of Ser-536 p65/RelA phosphorylation. J Biol Chem. 2006;281:26976-26984. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 116] [Cited by in F6Publishing: 125] [Article Influence: 6.9] [Reference Citation Analysis (0)] |
| 58. | Kravchenko VV, Mathison JC, Schwamborn K, Mercurio F, Ulevitch RJ. IKKi/IKKepsilon plays a key role in integrating signals induced by pro-inflammatory stimuli. J Biol Chem. 2003;278:26612-26619. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 100] [Cited by in F6Publishing: 104] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 59. | Wang N, Ahmed S, Haqqi TM. Genomic structure and functional characterization of the promoter region of human IkappaB kinase-related kinase IKKi/IKKvarepsilon gene. Gene. 2005;353:118-133. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 23] [Cited by in F6Publishing: 24] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 60. | Hoofnagle JH. Course and outcome of hepatitis C. Hepatology. 2002;36:S21-S29. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 387] [Cited by in F6Publishing: 355] [Article Influence: 16.1] [Reference Citation Analysis (0)] |
| 61. | Heim MH, Moradpour D, Blum HE. Expression of hepatitis C virus proteins inhibits signal transduction through the Jak-STAT pathway. J Virol. 1999;73:8469-8475. [PubMed] [Cited in This Article: ] |
| 62. | Larrea E, Aldabe R, Molano E, Fernandez-Rodriguez CM, Ametzazurra A, Civeira MP, Prieto J. Altered expression and activation of signal transducers and activators of transcription (STATs) in hepatitis C virus infection: in vivo and in vitro studies. Gut. 2006;55:1188-1196. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 55] [Cited by in F6Publishing: 63] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 63. | Melén K, Fagerlund R, Nyqvist M, Keskinen P, Julkunen I. Expression of hepatitis C virus core protein inhibits interferon-induced nuclear import of STATs. J Med Virol. 2004;73:536-547. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 66] [Cited by in F6Publishing: 78] [Article Influence: 3.9] [Reference Citation Analysis (0)] |
| 64. | Pavio N, Lai MM. The hepatitis C virus persistence: how to evade the immune system? J Biosci. 2003;28:287-304. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 63] [Cited by in F6Publishing: 69] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 65. | Foy E, Li K, Wang C, Sumpter R, Ikeda M, Lemon SM, Gale M. Regulation of interferon regulatory factor-3 by the hepatitis C virus serine protease. Science. 2003;300:1145-1148. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 624] [Cited by in F6Publishing: 649] [Article Influence: 30.9] [Reference Citation Analysis (0)] |
| 66. | De Francesco R, Tomei L, Altamura S, Summa V, Migliaccio G. Approaching a new era for hepatitis C virus therapy: inhibitors of the NS3-4A serine protease and the NS5B RNA-dependent RNA polymerase. Antiviral Res. 2003;58:1-16. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 149] [Cited by in F6Publishing: 148] [Article Influence: 7.0] [Reference Citation Analysis (0)] |
| 67. | Dumont S, Cheng W, Serebrov V, Beran RK, Tinoco I, Pyle AM, Bustamante C. RNA translocation and unwinding mechanism of HCV NS3 helicase and its coordination by ATP. Nature. 2006;439:105-108. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 293] [Cited by in F6Publishing: 277] [Article Influence: 15.4] [Reference Citation Analysis (0)] |
| 68. | Bartenschlager R. The NS3/4A proteinase of the hepatitis C virus: unravelling structure and function of an unusual enzyme and a prime target for antiviral therapy. J Viral Hepat. 1999;6:165-181. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 109] [Cited by in F6Publishing: 114] [Article Influence: 4.6] [Reference Citation Analysis (0)] |
| 69. | Foy E, Li K, Sumpter R, Loo YM, Johnson CL, Wang C, Fish PM, Yoneyama M, Fujita T, Lemon SM. Control of antiviral defenses through hepatitis C virus disruption of retinoic acid-inducible gene-I signaling. Proc Natl Acad Sci USA. 2005;102:2986-2991. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 437] [Cited by in F6Publishing: 422] [Article Influence: 22.2] [Reference Citation Analysis (0)] |
| 70. | Breiman A, Grandvaux N, Lin R, Ottone C, Akira S, Yoneyama M, Fujita T, Hiscott J, Meurs EF. Inhibition of RIG-I-dependent signaling to the interferon pathway during hepatitis C virus expression and restoration of signaling by IKKepsilon. J Virol. 2005;79:3969-3978. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 138] [Cited by in F6Publishing: 134] [Article Influence: 7.1] [Reference Citation Analysis (0)] |
| 71. | Li K, Foy E, Ferreon JC, Nakamura M, Ferreon AC, Ikeda M, Ray SC, Gale M, Lemon SM. Immune evasion by hepatitis C virus NS3/4A protease-mediated cleavage of the Toll-like receptor 3 adaptor protein TRIF. Proc Natl Acad Sci USA. 2005;102:2992-2997. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 847] [Cited by in F6Publishing: 869] [Article Influence: 45.7] [Reference Citation Analysis (0)] |
| 72. | Li XD, Sun L, Seth RB, Pineda G, Chen ZJ. Hepatitis C virus protease NS3/4A cleaves mitochondrial antiviral signaling protein off the mitochondria to evade innate immunity. Proc Natl Acad Sci USA. 2005;102:17717-17722. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 630] [Cited by in F6Publishing: 657] [Article Influence: 34.6] [Reference Citation Analysis (0)] |
| 73. | Ferreon JC, Ferreon AC, Li K, Lemon SM. Molecular determinants of TRIF proteolysis mediated by the hepatitis C virus NS3/4A protease. J Biol Chem. 2005;280:20483-20492. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 91] [Cited by in F6Publishing: 95] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 74. | Lamarre D, Anderson PC, Bailey M, Beaulieu P, Bolger G, Bonneau P, Bös M, Cameron DR, Cartier M, Cordingley MG. An NS3 protease inhibitor with antiviral effects in humans infected with hepatitis C virus. Nature. 2003;426:186-189. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 716] [Cited by in F6Publishing: 653] [Article Influence: 31.1] [Reference Citation Analysis (0)] |
| 75. | Lin K, Perni RB, Kwong AD, Lin C. VX-950, a novel hepatitis C virus (HCV) NS3-4A protease inhibitor, exhibits potent antiviral activities in HCv replicon cells. Antimicrob Agents Chemother. 2006;50:1813-1822. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 159] [Cited by in F6Publishing: 162] [Article Influence: 9.0] [Reference Citation Analysis (0)] |
| 76. | Bogen SL, Arasappan A, Bennett F, Chen K, Jao E, Liu YT, Lovey RG, Venkatraman S, Pan W, Parekh T. Discovery of SCH446211 (SCH6): a new ketoamide inhibitor of the HCV NS3 serine protease and HCV subgenomic RNA replication. J Med Chem. 2006;49:2750-2757. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 37] [Cited by in F6Publishing: 36] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 77. | Lindenbach BD, Evans MJ, Syder AJ, Wölk B, Tellinghuisen TL, Liu CC, Maruyama T, Hynes RO, Burton DR, McKeating JA. Complete replication of hepatitis C virus in cell culture. Science. 2005;309:623-626. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1843] [Cited by in F6Publishing: 1817] [Article Influence: 95.6] [Reference Citation Analysis (0)] |
| 78. | Wakita T, Pietschmann T, Kato T, Date T, Miyamoto M, Zhao Z, Murthy K, Habermann A, Kräusslich HG, Mizokami M. Production of infectious hepatitis C virus in tissue culture from a cloned viral genome. Nat Med. 2005;11:791-796. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 2241] [Cited by in F6Publishing: 2239] [Article Influence: 117.8] [Reference Citation Analysis (0)] |
| 79. | Zhong J, Gastaminza P, Cheng G, Kapadia S, Kato T, Burton DR, Wieland SF, Uprichard SL, Wakita T, Chisari FV. Robust hepatitis C virus infection in vitro. Proc Natl Acad Sci USA. 2005;102:9294-9299. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1427] [Cited by in F6Publishing: 1441] [Article Influence: 75.8] [Reference Citation Analysis (0)] |
| 80. | Yi M, Villanueva RA, Thomas DL, Wakita T, Lemon SM. Production of infectious genotype 1a hepatitis C virus (Hutchinson strain) in cultured human hepatoma cells. Proc Natl Acad Sci USA. 2006;103:2310-2315. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 306] [Cited by in F6Publishing: 327] [Article Influence: 18.2] [Reference Citation Analysis (0)] |
| 81. | Blight KJ, McKeating JA, Rice CM. Highly permissive cell lines for subgenomic and genomic hepatitis C virus RNA replication. J Virol. 2002;76:13001-13014. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 980] [Cited by in F6Publishing: 995] [Article Influence: 45.2] [Reference Citation Analysis (0)] |
| 82. | Sumpter R, Loo YM, Foy E, Li K, Yoneyama M, Fujita T, Lemon SM, Gale M. Regulating intracellular antiviral defense and permissiveness to hepatitis C virus RNA replication through a cellular RNA helicase, RIG-I. J Virol. 2005;79:2689-2699. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 736] [Cited by in F6Publishing: 701] [Article Influence: 36.9] [Reference Citation Analysis (0)] |
| 83. | Marques JT, Devosse T, Wang D, Zamanian-Daryoush M, Serbinowski P, Hartmann R, Fujita T, Behlke MA, Williams BR. A structural basis for discriminating between self and nonself double-stranded RNAs in mammalian cells. Nat Biotechnol. 2006;24:559-565. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 288] [Cited by in F6Publishing: 298] [Article Influence: 16.6] [Reference Citation Analysis (0)] |
| 84. | Loo YM, Owen DM, Li K, Erickson AK, Johnson CL, Fish PM, Carney DS, Wang T, Ishida H, Yoneyama M. Viral and therapeutic control of IFN-beta promoter stimulator 1 during hepatitis C virus infection. Proc Natl Acad Sci USA. 2006;103:6001-6006. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 316] [Cited by in F6Publishing: 318] [Article Influence: 17.7] [Reference Citation Analysis (0)] |
| 85. | Cheng G, Zhong J, Chisari FV. Inhibition of dsRNA-induced signaling in hepatitis C virus-infected cells by NS3 protease-dependent and -independent mechanisms. Proc Natl Acad Sci USA. 2006;103:8499-8504. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 99] [Cited by in F6Publishing: 104] [Article Influence: 5.8] [Reference Citation Analysis (0)] |
| 86. | Vilasco M, Larrea E, Vitour D, Dabo S, Breiman A, Regnault B, Riezu JI, Eid P, Prieto J, Meurs EF. The protein kinase IKKepsilon can inhibit HCV expression independently of IFN and its own expression is downregulated in HCV-infected livers. Hepatology. 2006;44:1635-1647. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 15] [Cited by in F6Publishing: 17] [Article Influence: 0.9] [Reference Citation Analysis (0)] |
| 87. | Boya P, Larrea E, Sola I, Majano PL, Jiménez C, Civeira MP, Prieto J. Nuclear factor-kappa B in the liver of patients with chronic hepatitis C: decreased RelA expression is associated with enhanced fibrosis progression. Hepatology. 2001;34:1041-1048. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 38] [Cited by in F6Publishing: 43] [Article Influence: 1.9] [Reference Citation Analysis (0)] |