Published online Dec 28, 2006. doi: 10.3748/wjg.v12.i48.7805
Revised: August 8, 2006
Accepted: August 29, 2006
Published online: December 28, 2006
AIM: To assess the level of undiagnosed coeliac disease (CD) in relatives of patients affected by the condition.
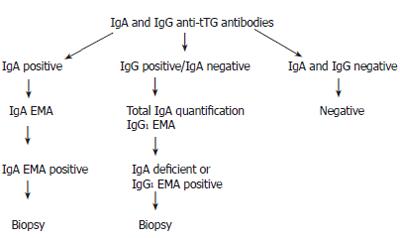
METHODS: We collected blood from 914 relatives of probands. We screened these individuals by ELISA for IgA and IgG tTG antibodies, confirming any positive IgA tTG results with an IgA EMA and looked for evidence of IgA deficiency in those who were IgG tTG positive alone, and performed IgG1 EMA in these individuals. We undertook HLA typing where positive screening was found, and this confirmed a strong prevalence of HLA-DQ2 in the coeliac population. Follow-up small intestinal biopsy was undertaken in cases with positive serological screening, wherever possible.
RESULTS: Use of this serological screening algorithm revealed a prevalence of undiagnosed CD in 5%-6% of first degree relatives of probands.
CONCLUSION: Our data suggests that first degree relatives of individuals with CD should be screened for this condition.
- Citation: Fraser JS, King AL, Ellis HJ, Moodie SJ, Bjarnason I, Swift J, Ciclitira PJ. An algorithm for family screening for coeliac disease. World J Gastroenterol 2006; 12(48): 7805-7809
- URL: https://www.wjgnet.com/1007-9327/full/v12/i48/7805.htm
- DOI: https://dx.doi.org/10.3748/wjg.v12.i48.7805
Coeliac disease (CD) is a disorder in which genetically predisposed individuals develop a small intestinal enteropathy on exposure to dietary gluten. The small bowel abnormalities are reversed on withdrawal of gluten from the diet. Recent population studies and serological testing of at-risk groups reveal a much higher prevalence of CD than previous studies. Whereas the previous prevalence was thought to be in the order of 1 in 1500 in Europeans, it is now thought to be in the order of 1 in 100 to 1 in 250. In the largest population screening study[1], 17 000 Italian school children, aged 6-15 years, were screened using a stepwise protocol with anti-gliadin antibodies (AGA), anti-endomysial antibodies (EMA), and finally duodenal biopsies in those who screened positive for these two serological tests. A prevalence rate of 1 in 184 was found. A Swedish study involving healthy blood donors found a prevalence of 1 in 256, confirmed by small bowel biopsy[2], and an American study[3] using EMA in blood donors found a rate of 1 in 250, although this was not confirmed by biopsy. In Ireland, the rates are even higher, with a reported prevalence rate of 1 in 122 determined in a screening study [4].
The incidence and prevalence of CD are therefore similar in populations with a similar genetic background. CD is thought to occur rarely in people from an Afro-Caribbean background, though there are reports of the condition in Asians from the Indian sub-continent[5]. CD is a familial condition, and the main risk factor for development of the condition is the presence of HLA DQ2 or DQ8. Most Northern European patients express the DQ2 heterodimer HLA-DQA1*0501 and DQB1*0201. Those who do not express this heterodimer, most commonly, have the HLA-DR4, DQ8 haplotype. In Italian and Tunisian patients there is also a significant association with DR53 heterodimers[6,7]. Further susceptibility genes, such as the CTLA-4 gene on chromosome 2q33[8], are thought to reside both inside and outside the HLA region and are currently being evaluated, although the disease is not expressed in the absence of the HLA genes. CD is thought to occur in 10%-15% of first degree relatives of probands[9], with 40% of HLA identical siblings being affected, and a concordance of 75% in monozygotic twins[10]. In some countries, such as Italy, first-degree relatives of patients with CD are screened routinely.
There are various serological screening tests available, which have different sensitivity and specificities. Circulating antibodies to gliadin were previously used for screening, but have largely been superseded due to their low specificity. Anti-endomysial antibodies (EMA) of the IgA class are considered highly specific markers of coeliac disease. Using human umbilical cord (HUC), the reported sensitivity is 90%, with a specificity of 99% in adults with untreated coeliac disease (Table 1)[11]. However, this test is labour intensive and somewhat subjective, relying on the interpretation of a staining pattern on connective tissue, using a fluorescent microscope. The discovery of tissue transglutaminase (tTG) as the antigen for EMA[12], allowed the development of a simple ELISA to detect this antibody, the sensitivity and specificity of IgA tTG test are reported to be 94.5% and 93.7% respectively[13]. However, there are pitfalls in serological screening for CD. Selective IgA deficiency occurs in 2.6% of patients with CD[14], which is a 10-15 fold increase in prevalence of IgA deficiency over that in the general population. Testing for IgA antibodies only would cause false negative results and missed diagnoses. Additionally, a new sub-group of CD patients has recently been described, who develop only IgG class antibodies (specifically IgG1) to endomysium, in the absence of IgA class antibodies, and with normal quantities of total serum IgA[15].
| IgA-AGA | IgG-AGA | IgA-EMA | |
| Sensitivity | 83 | 86 | 90 |
| Specificity | 82 | 76 | 99 |
We wished to establish an accurate screening protocol to assess the prevalence of undiagnosed coeliac disease in relatives of probands.
We collected details of families where either one or more individuals were affected by CD. The families were identified either in the Gastroenterology Outpatient Clinic at St Thomas’ Hospital, or by consultant colleagues at other hospitals. A further recruitment drive involved a short article and request for volunteers printed in the ‘Crossed Grain’ magazine, published by Coeliac UK for its members. In this way we were able to recruit a total of 151 families into the study. Of these 73 families had only one affected member and were referred to as single affected families, 78 families had more than one affected family member (range 2-7) and were referred to as multiply affected families. Full ethical approval was obtained from the Local Research Ethics Committee of St Thomas’ Hospital (Ref. No.EC00/233). We recorded family relationships and collected blood for serology from as many relatives as were willing to consent. Serum was stored at -20°C. DNA was extracted from heparinized blood using the Nucleon BACC3 kit and stored at -20°C.
In order to set up the parameters for serological testing we first took a group of normal controls. These comprised laboratory staff and their relatives who were healthy and symptom-free. The age range was 24-60 years. Volunteers were questioned about the health of other family members. Those with any family history of gastrointestinal problems, diabetes and auto-immune thyroid disease were excluded from the study.
Two individuals with biopsy-confirmed CD and high titre for IgA and IgG-tTG antibody respectively were selected as positive controls. Sera of these individuals were made at 1:100 dilution, aliquoted and sterilised by gamma irradiation. ELISA was performed according to the method of Sulkanen[13]. Microtitre plates were coated with guinea pig liver tTG (Sigma T 5398), 1 μg per well in 100 μL of 0.05 mol/L tris buffered saline, with 5 mmol/L calcium chloride. The plates were left over night at 4°C, then washed three times with 0.05 mol/L TBS, 0.01 mol/L EDTA and 0.1% Tween 20 (TTBS). Test sera were diluted to 1:100 in TTBS, 100 μL of the test and positive control sera was added in duplicate to two plates. The plates were covered and incubated for 1 h at room temperature, then washed three times. Peroxidase-conjugated rabbit anti-human IgA (Dako P-0216) or anti-human IgG (Dako P-0214) was diluted to 1:2000 in TTBS, and added to the plates at 100 μL per well. This was incubated for 1 h at room temperature, and washed three times. The reaction was developed by adding OPD as substrate (Dako S2045), prepared according to the manufacturer's instructions. One hundred microlitres was placed in each well, and the plates were incubated in the dark at room temperature for 30 min. The plates were read on a Titertek Multiskan MCC/340 ELISA plate reader at 450 nm. The end point was reached when the IgA and IgG positive controls reached optical density of 1.2-1.3. The cut off value for a positive result was established as 0.3 for IgA and 0.325 for IgG. These values were calculated from the mean plus 2 standard deviations for our normal population. Individuals whose IgA-tTG was above the cut-off value were further investigated by IgA-EMA. Those without IgA-tTG antibodies, but with IgG-tTG antibodies went on to have total IgA quantification and IgG1-EMA. Those with negative results for both antibodies were considered negative for screening, and no further action was taken.
The method used was described by Ladinser et al[16]. Human umbilical cord (HUC) was cut into 5-μm cryostat sections on 4 well coated slides. Each section was blocked with 100 μL of 1% BSA in PBS for 30 min. Test sera were diluted to a concentration of 1:5, and added to each well. Each experiment also contained a positive and a negative control. The sections were incubated for 30 min, and washed twice in a PBS bath. FITC-conjugated rabbit anti-human IgA (specific for alpha chains) immunofluorescent antibody (DAKO F0204) or FITC-conjugated mouse anti-human IgG1 (Sigma Monoclonal anti-human IgG1 clone 8c/6-39, product number F0767) was diluted to 1:40 using PBS. Fifty microlitres was added to each well and the sections were incubated at room temperature for 30 min in a humid chamber, and immersed in a PBS bath as before. Fluorescent mounting medium (DAKO, S3023) was added and the sections were examined immediately under a fluorescent microscope. The test was considered positive if the antibody stained the endomysium of umbilical arteries in a defined reticular pattern at a dilution of 1:5.
This was performed by a competitive ELISA assay. Microtitre plates were coated with 100 μL of 2 μL/mL whole molecule human IgA (Harlan Sera-Lab PP-17-01) in PBS, and left overnight at 4°C. The plates were then washed three times in PBS with 0.05% Tween 20, blocked with 100 μL of 1% BSA, incubated at 37°C for 1 h, and then drained.
The serum samples were diluted in peroxidase-conjugated rabbit anti-human IgA at a concentration of 1:4000. A standard curve was produced by diluting known amounts of human IgA in peroxidase-conjugated rabbit anti-human IgA. Dilutions and sera were pre-incubated for 30 min at room temperature, then added to the plates and incubated for a further 30 min. The plates were washed three times in PBS/Tween, the reaction was developed by adding OPD as a substrate (Dako S2045), prepared according to the manufacturer’s instructions. One hundred microlitres was placed in each well. The plates were incubated in the dark at room temperature for 30 min and read on a Titertek Multiskan MCC/340 ELISA plate reader at 450 nm. The concentrations of IgA in the serum were calculated from the standard curve.
DNA was extracted from whole blood by the following protocol, using the Nucleon BACC kit (SL-8512). In brief, primer sequences were chosen to detect the presence of HLA-DR3, -DR7, -DR5 and -DR4, which are the most common haplotypes in patients with coeliac disease, being present in > 98% of European individuals with the condition. The products were loaded onto 1% agarose gel containing ethidium bromide. The gels were run for 22 min at 300 V, and visualised under UV light.
Nine hundred and thirteen serum samples were tested for anti-tTG antibodies, which were from first, second and third degree relatives, as well as some individuals who were not blood relatives of coeliac disease probands, including individuals with coeliac disease such as husbands or wives and their relatives.
IgA-tTG antibodies were found to be present in 60 individuals. When these were followed up with IgA-EMA, 36 were found to have IgA-EMA antibodies, and 24 individuals were deemed to be false positives. The individuals who were positive for IgA-EMA had IgA-tTG levels of 0.31 to 2.34, whilst those who were negative for IgA-EMA had IgA-tTG levels of 0.31-1.1. IgG-tTG antibodies in the absence of IgA-tTG antibodies were found in 194 individuals. Samples from all of these individuals were tested for IgG1-EMA. Of these, only 3 were found to be positive for IgG1 EMA. In total, 194 individuals had IgA quantification. Of these, only 2 out of the 3 IgG1 positive individuals were IgA deficient (Table 2).
| Type of family | Coeliacs | Relatives | Positive | 1st degree | 2nd degree | Not |
| (n) | (n) | screenings | relatives | relatives | related | |
| Singly affected family | 73 | 223 | 11 | 11 | 0 | 0 |
| Multiply affected family | 232 | 691 | 28 | 22 | 2 | 4 |
Of all the relatives in the single affected families, those newly diagnosed with coeliac disease are shown in Table 3. Five point four seven percent of first-degree relatives were found to have positive anti-endomysial antibodies, no second-degree relatives were found to have positive anti-endomysial antibodies. These data were analysed to give a percentage factor of those affected in each category of relative. There were not many children of individuals with CD in this group, as the proband was a child in the majority of volunteer families.
| Relationship | Numbertested (n) | Numberaffected (n) | Percentageaffected (%) |
| Mother | 73 | 5 | 6.85 |
| Father | 73 | 4 | 5.48 |
| Sibling | 37 | 2 | 5.41 |
| Child | 18 | 0 | 0 |
| Uncle/Aunt | 5 | 0 | 0 |
| Grandparent | 6 | 0 | 0 |
| Grandchild | 2 | 0 | 0 |
| Nephew/Niece | 3 | 0 | 0 |
| Husband/Wife | 6 | 0 | 0 |
Of all the relatives in the multiply affected families, those newly diagnosed with coeliac disease are shown in Table 4. Five point forty-one percent of first degree relatives were found to have anti-endomysial antibodies, and 1.62% of second degree relatives we found to have positive coeliac antibodies. These data were analysed to give a percentage factor of those affected in each category of relative.
| Relationship | Numbertested (n) | Numberaffected (n) | Percentageaffected (%) |
| Mother | 51 | 3 | 5.88 |
| Father | 31 | 3 | 9.68 |
| Sibling | 165 | 9 | 5.4 |
| Child | 137 | 7 | 5.11 |
| Uncle/Aunt | 6 | 1 | 16.67 |
| Grandparent | 11 | 0 | 0 |
| Grandchild | 39 | 0 | 0 |
| Nephew/Niece | 67 | 1 | 1.49 |
| 3rd degree or more | 24 | 0 | 0 |
| Husband/Wife | 159 | 4 | 2.52 |
Four (2.52%) of the 159 individuals who were related only by marriage to the person with coeliac disease were found to have positive coeliac antibodies. This rate was significantly higher than would be expected for the general population (1%).
The 39 individuals with positive anti-endomysial antibodies were further investigated by HLA-typing. Twenty-nine of these were successfully typed. Reasons for failure in the other 10 individuals included: inability to locate EDTA blood for extraction, poor DNA extraction and poor PCR or uninterpretable gels. Our HLA-typing revealed the same distribution of HLA-types, as would be expected in a population of patients with coeliac disease. The HLA types are shown in Table 5.
| HLA-type | DR3/DRx | DR3/DR7 | DR5/DR7 | DR4 | DR3/DR4 | DR4/DR7 |
| Number | 25 | 4 | 0 | 0 | 0 | 0 |
| Percentage | 86.2% | 13.8% | 0 | 0 | 0 | 0 |
We attempted to contact all those individuals with a positive screening result, to arrange a small intestinal biopsy. Thirty-five underwent a small intestinal biopsy. Of these, 32 were positive, having increased intra-epithelial lymphocytes with partial or sub-total villous atrophy. Three biopsies were reported as normal.
We used a two-tier screening system for coeliac disease. The initial anti-tTG ELISA test was used as a highly sensitive, cheap and simple initial screening test, rather than as a specific diagnostic test. The limits for a positive result were deliberately set low in order to avoid missing any cases, but this did have a major impact on the specificity, hence the number of positives were subsequently found to be EMA negative. While we would not propose this two-tier method for use in a diagnostic laboratory, we found it useful for rapid large scale screening, avoiding EMA testing on a great number of samples.
Our prevalence rates in relatives of probands with coeliac disease were significantly lower than those previously estimated by other groups (10%-15% for first degree relatives). This is perhaps surprising, since our families were recruited through voluntary self-referral. Thus, one may have expected increased rates, as suspicious symptoms may have encouraged some families to be volunteers. However, one would expect these lower rates in the multiply affected families since by definition, relatives of probands have already been diagnosed, and our figures only indicate the existence of undiagnosed cases. We used guinea-pig liver tissue transglutaminase as our detection antigen, on a cost basis. However, human recombinant tTg is now available, and might have given a higher rate of positive detection.
In the two IgA deficient individuals, one had a normal duodenal biopsy, while the other had an increase in intra-epithelial lymphocytes (IEL) only. The patient with increased IELs was investigated for ataxia when she joined our screening study. After having a gluten free diet (GFD) for 24 mo, her symptoms improved slightly.
It could be argued that volunteers in the study may be more likely to have been symptomatic, although we have no evidence for this. Our study raises the question whether we should screen first-degree relatives of patients with CD, since they carry a high-risk of being similarly affected. The screening method we proposed is sensitive, specific and non-invasive. The general well being of individuals with sub-clinical coeliac disease appears to improve on a GFD. It has been shown that there is a long-term health benefit to these people if a GFD is instituted with a reduction in the otherwise significantly increased mortality.
In conclusion, we propose the algorithm shown in Figure 1 for screening family members for coeliac disease, as we believe it is important that these individuals should be picked up, diagnosed and offered appropriate treatment.
The authors thank the following for financial support: Coeliac UK and Action Research (JSF), The German Federal Ministry of Education and Research (HJE) and the European Union (SJM).
S- Editor Wang GP L- Editor Wang XL E- Editor Bai SH
| 1. | Catassi C, Fabiani E, Rätsch IM, Coppa GV, Giorgi PL, Pierdomenico R, Alessandrini S, Iwanejko G, Domenici R, Mei E. The coeliac iceberg in Italy. A multicentre antigliadin antibodies screening for coeliac disease in school-age subjects. Acta Paediatr Suppl. 1996;412:29-35. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 245] [Cited by in F6Publishing: 240] [Article Influence: 8.6] [Reference Citation Analysis (0)] |
| 2. | Grodzinsky E, Franzen L, Hed J, Ström M. High prevalence of celiac disease in healthy adults revealed by antigliadin antibodies. Ann Allergy. 1992;69:66-70. [PubMed] [Cited in This Article: ] |
| 3. | Fasano A. Where have all the American celiacs gone. Acta Paediatr Suppl. 1996;412:20-24. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 49] [Cited by in F6Publishing: 53] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 4. | Johnston SD, Watson RG, McMillan SA, Sloan J, Love AH. Coeliac disease detected by screening is not silent--simply unrecognized. QJM. 1998;91:853-860. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 87] [Cited by in F6Publishing: 86] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 5. | Butterworth JR, Iqbal TH, Cooper BT. Coeliac disease in South Asians resident in Britain: comparison with white Caucasian coeliac patients. Eur J Gastroenterol Hepatol. 2005;17:541-545. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 15] [Cited by in F6Publishing: 17] [Article Influence: 0.9] [Reference Citation Analysis (0)] |
| 6. | King AL, Ciclitira PJ. Celiac disease: strongly heritable, oligogenic, but genetically complex. Mol Genet Metab. 2000;71:70-75. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 26] [Cited by in F6Publishing: 26] [Article Influence: 1.1] [Reference Citation Analysis (0)] |
| 7. | Sollid LM. Molecular basis of celiac disease. Annu Rev Immunol. 2000;18:53-81. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 465] [Cited by in F6Publishing: 443] [Article Influence: 18.5] [Reference Citation Analysis (0)] |
| 8. | King AL, Moodie SJ, Fraser JS, Curtis D, Reid E, Dearlove AM, Ellis HJ, Ciclitira PJ. CTLA-4/CD28 gene region is associated with genetic susceptibility to coeliac disease in UK families. J Med Genet. 2002;39:51-54. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 32] [Cited by in F6Publishing: 33] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 9. | Marsh MN. Gluten, major histocompatibility complex, and the small intestine. A molecular and immunobiologic approach to the spectrum of gluten sensitivity ('celiac sprue'). Gastroenterology. 1992;102:330-354. [PubMed] [Cited in This Article: ] |
| 10. | Greco L, Romino R, Coto I, Di Cosmo N, Percopo S, Maglio M, Paparo F, Gasperi V, Limongelli MG, Cotichini R. The first large population based twin study of coeliac disease. Gut. 2002;50:624-628. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 287] [Cited by in F6Publishing: 302] [Article Influence: 13.7] [Reference Citation Analysis (0)] |
| 11. | Stern M. Comparative evaluation of serologic tests for celiac disease: a European initiative toward standardization. J Pediatr Gastroenterol Nutr. 2000;31:513-519. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 100] [Cited by in F6Publishing: 108] [Article Influence: 4.5] [Reference Citation Analysis (0)] |
| 12. | Dieterich W, Ehnis T, Bauer M, Donner P, Volta U, Riecken EO, Schuppan D. Identification of tissue transglutaminase as the autoantigen of celiac disease. Nat Med. 1997;3:797-801. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1371] [Cited by in F6Publishing: 1299] [Article Influence: 48.1] [Reference Citation Analysis (0)] |
| 13. | Sulkanen S, Halttunen T, Laurila K, Kolho KL, Korponay-Szabó IR, Sarnesto A, Savilahti E, Collin P, Mäki M. Tissue transglutaminase autoantibody enzyme-linked immunosorbent assay in detecting celiac disease. Gastroenterology. 1998;115:1322-1328. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 392] [Cited by in F6Publishing: 373] [Article Influence: 14.3] [Reference Citation Analysis (0)] |
| 14. | Cataldo F, Marino V, Ventura A, Bottaro G, Corazza GR. Prevalence and clinical features of selective immunoglobulin A deficiency in coeliac disease: an Italian multicentre study. Italian Society of Paediatric Gastroenterology and Hepatology (SIGEP) and "Club del Tenue" Working Groups on Coeliac Disease. Gut. 1998;42:362-365. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 208] [Cited by in F6Publishing: 227] [Article Influence: 8.7] [Reference Citation Analysis (0)] |
| 15. | Picarelli A, di Tola M, Sabbatella L, Mastracchio A, Trecca A, Gabrielli F, di Cello T, Anania MC, Torsoli A. Identification of a new coeliac disease subgroup: antiendomysial and anti-transglutaminase antibodies of IgG class in the absence of selective IgA deficiency. J Intern Med. 2001;249:181-188. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 42] [Cited by in F6Publishing: 44] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 16. | Ladinser B, Rossipal E, Pittschieler K. Endomysium antibodies in coeliac disease: an improved method. Gut. 1994;35:776-778. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 186] [Cited by in F6Publishing: 192] [Article Influence: 6.4] [Reference Citation Analysis (0)] |