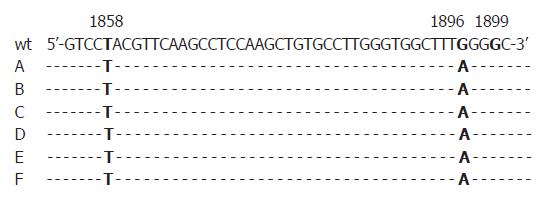Published online Apr 14, 2006. doi: 10.3748/wjg.v12.i14.2235
Revised: October 1, 2005
Accepted: November 18, 2005
Published online: April 14, 2006
AIM: To find out whether there is a significant difference in the prevalence of the precore stop codon mutation between HBeAg positive and anti-HBe positive children.
METHODS: We investigated a large pediatric population of 155 European children (mean age 10.9 years) with chronic hepatitis B by PCR and direct sequencing. Ninety were HBeAg positive and 65 had seroconversion to anti-HBe. Additionally genotyping was performed.
RESULTS: Seventy-four (48%) of the sequenced HBV strains were attributed to genotype D and 81 (52%) to genotype A. In the group of 90 HBeAg positive patients, 2 (2.2%) 1896-G-to-A transitions leading to precore stop codon mutation were found, and in the group of 65 anti-HBe positive children, 5 (7.7%) were identified harbouring HBeAg-minus mutants. The difference was not statistically significant (P= 0 .13).
CONCLUSIONS: HBeAg minus variants as predominant viral HB strains play a minor role in the course of chronic hepatitis B in European children.
- Citation: Wintermeyer P, Gerner P, Gehring S, Karimi A, Wirth S. Prevalence of hepatitis B virus precore stop codon mutations in chronically infected children. World J Gastroenterol 2006; 12(14): 2235-2238
- URL: https://www.wjgnet.com/1007-9327/full/v12/i14/2235.htm
- DOI: https://dx.doi.org/10.3748/wjg.v12.i14.2235
Chronic hepatitis B (HB)-infection is a serious health problem worldwide and the leading cause of liver cirrhosis and hepatocellular carcinoma (HCC). In Germany 0.3%-0.5% of the population are HBsAg carriers and it is estimated that approximately 5% of them are children. Mutations preventing the expression of HBeAg, so-called precore stop codon mutations, have been reported to aggravate liver disease and to cause fulminant hepatitis in children as well as in adults[1-3]. Currently it is not yet clear whether these mutations influence the response to antiviral treatment. However, a recent study has shown a better outcome after interferon-alpha treatment for chronic hepatitis in individuals without precore mutant strains before treatment[4]. The most common stop-mutation is the G1896A-substitution in the precore region of HB-virus. A guanine (G) to adenine (A) mutation of the HBV precore gene at nucleotide 1896 (numbered from the EcoRI site) leads to a conversion of codon 28 from TGG (tryptophan) to TAG, which is a stop codon, thereby rendering HBV incapable of producing HBeAg[5]. In adults the prevalence of this mutation ranges between 20%-95%, whereas it is more common in HBeAg-negative than in HBeAg-positive patients[6]. Results of a cross-sectional multicenter study of adults with chronic HBV infection in the United States show an association of the presence of precore variants with higher serum HBV DNA levels in HBeAg-negative but not in HBeAg-positive patients[7]. The selection of precore variants is dependent on HBV genotype. It is most common in patients with genotype D and rare in patients with genotype A[8]. This phenomenon is related to base pairing in the stem-loop structure of the pregenome encapsidation sequence[9,10].
Fukuda et al[11] have postulated that hepatitis B virus exists mainly as a quasi species. On account of this, a correlation of nucleotide sequences with clinical and serological findings has to be made with caution. Available data about prevalence and significance of precore mutations in childhood are limited. A study of children with chronic HBV infection (n = 60) has shown the 1896-G-A-transition in 93% as mixed infection, with a similar prevalence of mixed viral populations in responders and non-responders to interferon treatment[12]. In the course of chronic HBV infection in children the frequency of precore mutant is increased after seroconversion from HBeAg to anti-HBe. It has been postulated that the higher the aminotransferase levels are, the higher and the earlier the mutant emerges[13].
The aim of this study was to evaluate the prevalence of precore mutations in a large cohort of European children with chronic hepatitis B.
A cross sectional testing of sera from 155 randomly assigned HBsAg positive chronic hepatitis B virus carriers was performed. The median age of the children at the time of blood sampling was 10.9 years. Ninety were HBeAg positive and 65 had seroconversion to anti-HBe 1 - 3 years before blood sampling. Fifty-four (35%) were female, 101 (65%) male; 108 (70%) of Caucasian, 42 (27%) of Mediterranean and 5 (3%) of Southeast Asian origin. ALT was elevated in 66 (73%) of HBeAg positive individuals (mean: 1100 nkat/L) and in 18 (28%) of anti-HBe positive individuals (mean: 367 nkat/L; normal <417 nkat/L).
DNA was extracted from sera with the QiaAmp blood kit (Qiagen, Chatsworth, CA, USA) and eluted with 50 µL distilled water according to the manufacturer’s recommendations. The precore region of the HBV genome was amplified and re-amplified with a proof reading of expand-polymerase (Expand High Fidelity PCR System) by nested PCR. The following primers were used for amplification (nucleotide positions are according to the unique EcoRI site[14]: Sense P1: 5’-TGTCAACGACCGACCTTGAG-3’ (nt 1683-1702), anti-sense P2: 5’-CAATGCTCAGGAGACTCTAAGGC-3’ (nt 2045-2023); nested PCR: sense P3: 5’-GAGGAGTTGGGGTAGGACATT-3’ (nt 1736-1756), anti-sense P4: 5’-TAGCTCCAAATTCTTTATA-3’ (nt 1936-1918). PCR was performed in a 50-µL mixture with 20 pmol of each primer (Roth, Germany), 100 mmol/L of each dNTP, and 2.5 units of a Taq-Tgo polymerase mixture (Expand High Fidelity PCR System, Roche, Germany) diluted in 10 × expand polymerase buffer (Expand High Fidelity PCR System, Roche, Germany) in a DNA thermal cycler (Eppendorf Mastercycler personal). The amplified products were visualized by 20 g/L agarose electrophoresis and ethidium bromide staining.
The amplified PCR products were purified by QIA Quick PCR-purification kit (Quiagen, Chatsworth, CA) according to the manufacturer’s recommendations and precipitated with isopropanol to remove residual dNTPs and primers and re-suspended in a final volume of 10 μL distilled water. Nucleotide sequences of the PCR products were determined using a dye terminator cycle sequencing kit (big dye vs 3.1) in an automated sequencer (ABI PRISM, Foster City, CA, USA). The sequencing primers were the same as those used for DNA amplification.
The HBV-DNA sequences were assigned to the appropriate genotype based on the restriction fragment length polymorphism (RFLP) created by Ava2 and Dpn2 action on an amplified segment of the pres-S-region according to Lindh et al[15]. PCR of certain serum samples was performed under conditions as previously described with the following primers: Sense P1: 5’-CGAGGCAGGTCCCCTAGAAGAAGAA-3’ (nt 2356-2380), anti-sense P2: 5’-GTCCTAGGAATCCTGAA-3’ (nt 187-171); nested PCR: sense P3: 5’-TCACCATATTCTTGGGAACAAGA-3’ (nt 2819-2841), anti-sense P4: 5’-TTCCTGAACTGGAGCCACCA-3’ (nt 82-63). The PCR products were incubated with restriction enzymes Ava II and Dpn II (New England Biolabs, Inc., USA) for 3 h at 37 °C in a 15 µL reaction sample according to the manufacturer’s recommendations. The products were visualized by 2 g/L agarose electrophoresis and ethidium bromide staining.
Fisher’s exact test was used for statistical analysis P < 0.005 was taken as significant.
A total of 155 patients were enrolled. Seventy-four (48%) of the sequenced HBV strains were attributed to genotype D and 81 (52%) to genotype A. In the group of 90 HBeAg positive patients, 2 (2.2%) 1896-G-to-A transitions leading to precore stop codon were found, and in the group of 65 anti-HBe positive children, 5 (7.7%) were identified harbouring HBeAg-minus mutants. The difference was not statistically significant (P = 0 .13).
First, all sera were randomly selected for analysis without association with patients’ identities. We were then able to additionally investigate four HBeAg positive sera of the five patients who showed precore-mutations in anti-HBe positive status of the disease in order to find out if mutations already emerged in the HBeAg positive status. One of them was also positive for the G1896A-substitution in the HBeAg positive phase. All mutations identified were found in HBV DNA sequences of genotype D (Figures 1, 2).
The ALT levels in the two HBeAg positive sera with precore-mutation were 883 and 2983 nkat/L, reflecting a considerable elevation in one patient compared to the wild type sera of HBeAg positive status (mean: 1100 nkat/L). In the five anti-HBe positive individuals with the G1896A-substitution, the ALT levels were mildly elevated (mean: 467 nkat/L) compared to the group of 60 patients with wild type virus infection (mean: 367 nkat/L).
In contrast to previous studies with adults in particular, which reported HBeAg stop codon mutants in 20-95% of investigated patients (6, 16), we found a very low prevalence in children, both in the HBeAg (2.2%) and anti-HBe positive (8%) phase of the disease. However, our results demonstrated consistently that precore variants were more common in anti-HBe positive children, but the difference did not reach statistical significance. We were not able to show that mutations in anti-HBe positive sera have necessarily emerged already in the HBeAg positive phase of the disease. Thus, our data suggest that there is at least no strong evidence for the hypothesis that the mutant is selected by host immune pressure[13].
Due to the fact that the occurrence of the G1896A mutation is restricted to HBV genotypes with T at nucleotide position 1858 (genotype D)[17], it was not surprising that no mutations were identified in the group of genotype A virus strains. It has to be conceded that the existence of viral quasi-species including small amounts of precore stop mutations could not be excluded in our survey due to the method of direct sequencing. However, considering the high proportion of genotype D in our survey (48%), the postulation of a high prevalence of the G1896A precore mutation in these HBV strains[16] was not confirmed in chronically infected children.
It was reported that the precore mutation is common in Mediterranean and Asian populations, and two studies have shown a high prevalence of 16%-40% of the HBeAg minus variants in German adult populations as well[18,19]. It seems obvious that the age of the patients is more important than their ethnic origin.
Due to the elevated levels of transaminases in HBeAg positive sera, precore mutations may influence the inflammatory activity of the disease. But indeed, evidence is limited due to the small number of patients. In contrast to former studies, wherein a difference in clinical courses between genotypes B and C was described[20], this seems to be different for the most prevalent genotypes A and D in Europe, because they were equally distributed in our patients.
In summary, HBeAg minus variants as predominant viral HB strains play a minor role in the course of chronic disease in European children. Our results confirm the recent study of Söderström and colleagues who postulated that the most influential factors of HB-infection in childhood are epidemiologic parameters and the route of transmission[21].
S- Editor Pan BR L- Editor Wang XL E- Editor Bi L
| 1. | Papatheodoridis GV, Hadziyannis SJ. Diagnosis and management of pre-core mutant chronic hepatitis B. J Viral Hepat. 2001;8:311-321. [PubMed] [DOI] [Cited in This Article: ] |
| 2. | Omata M, Ehata T, Yokosuka O, Hosoda K, Ohto M. Mutations in the precore region of hepatitis B virus DNA in patients with fulminant and severe hepatitis. N Engl J Med. 1991;324:1699-1704. [PubMed] [DOI] [Cited in This Article: ] |
| 3. | Friedt M, Gerner P, Lausch E, Trübel H, Zabel B, Wirth S. Mutations in the basic core promotor and the precore region of hepatitis B virus and their selection in children with fulminant and chronic hepatitis B. Hepatology. 1999;29:1252-1258. [PubMed] [DOI] [Cited in This Article: ] |
| 4. | Seo Y, Yoon S, Hamano K, Nakaji M, Yano Y, Katayama M, Ninomiya T, Hayashi Y, Kasuga M. Response to interferon-alpha in chronic hepatitis B with and without precore mutant strain detected by mutation site-specific assay. J Clin Gastroenterol. 2004;38:460-464. [PubMed] [DOI] [Cited in This Article: ] |
| 5. | Carman WF, Jacyna MR, Hadziyannis S, Karayiannis P, McGarvey MJ, Makris A, Thomas HC. Mutation preventing formation of hepatitis B e antigen in patients with chronic hepatitis B infection. Lancet. 1989;2:588-591. [PubMed] [DOI] [Cited in This Article: ] |
| 6. | Günther S, Fischer L, Pult I, Sterneck M, Will H. Naturally occurring variants of hepatitis B virus. Adv Virus Res. 1999;52:25-137. [PubMed] [DOI] [Cited in This Article: ] |
| 7. | Chu CJ, Keeffe EB, Han SH, Perrillo RP, Min AD, Soldevila-Pico C, Carey W, Brown RS, Luketic VA, Terrault N. Prevalence of HBV precore/core promoter variants in the United States. Hepatology. 2003;38:619-628. [PubMed] [DOI] [Cited in This Article: ] |
| 8. | Rodriguez-Frias F, Buti M, Jardi R, Cotrina M, Viladomiu L, Esteban R, Guardia J. Hepatitis B virus infection: precore mutants and its relation to viral genotypes and core mutations. Hepatology. 1995;22:1641-1647. [PubMed] [DOI] [Cited in This Article: ] |
| 9. | Junker-Niepmann M, Bartenschlager R, Schaller H. A short cis-acting sequence is required for hepatitis B virus pregenome encapsidation and sufficient for packaging of foreign RNA. EMBO J. 1990;9:3389-3396. [PubMed] [Cited in This Article: ] |
| 10. | Lok AS, Akarca U, Greene S. Mutations in the pre-core region of hepatitis B virus serve to enhance the stability of the secondary structure of the pre-genome encapsidation signal. Proc Natl Acad Sci U S A. 1994;91:4077-4081. [PubMed] [DOI] [Cited in This Article: ] |
| 11. | Fukuda R, Mohammad R, Hamamoto S, Ishimura N, Ishihara S, Akagi S, Watanabe M, Kinoshita Y. Clinical relevance of precore and basal core promoter variants of hepatitis B virus during natural hepatitis B e antigen seroconversion may be overstated. J Pediatr Gastroenterol Nutr. 2001;33:301-306. [PubMed] [DOI] [Cited in This Article: ] |
| 12. | Cabrerizo M, Bartolomé J, Ruiz-Moreno M, Otero M, López-Alcorocho JM, Carreño V. Distribution of the predominant hepatitis B virus precore variants in hepatitis B e antigen-positive children and their effect on treatment response. Pediatr Res. 1996;39:980-984. [PubMed] [DOI] [Cited in This Article: ] |
| 13. | Chang MH, Hsu HY, Ni YH, Tsai KS, Lee PI, Chen PJ, Hsu YL, Chen DS. Precore stop codon mutant in chronic hepatitis B virus infection in children: its relation to hepatitis B e seroconversion and maternal hepatitis B surface antigen. J Hepatol. 1998;28:915-922. [PubMed] [DOI] [Cited in This Article: ] |
| 14. | Galibert F, Mandart E, Fitoussi F, Tiollais P, Charnay P. Nucleotide sequence of the hepatitis B virus genome (subtype ayw) cloned in E. coli. Nature. 1979;281:646-650. [PubMed] [DOI] [Cited in This Article: ] |
| 15. | Lindh M, Gonzalez JE, Norkrans G, Horal P. Genotyping of hepatitis B virus by restriction pattern analysis of a pre-S amplicon. J Virol Methods. 1998;72:163-174. [PubMed] [DOI] [Cited in This Article: ] |
| 16. | Funk ML, Rosenberg DM, Lok AS. World-wide epidemiology of HBeAg-negative chronic hepatitis B and associated precore and core promoter variants. J Viral Hepat. 2002;9:52-61. [PubMed] [DOI] [Cited in This Article: ] |
| 17. | Li JS, Tong SP, Wen YM, Vitvitski L, Zhang Q, Trépo C. Hepatitis B virus genotype A rarely circulates as an HBe-minus mutant: possible contribution of a single nucleotide in the precore region. J Virol. 1993;67:5402-5410. [PubMed] [Cited in This Article: ] |
| 18. | Knoll A, Rohrhofer A, Kochanowski B, Wurm EM, Jilg W. Prevalence of precore mutants in anti-HBe-positive hepatitis B virus carriers in Germany. J Med Virol. 1999;59:14-18. [PubMed] [DOI] [Cited in This Article: ] |
| 19. | Tillmann H, Trautwein C, Walker D, Michitaka K, Kubicka S, Böker K, Manns M. Clinical relevance of mutations in the precore genome of the hepatitis B virus. Gut. 1995;37:568-573. [PubMed] [DOI] [Cited in This Article: ] |
| 20. | Orito E, Mizokami M, Sakugawa H, Michitaka K, Ishikawa K, Ichida T, Okanoue T, Yotsuyanagi H, Iino S. A case-control study for clinical and molecular biological differences between hepatitis B viruses of genotypes B and C. Japan HBV Genotype Research Group. Hepatology. 2001;33:218-223. [PubMed] [DOI] [Cited in This Article: ] |
| 21. | Söderström A, Norkrans G, Conradi N, Krantz M, Horal P, Lindh M. Histologic activity of childhood chronic hepatitis B related to viremia levels, genotypes, mutations, and epidemiologic factors. J Pediatr Gastroenterol Nutr. 2002;35:487-494. [PubMed] [DOI] [Cited in This Article: ] |