Published online Feb 7, 2005. doi: 10.3748/wjg.v11.i5.756
Revised: April 12, 2004
Accepted: May 9, 2004
Published online: February 7, 2005
AIM: To investigate the survivin gene expression in human hepatocellular carcinoma cell line SMMC-7721 and the effects of survivin gene RNA interference (RNAi) on cell apoptosis and biological behaviors of SMMC-7721 cells.
METHODS: Eukaryotic expression vector of survivin gene RNAi and recombinant plasmid pSuppressorNeo-survivin(pSuNeo-SVV), were constructed by ligating into the vector, pSupperssorNeo (pSuNeo) digested with restriction enzymes Xba I and Sal I and the designed double-chain RNAi primers. A cell model of SMMC-7721 after treatment with RNAi was prepared by transfecting SMMC-7721 cells with the lipofectin transfection method. Strept-avidin- biotin-complex (SABC) immunohistochemical staining and RT-PCR were used to detect survivin gene expressions in SMMC-7721 cells. Flow cytometry was used for the cell cycle analysis. Transmission electron microscopy was performed to determine whether RNAi induced cell apoptosis, and the method of measuring the cell growth curve was utilized to study the growth of SMMC-7721 cells before and after treatment with RNAi.
RESULTS: The eukaryotic expression vector of survivin gene RNAi and pSuNeo-SVV, were constructed successfully. The expression level of survivin gene in SMMC-7721 cells was observed. After the treatment of RNAi, the expression of survivin gene in SMMC-7721 cells was almost absent, apoptosis index was increased by 15.6%, and the number of cells was decreased in G2/M phase and the cell growth was inhibited.
CONCLUSION: RNAi can exert a knockdown of survivin gene expression in SMMC-7721 cells, and induce apoptosis and inhibit the growth of carcinoma cells.
- Citation: Cheng SQ, Wang WL, Yan W, Li QL, Wang L, Wang WY. Knockdown of survivin gene expression by RNAi induces apoptosis in human hepatocellular carcinoma cell line SMMC-7721. World J Gastroenterol 2005; 11(5): 756-759
- URL: https://www.wjgnet.com/1007-9327/full/v11/i5/756.htm
- DOI: https://dx.doi.org/10.3748/wjg.v11.i5.756
Survivin gene is a new member of inhibitors of the apoptosis protein (IAP) family, and the most powerful apoptosis inhibitory factor as far as we know. Its distribution characteristics in tissues are different from other apoptosis inhibitory factors. It is selectively overexpressed in embryonic and fetal development, as well as in transformed cells and human carcinoma tissues, but not in adult differentiated tissues (with the exception of thymus and genital gland), and is associated with the aggressiveness of diseases and unfavorable outcomes[1-13]. The expression of survivin gene directly relates to the histological classification of carcinoma, its relapse, metastasis, and growth index and inversely relates to apoptotic index[1-4,6-8,11]. In contrast, some studies have demonstrated that the expression of survivin gene is not related to the histological classification of carcinoma, its size, infiltration depth, relapse, metastasis and prognosis[8-10].
RNA interference (RNAi) is a genetic interference phenomenon directed by the double-stranded RNA (dsRNA). It could specifically and efficiently degrade mRNA, resulting in post-transcriptional gene silencing (PTGS)[14-16], which is a natural mechanism in organisms underlying the resistance to virus invasion and inhibition of transposon mobility. Its blocking action on gene expression has been successfully observed in rat and human cells cultured in vitro, and the knockdown of genes in cells has been achieved[15,16].A latest study[17] has shown that 21-25 nt small interference RNA (siRNA) can mediate specific gene silencing in mammal cells. Being effective and highly specific, RNAi probably becomes a new technique in knocking gene down and plays an important role in gene function study and gene therapy of diseases. We constructed the survivin gene eukaryotic expression vector of dsRNAi, and transfected SMMC-7721 cells, to observe the survivin gene expression following RNAi and its effects on cell apoptosis and growth, which has laid a foundation for further studies on the functions of survivin gene and genetic therapy involved in human hepatocellular carcinoma (HCC).
Trizol reagent and M-MLV were purchased from Gibco BRL. Taq DNA polymerase and dNTPs were obtained from Promega. DNA Marker DL-2000 and DL-15000, T4 DNA ligase, BamH, Xho I, Xba, Sal I and Sca I were bought from Takara. Lipfectamine 2000 was purchased from Invitrogen. Competent bacteria (E.coli DH5α) were preserved in our laboratory. Polyclonal rabbit anti-human survivin antibody (sc-10811) was purchased from Santa Cruz. pSuppressorNeo (pSuNeo) was a gift of YAN-Yan (New York University, USA).
A survivin sense primer corresponding to nucleotides, 5’-TAGGATCCATGGGTGCCCCGACG-3’ was added to compatible restriction sites BamH I at the 5’end of the primer sequences. A survivin antisense primer complementary to nucleotides, 5’-ACCTCGAGCTCAATCCATGGCAGCC-3’ was added to compatible restriction sites Xho I at the 3’end of the primer sequences. The length of amplified fragments was 445 bp.
A forward primer of dsRNAi corresponding to nucleotideswas 5’-TCGAGgagaacgagccagacttggccGAGTACTGggccaagtctggctcgttctcTTTTT-3. The Xho I and Sca I overhang sites were underlined. A reverse primer of dsRNAi corresponding to nucleotides was 3’-cctcttgctcggtctga accggCTCATGAC ccggttcagaccgagcaagagAA AAAGATC-5’. The Sca I and Xba I overhang sites were underlined. The primers designed were synthesized by Shanghai Sangon Biological Co. (China).
Human hepatocellular carcinoma cell line SMMC-7721 was maintained in our laboratory. The cells were grown in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 100 mL/L fetal bovine serum (FBS) and incubated in a humidified incubator containing 50 mL/L CO2 in at 37 °C.
Primer annealing (siRNA insert) Forward primer, reverse primer and annealing buffer were incubated at 95 °C for 10 min, and the tube was gradually cooled to room temperature.
Ligation and insertion of vector pSuNeo One microliter of linearized vector pSuNeo (completely digested with Xba I and Sal I), 1 μL of insert DNA, 1 μL of T4 DNA ligase, and 2 μL of 10×ligase buffer were incubated at 16 °C overnight. Following the ligation reaction, the ligated plasmid DNA was transformed into 200 μL of competent cells of an appropriate host strain (DH5α), and then plated on LB plates containing 40 ng/mL of kanamycin.
The gene transfection mediated by lipofectin was used to introduce the plasmid pSuNeo-SVV and empty the vector pSuNeo into human hepatocellular carcinoma cell line SMMC-7721, respectively. After selected with G418, resistant colonies were obtained. Immunohistochemical staining and RT-PCR were performed to confirm the transfection.
Immunohistochemical staining was performed by the a SABC method using SABC kit (Wuhan Boster Biological Co., China). Adherent cell sections were treated with 3 mL/L H2O2 in methanol for 30 min to abolish the endogenous peroxidase activity. Sections were blocked with normal goat serum for 30 min at room temperature , and incubated with anti-survivin antibody (1:200) overnight at 4 °C. The sections were incubated with biotinylated anti-rabbit IgG antibody, followed by avidin-biotin-peroxidase complex. Color was developed in a substrate solution of 0.1 mL/L diaminobenzidine-hydrogen peroxide and counterstained with hematoxylin.
Total RNA was extracted from human hepatocellular carcinoma cell line SMMC-7721 using the Trizol reagent according to the manufacturer’s instructions. Complementary DNA (cDNA) was generated from total RNA using M-MLV. PCR of the cDNA was performed in a final volume of 50 μL containing 4 μL of 4×dNTPs, 2 units of Taq DNA polymerase, and 20 mmol/L of each primer. The samples were amplified 28 cycles at 95 °C for 1 min, at 58 °C for 30 s, and at 72 °C for 40 s, and finally at 72 °C for 10 min. The PCR products were separated by electrophoresis on 10 g/L agarose gels and visualized by ethidium bromide staining. Amplification of human β-actin served as a control for a sample loading and integrity. A β-actin sense primer corresponding to nucleotides was 5’-ACACTGTGCCCATCTACGAGG-3’, and an antisense primer complementary to nucleotides was 5’-AGGGGCCGGACTCG-CATTACT-3’, the length of amplified fragments was 621 bp.
The SMMC-7721 cells were digested with 2.5 g/L trypsinase and collected. After rinsed with PBS, the cells were prefixed with 30 g/L glutaraldehyde for 30 min, post-fixed with 10 mL/L osmic acid, dehydrated in graded ethanol, embedded in Epon 812 mixture, and cut into 0.05 μm thick sections on an ultramicrotome. The cells were observed under Hitachi JEM-2000EX electron microscopy.
Cell cycle distributions were determined by measuring the cellular DNA content using flow cytometry. Cells were washed with PBS, fixed with 700 mL/L ethanol for 20 min and stored at 4 °C overnight, then washed with PBS, and stained with 100 μL of 50 mg/L PI at 4 °C for 30 min. Apoptotic cells were assayed using the Elite ESP flow cytometer at 488 nm, and data were analyzed with the CELLQuest software.
A total of 2×104 cells from transfected plasmid pSuNeo-SVV, vector pSuNeo and SMMC-7721 cell line were seeded in triplicate in 24-well plates and cultured in DMEM supplemented with 100 mL/L FBS. At each time point, cells were trypsinized to a single cell suspension and counted on a Coulter counter set at >10 μm in diameter.
Data were expressed as mean±SD. Statistical significance was determined by the Students’t-test. P<0.05 was considered statistically significant.
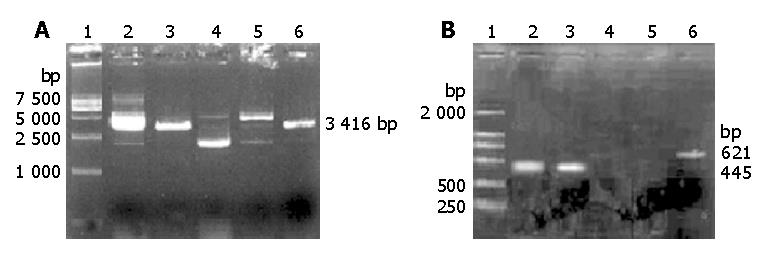
Digested results of the empty vector pSuNeo and recombinant plasmid pSuNeo-SVV showed 2 to 3 bands in pSuNeo, pSuNeo-SVV, and pSuNeo-SVV digested by Sal I. After pSuNeo was digested by Sal I and pSuNeo-SVV was digested by Scal, a 3.4 kb band appeared in both of them (Figure 1A).
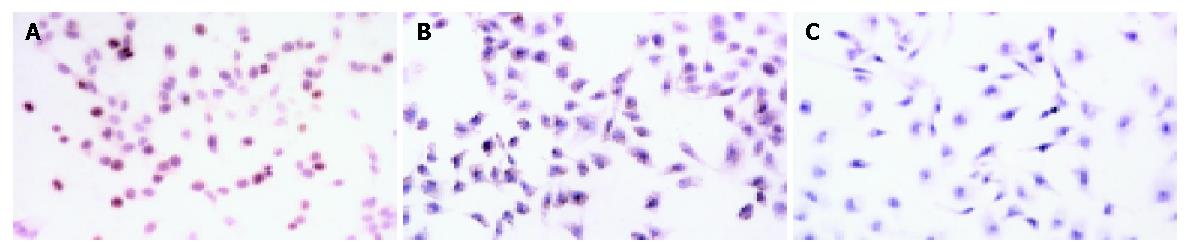
The RT-PCR findings revealed that the SMMC-7721 cells in the control groups (SMMC-7721 and SMMC-7721 transfected with empty vector pSuNeo) presented a bright strip at 445 bp, while SMMC-7721 transfected with pSuNeo-SVV did not (Figure 1B). By SABC immunohistochemical staining, we found that the cell nuclei of SMMC-7721 and SMMC-7721 transfected with empty vector pSuNeo were yellow colored, while the cell nuclei of SMMC-7721 transfected with pSuNeo-SVV were not colored (Figure 2).
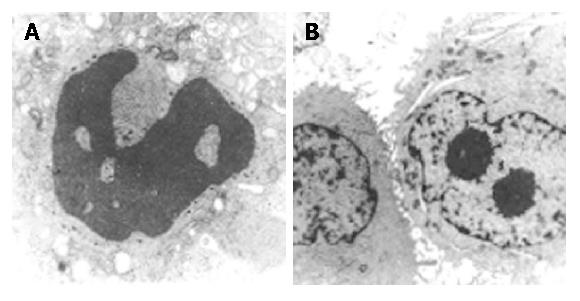
Transmission electron microscopy revealed that in SMMC-7721 cells transfected with pSuNeo-SVV, dense chromatin appeared near the nucleus membranes, which presented some typical manifestations of cell apoptosis (Figure 3A), while no typical manifestation of cell apoptosis was observed in the control groups and the nuclei appeared to undergo karyokinesis, which showed a vigorous cell proliferation (Figure 3B).
Flow cytometry analysis of cell cycle revealed that SMMC-7721 cells transfected with pSuNeo-SVV showed a marked reduction in G2/M phase by 9.5%, the apoptosis index was as high as 15.6%,17.1% and 17.1% in the control groups respectively.
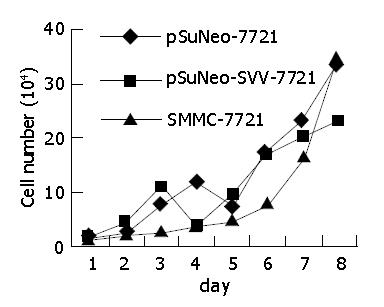
The cell growth curve shows that SMMC-7721 cells transfected with pSuNeo-SVV grew slowly, reached a stage of platform on the 7th d, and the number of cells was much less than that of the two control groups, while SMMC-7721 cells in the control groups proliferated rapidly on the 6th d and reached a stage of platform on the 8th d (Figure 4).
In recent years, researches on the functions of gene and gene therapeutic technologies for cancers, have been designed to identify these functions with gene transduction, antisense technology, transgene and gene knockout, and artificial chromosome transduction to deprive the functions of particular genes. RNAi possesses a high ability to specifically silence particular genes. Therefore, it can be used as a powerful tool in researches on the functions of genes and genetic therapy for carcinoma[15,16]. Attention has been paid to RNAi in the field of researches on gene functions.
Our study demonstrated that the recombinant pSuNeo-SVV was not linearized when digested with Xba I, Xho I, or Sal I, but was linearized only with Sca I. SABC immunohistochemical staining showed that the nuclei of SMMC-7721 cells and SMMC-7721 cells transfected with empty vector pSuNeo revealed positive staining for survivin, while the nuclei of SMMC-7721 cells transfected with pSuNeo-SVV were almost not stained, suggesting that survivin gene is blocked at the level of protein. RT-PCR findings demonstrated that survivin gene was blocked in SMMC-7721 cells transfected with pSuNeo-SVV at the level of transcription. Flow cytometry analysis revealed that the SMMC-7721 cells transfected with pSuNeo-SVV presented a marked reduction in G2/M phase, an obvious AP peak, and an increase in apoptosis index compared to the control groups. Transmission electron microscopy showed that in SMMC-7721 cells transfected with pSuNeo-SVV, dense chromatin appeared near the nuclei membranes which presents some typical manifestations of cell apoptosis, while no typical manifestation of cell apoptosis was observed in the control groups and the nuclei underwent karyokinesis, indicating a vigorous cell proliferation. We also observed that SMMC-7721 cells transfected with pSuNeo-SVV grew slowly, as compared with the control groups.
The above-mentioned findings comfirm that chemically synthesized siRNAs can specifically block survivin gene expression, induce cell apoptosis, and inhibit the growth of carcinoma cells. Therefore, our study has laid a foundation for further studies on the use of RNAi in treating liver cancer.
It has been reported that the possible anti-apoptosis mechanisms of survivin gene include the direct inhibitory effect of survivin gene on the activities of caspases, especially caspase-3 and caspase-7 to block cell apoptosis; interactions of survivin gene with cyclin-dependent kinase 4 (CDK4), leading to CDK2/cyclin E activation and Rb phosphorylation; the release of p21waf1/cip from survivin-CDK4 complex and its combination with the procaspase-3 of mitochondria, leading to the inhibition of its activity and the blockage of cell apoptosis blocked[18,19]. Survivin is characterized by cell cycle-dependent expression, that is, it is expressed only in G2/M. Ito et al[4] transfected survivin into 4 human hepatocellular carcinoma cell lines and found that the number of cells in G0/G1 was remarkably reduced, and the number of cells in S or G2/M increased. It has been reported that survivin is involved in the formation of blood vessels, as a possible protective gene induced by growth factor in the formation of blood vessels to protect the normal proliferation of endothelial cells[20]. In the formation of blood vessels, the action of angiopoietin-1 (the key factor to maintain the vascular stability and to form the cavity) depends on the up-regulation of survivin gene expression[21]. In the carcinoma cell lines transfected with antisense oligonucleotide target survivin, cell apoptosis is increased and cell proliferation is inhibited[22,23]. A study reported that apoptosis induced by tumor necrosis factors-related apoptosis inducing ligand (TRAIL) was related to its inhibitory action on survivin gene expression. Survivin expression might resist apoptosis induced by chemotherapy agents in cholangiocarcinomas.
The results of our study confirm that the inhibitory activity of survivin gene on the growth of liver cancer cells could be realized by inducing cell apoptosis. Besides, RNAi alone could block survivin gene expression to induce a remarkable increase in cell apoptosis. This unique effect of survivin provides new evidence for its antiapoptotic effects on liver cancer cells.
In summary, survivin gene can be regarded as a very good target gene in genetic therapy for carcinomas. RNAi of survivin gene is a promising approach in treating carcinomas.
Edited by Wang XL and Kumar M
| 1. | Sarela AI, Macadam RC, Farmery SM, Markham AF, Guillou PJ. Expression of the antiapoptosis gene, survivin, predicts death from recurrent colorectal carcinoma. Gut. 2000;46:645-650. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 227] [Cited by in F6Publishing: 250] [Article Influence: 10.4] [Reference Citation Analysis (0)] |
| 2. | Mori A, Wada H, Nishimura Y, Okamoto T, Takemoto Y, Kakishita E. Expression of the antiapoptosis gene survivin in human leukemia. Int J Hematol. 2002;75:161-165. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 36] [Cited by in F6Publishing: 43] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 3. | Das A, Tan WL, Teo J, Smith DR. Expression of survivin in primary glioblastomas. J Cancer Res Clin Oncol. 2002;128:302-306. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 43] [Cited by in F6Publishing: 49] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 4. | Ito T, Shiraki K, Sugimoto K, Yamanaka T, Fujikawa K, Ito M, Takase K, Moriyama M, Kawano H, Hayashida M. Survivin promotes cell proliferation in human hepatocellular carcinoma. Hepatology. 2000;31:1080-1085. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 242] [Cited by in F6Publishing: 267] [Article Influence: 11.1] [Reference Citation Analysis (0)] |
| 5. | Sarela AI, Verbeke CS, Ramsdale J, Davies CL, Markham AF, Guillou PJ. Expression of survivin, a novel inhibitor of apoptosis and cell cycle regulatory protein, in pancreatic adenocarcinoma. Br J Cancer. 2002;86:886-892. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 123] [Cited by in F6Publishing: 142] [Article Influence: 6.5] [Reference Citation Analysis (0)] |
| 6. | Wakana Y, Kasuya K, Katayanagi S, Tsuchida A, Aoki T, Koyanagi Y, Ishii H, Ebihara Y. Effect of survivin on cell proliferation and apoptosis in gastric cancer. Oncol Rep. 2002;9:1213-1218. [PubMed] [Cited in This Article: ] |
| 7. | Cohen C, Lohmann CM, Cotsonis G, Lawson D, Santoianni R. Survivin expression in ovarian carcinoma: correlation with apoptotic markers and prognosis. Mod Pathol. 2003;16:574-583. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 76] [Cited by in F6Publishing: 81] [Article Influence: 3.9] [Reference Citation Analysis (0)] |
| 8. | Lu CD, Altieri DC, Tanigawa N. Expression of a novel antiapoptosis gene, survivin, correlated with tumor cell apoptosis and p53 accumulation in gastric carcinomas. Cancer Res. 1998;58:1808-1812. [PubMed] [Cited in This Article: ] |
| 9. | Kawasaki H, Altieri DC, Lu CD, Toyoda M, Tenjo T, Tanigawa N. Inhibition of apoptosis by survivin predicts shorter survival rates in colorectal cancer. Cancer Res. 1998;58:5071-5074. [PubMed] [Cited in This Article: ] |
| 10. | Tanaka K, Iwamoto S, Gon G, Nohara T, Iwamoto M, Tanigawa N. Expression of survivin and its relationship to loss of apoptosis in breast carcinomas. Clin Cancer Res. 2000;6:127-134. [PubMed] [Cited in This Article: ] |
| 11. | Trieb K, Lehner R, Stulnig T, Sulzbacher I, Shroyer KR. Survivin expression in human osteosarcoma is a marker for survival. Eur J Surg Oncol. 2003;29:379-382. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 87] [Cited by in F6Publishing: 90] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 12. | Sui L, Dong Y, Ohno M, Watanabe Y, Sugimoto K, Tokuda M. Survivin expression and its correlation with cell proliferation and prognosis in epithelial ovarian tumors. Int J Oncol. 2002;21:315-320. [PubMed] [Cited in This Article: ] |
| 13. | Zhu XD, Lin GJ, Qian LP, Chen ZQ. Expression of survivin in human gastric carcinoma and gastric carcinoma model of rats. World J Gastroenterol. 2003;9:1435-1438. [PubMed] [Cited in This Article: ] |
| 14. | Fire A, Xu S, Montgomery MK, Kostas SA, Driver SE, Mello CC. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature. 1998;391:806-811. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 10522] [Cited by in F6Publishing: 9727] [Article Influence: 374.1] [Reference Citation Analysis (1)] |
| 15. | Konnikova L, Kotecki M, Kruger MM, Cochran BH. Knockdown of STAT3 expression by RNAi induces apoptosis in astrocytoma cells. BMC Cancer. 2003;3:23. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 170] [Cited by in F6Publishing: 178] [Article Influence: 8.5] [Reference Citation Analysis (0)] |
| 16. | Tuschl T, Borkhardt A. Small interfering RNAs: a revolutionary tool for the analysis of gene function and gene therapy. Mol Interv. 2002;2:158-167. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 122] [Cited by in F6Publishing: 131] [Article Influence: 6.6] [Reference Citation Analysis (0)] |
| 17. | Elbashir SM, Lendeckel W, Tuschl T. RNA interference is mediated by 21- and 22-nucleotide RNAs. Genes Dev. 2001;15:188-200. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 2287] [Cited by in F6Publishing: 2246] [Article Influence: 97.7] [Reference Citation Analysis (0)] |
| 18. | Suzuki A, Ito T, Kawano H, Hayashida M, Hayasaki Y, Tsutomi Y, Akahane K, Nakano T, Miura M, Shiraki K. Survivin initiates procaspase 3/p21 complex formation as a result of interaction with Cdk4 to resist Fas-mediated cell death. Oncogene. 2000;19:1346-1353. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 166] [Cited by in F6Publishing: 186] [Article Influence: 7.8] [Reference Citation Analysis (0)] |
| 19. | Shin S, Sung BJ, Cho YS, Kim HJ, Ha NC, Hwang JI, Chung CW, Jung YK, Oh BH. An anti-apoptotic protein human survivin is a direct inhibitor of caspase-3 and -7. Biochemistry. 2001;40:1117-1123. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 510] [Cited by in F6Publishing: 516] [Article Influence: 22.4] [Reference Citation Analysis (0)] |
| 20. | O'Connor DS, Schechner JS, Adida C, Mesri M, Rothermel AL, Li F, Nath AK, Pober JS, Altieri DC. Control of apoptosis during angiogenesis by survivin expression in endothelial cells. Am J Pathol. 2000;156:393-398. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 258] [Cited by in F6Publishing: 258] [Article Influence: 10.8] [Reference Citation Analysis (0)] |
| 21. | Papapetropoulos A, Fulton D, Mahboubi K, Kalb RG, O'Connor DS, Li F, Altieri DC, Sessa WC. Angiopoietin-1 inhibits endothelial cell apoptosis via the Akt/survivin pathway. J Biol Chem. 2000;275:9102-9105. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 468] [Cited by in F6Publishing: 475] [Article Influence: 19.8] [Reference Citation Analysis (0)] |
| 22. | Ambrosini G, Adida C, Sirugo G, Altieri DC. Induction of apoptosis and inhibition of cell proliferation by survivin gene targeting. J Biol Chem. 1998;273:11177-11182. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 317] [Cited by in F6Publishing: 327] [Article Influence: 12.6] [Reference Citation Analysis (0)] |
| 23. | Grossman D, McNiff JM, Li F, Altieri DC. Expression of the apoptosis inhibitor, survivin, in nonmelanoma skin cancer and gene targeting in a keratinocyte cell line. Lab Invest. 1999;79:1121-1126. [PubMed] [Cited in This Article: ] |