Published online Nov 1, 2004. doi: 10.3748/wjg.v10.i21.3205
Revised: February 28, 2004
Accepted: March 6, 2004
Published online: November 1, 2004
AIM: To construct a recombinant E. coli strain that would highly express the proinflammatory outer membrane protein of human Helicobacter pylori (H pylori).
METHODS: The oipA DNA was amplified by PCR, inserted into pET-32a, and transformed into Top10 E. coli strain. This recombinant plasmid of Top10 was sent out for nucleotide sequence analysis. Finally this sequence AF479754 was compared with HP0638 and JHP0581.
RESULTS: The sequence of the aim gene was obtained. It had 924 base pairs. The identity was 95.32% against HP0638, 95.02% against JHP0581, which was higher than the identity between HP0638 and JHP0581.
CONCLUSION: Although the aim gene was obtained, but it was different from the published sequence of GenBank. It is not clear what makes this difference. Maybe it is because different strain was used or because there were some variations. So more researches are required to prove it.
-
Citation: Chen DR, Huang AL, Tao XH, Wang PL, Jiang Z. Cloning and sequence analysis of gene oipA encoding an outer membrane protein of human
Helicobacter pylori . World J Gastroenterol 2004; 10(21): 3205-3207 - URL: https://www.wjgnet.com/1007-9327/full/v10/i21/3205.htm
- DOI: https://dx.doi.org/10.3748/wjg.v10.i21.3205
In order to find a good vaccine to end the coexistence of human and Helicobacter pylori (H pylori), some scientists have focused on exploring some outer membrane proteins in these 10 years. Yamaoka et al[1-5] found six bands of the outer membrane proteins in H pylori on gels by immunoblot assay kits, 116 ku (CagA), 89 ku (VacA), 35 ku, 30 ku, 26.5 ku and 19.5 ku (Lpp20). In which 35 ku band is broader than others. It always appears at 33-35 ku. Now a few scientists are studying the 35 ku protein including Yamaoka. They found that 33-35 ku protein was positive in 97.5% patients with peptic ulcer, 70% patients with chronic gastritis. The results showed that it was related to the presence of peptic ulcer. Yamaoka deduced that Hpo638 encoding outer membrane protein oipA had the most possibility, because it had great antigen characteristics, also could increase the serum level of IL-8. Later studies have shown that oipA is the only index that can differ duodenum ulcer from gastritis, respond to high location of H pylori, serious mucosal infiltration with neutrophils cells, and high level of IL-8.
As so far, there is no study on the oipA in our country. We used PCR techinology to amplify oipA gene, to construct the recombinant plasmid, and analyse the DNA sequence. We found that the target sequence AF479754 had some difference with HP0638 and JHP0581.
H pylori was afforded by the Department of Microbiology, Chongqing University of Medical Sciences. Top10 E. coli strain and pET32a(+) plasmid were presented by the Institute of Viral Hepatitis of Chongqing University of Medical Sciences. Endonucleases, T4 ligase and Pfu-Taq DNA polymerase were purchased from Promega. PCR marker was purchased from Shanghai Sangon. The kits for purification of plasmids and PCR products were obtained from Omega. Primers were synthesized in Shanghai Sangon.
Each gastric biopsy specimen was inoculated on H pylori medium plates. The plates were incubated at 37 °C under microaerobic conditions (50 mL/L O2, 100 mL/L CO2 and 850 mL/L N2) for 3 to 5 d. A bacterial isolate was identified as H pylori according to typical Gram staining morphology, biochemical tests positive for urease and oxidase, and agglutination with commercial rabbit antibody against whole cell of the microbe. All H pylori isolates were stored at -70 °C.
Genomic DNA of H pylori were extracted by the conventional phenol-chloroform method and DNase-free RNase treatment. The obtained DNA was dissolved in TE buffer, and its concentration and purity were determined by ultraviolet spectrophotometry.
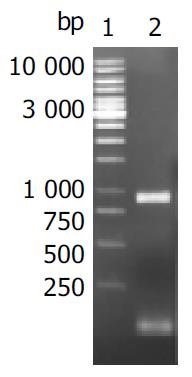
Oligonucleotide primers were designed to amplify the whole sequence of oipA gene from H pylori strain 26 695 based on the published corresponding genomic sequences. The sequence of oipA sense primer with an endonuclease site of BamH I was 5’-CCGGATCCATGAAAAAAGCTCTCTTACT-3’. The sequence of oipA antisense primer with an endonuclease site of XhoI was 5’- CGCGGCTCGAGTTAATGTTTGTTTTTAAAGTT -3’. The total volume per PCR was 100 L containing 2.5 mol/L each dNTP, 500 nmol/L each of the two primers, 15 mol/L MgCl2, 3.0 U Pfu-Taq polymerase, 100 ng DNA template and 1 × PCR buffer (pH8.8). The parameters for PCR were at 94 °C for 4 min, × 1; at 94 °C for 60 s, at 52 °C for 50 s, at 72 °C for 60 s, × 30; then at 72 °C for 7 min, × 1. The results of PCR were observed under UV light after electrophoresis in 10 g. l-1 agarose pre-stained with ethidium bromide.
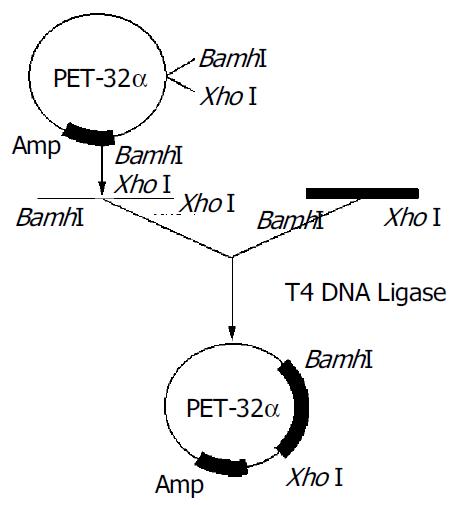
PCR products were digested by endonucleases BamH I and XhoI, meanwhile pET32a(+) plasmid was cut by BamH I and XhoI. After purification by the kits for purification of PCR products, these two fragments were ligated with cohesive ends. Then the recombinant plasmid was transformed into E. coli line Top10. The E. coli Top10 containing the recombinant plasmid was amplified in LB medium containing AMP. Clones were picked out randomly through blue/white screening. Finally the plasmids were extracted with a plasmid extraction kit according to the manufacturer’s instructions and identified by PCR and digestion endonucleases of BamH I and XhoI.
Top10 E.coli strains containing recombinant plasmid were sent to Shanghai GeneCore Biotechnologies Co. for DNA sequence analysis.
Target fragments of oipA genes with expected sizes amplified from DNA template of H pylori stains are shown in Figure 1.
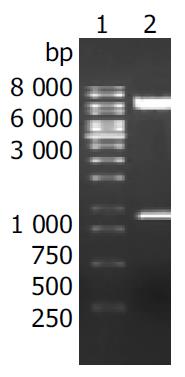
To construct the recombinant plasmid of H pylori oipA, purified PCR products were ligated with endonuclease-digested pET32a (+), and the recombinant was transformed into E. coli line Top10. White clones were picked out and confirmed by PCR and dual endonuclease digestion with BamH I /XhoI. The technologic route is shown in Figure 2. Electrophoresis showed that a fragment of about 924 bp was released (Figure 3).
Samples were analyzed with an automatic sequence analyzer. Sequencing results are published in the GenBank. The accession number is AF479754. The identity was 95.32% against HP0638 and 95.02% against JHP0581, which was higher than the identity between HP0638 and JHP0581. HP0638 only was 918 bp while JHP0581 was 924 bp (6 bp TCTCTC at 37th site) and AF479754 also was 924 bp (6 bp CGAACA at 190th site).
How to prevent and eradicate H pylori infection is the core of the clinical therapy. The native immunity has little effect on it. Some experiments concluded that vaccine could decrease the infection, especial the incidence rate of piptic ulcer and gastric cancer. So an optimal vaccine can end the coexistence of human and H pylori. In recent 10 years, studies have gone deep in this wind of proteins, such as CagA, VacA, UreA, UreB, HSP, PldA, NAp.
Yamaoka deduced that Hp0638 encoding outer membrane protein oipA was the best candidate, because it had great antigen characteristics, and could increase the serum level of IL-8 also. Later studies showed that oipA was the only index that could differ duodenum ulcer from gastritis, respond to high location of H pylori, serious mucosal infiltration with neutrophil cells, and high level of IL-8. So far, there is no study on the oipA in our country. So we chose this subject. Our results showed that the identity was 95.32% against HP0638 and 95.02% against JHP0581, which was higher than the identity between HP0638 and JHP0581. The difference could be explained in two ways. One is that the strain we used was different from 26695 and J99. Different strains have different gene fragments, just as the difference between HP0638 and JHP0581. The other is that variation of the same strain induced the difference. Compared with JHP0581, AF479754 had the same base pairs. Only the insert site of 6 bp was different. It maybe one subtype of the same strain as 26695 or J99.
The value of the difference lay in three points. First, it contributes to the finding of new strains. Some surveys should be done in the whole country, to show the region characteristics of H pylori, and to confirm the main strains in our country. Second, it contributes to the analysis of the changes in pathogenicity and antigen activity, which are very important in elucidating the immune pathogeny. Third, it contributes to the development of new vaccines. The immunity study can provide some help to design the vaccine and optimal immune route. But a variety of strains would bring some difficulties to develop vaccines. So more work should be done to certify the pathogeny of H pylori, to develop effective vaccines for the eradication of H pylori.
H pylori oipA sequence described in this paper was accepted by GenBank, GenBank accession number for it is AF479754.
Edited by Wang XL and Xu FM
| 1. | Yamaoka Y, Kodama T, Graham DY, Kashima K. Search for putative virulence factors of Helicobacter pylori: the low-molecular-weight (33-35 K) antigen. Dig Dis Sci. 1998;43:1482-1487. [PubMed] [DOI] [Cited in This Article: ] |
| 2. | Shiesh SC, Sheu BS, Yang HB, Tsao HJ, Lin XZ. Serologic response to lower-molecular-weight proteins of H. pylori is related to clinical outcome of H. pylori infection in Taiwan. Dig Dis Sci. 2000;45:781-788. [PubMed] [DOI] [Cited in This Article: ] |
| 3. | Yamaoka Y, Kwon DH, Graham DY. A M(r) 34,000 proinflammatory outer membrane protein (oipA) of Helicobacter pylori. Proc Natl Acad Sci U S A. 2000;97:7533-7538. [PubMed] [DOI] [Cited in This Article: ] |
| 4. | Yamaoka Y, Kikuchi S, el-Zimaity HM, Gutierrez O, Osato MS, Graham DY. Importance of Helicobacter pylori oipA in clinical presentation, gastric inflammation, and mucosal interleukin 8 production. Gastroenterology. 2002;123:414-424. [PubMed] [DOI] [Cited in This Article: ] |
| 5. | Ferrero RL, Labigne A. Helicobacter pylori vaccine development in the post-genomic era: can in silico translate to in vivo. Scand J Immunol. 2001;53:443-448. [PubMed] [DOI] [Cited in This Article: ] |