Copyright
©The Author(s) 2022.
World J Gastroenterol. Sep 21, 2022; 28(35): 5188-5202
Published online Sep 21, 2022. doi: 10.3748/wjg.v28.i35.5188
Published online Sep 21, 2022. doi: 10.3748/wjg.v28.i35.5188
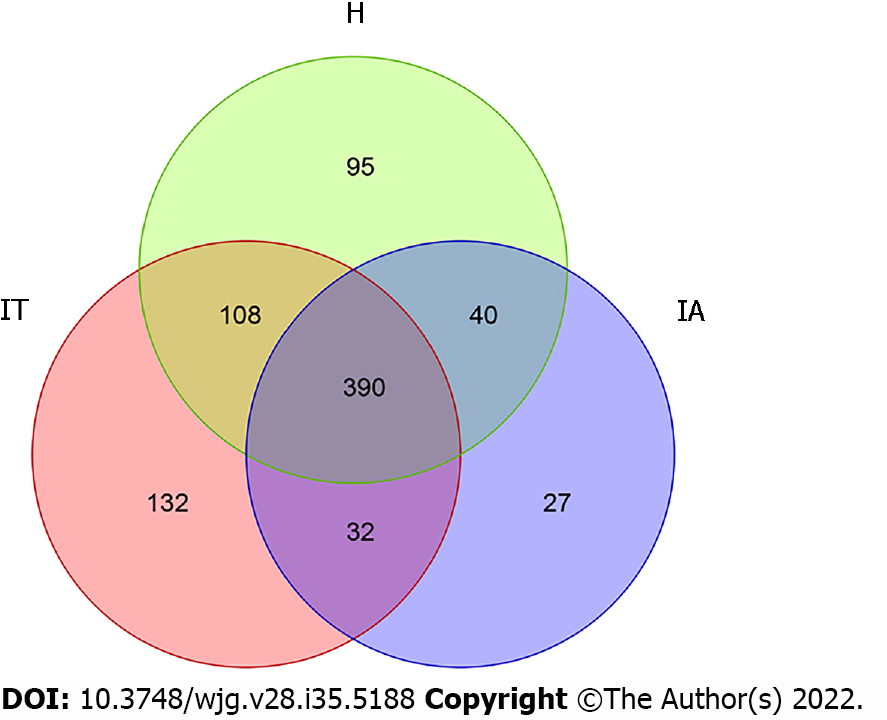
Figure 1 Operational taxonomic units in healthy controls, immune-tolerant phase patients, and immune-active phase patients.
IT: Immune-tolerant; IA: Immune-active; H: Healthy.
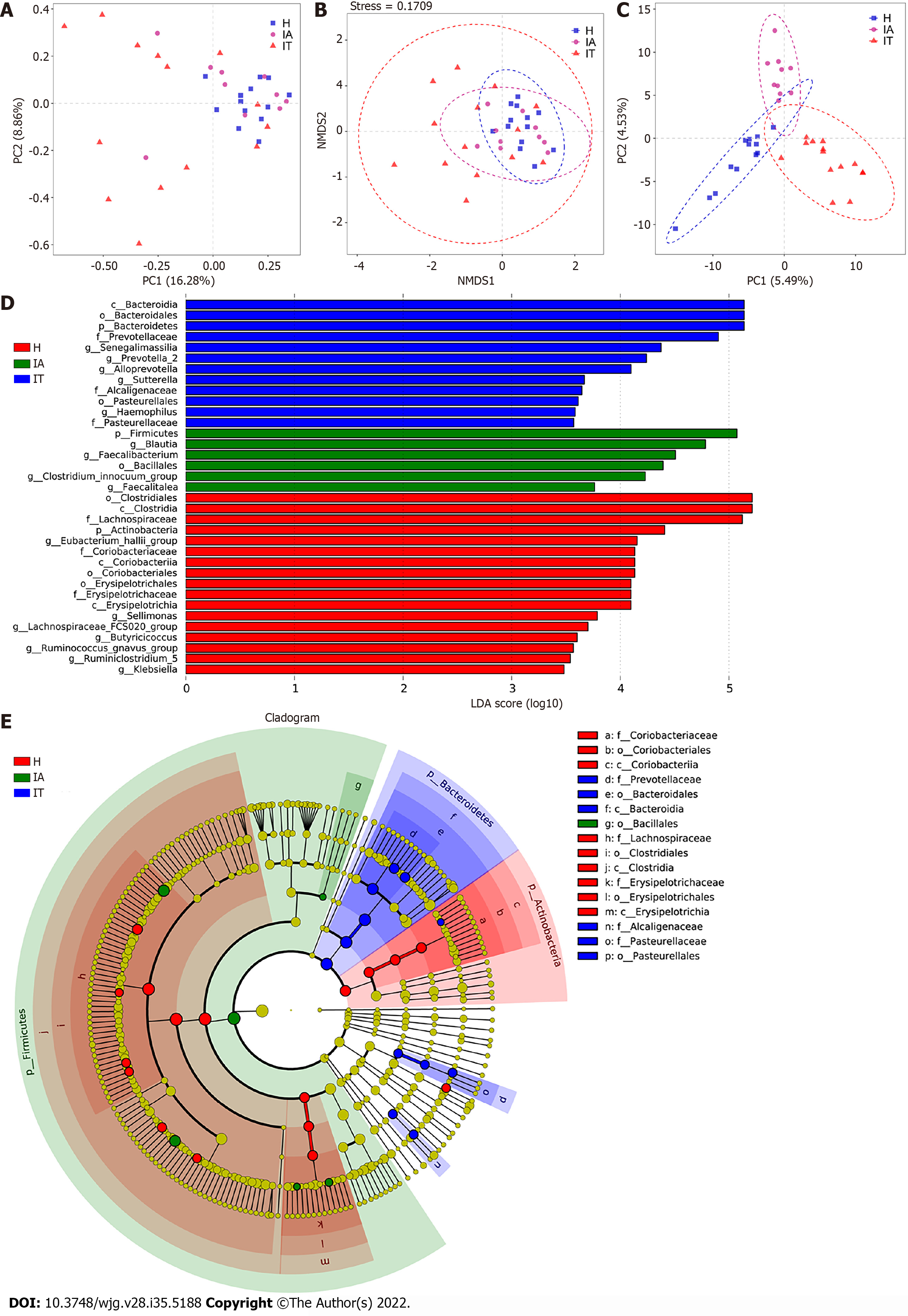
Figure 2 Beta-diversity analysis and comparison of variation in microbiota in the three groups using the linear discriminant analysis effect size online tool.
A: Principal component analysis on the relative abundance. Each point represents a sample, plotted by the second principal component on the Y-axis and the first principal component on the X-axis and colored by group; B: Comparison of the sample distribution of different subgroups using weighted non-metric multidimensional scaling analysis. Each sample is represented by a dot; C: Partial least squares discrimination analysis. Each point represents a sample; D: Histogram of the linear discriminant analysis (LDA) scores for differentially abundant genera between groups (a logarithmic LDA score > 3 indicated a higher relative abundance in the corresponding group compared to the other group); E: The taxonomic cladogram obtained from the LDA effect size analysis of 16S sequences and taxonomic representation of statistically significant differences between groups. The diameter of each circle is proportional to the taxon abundance. LDA: Linear discriminant analysis; IT: Immune-tolerant; IA: Immune-active; H: Healthy.
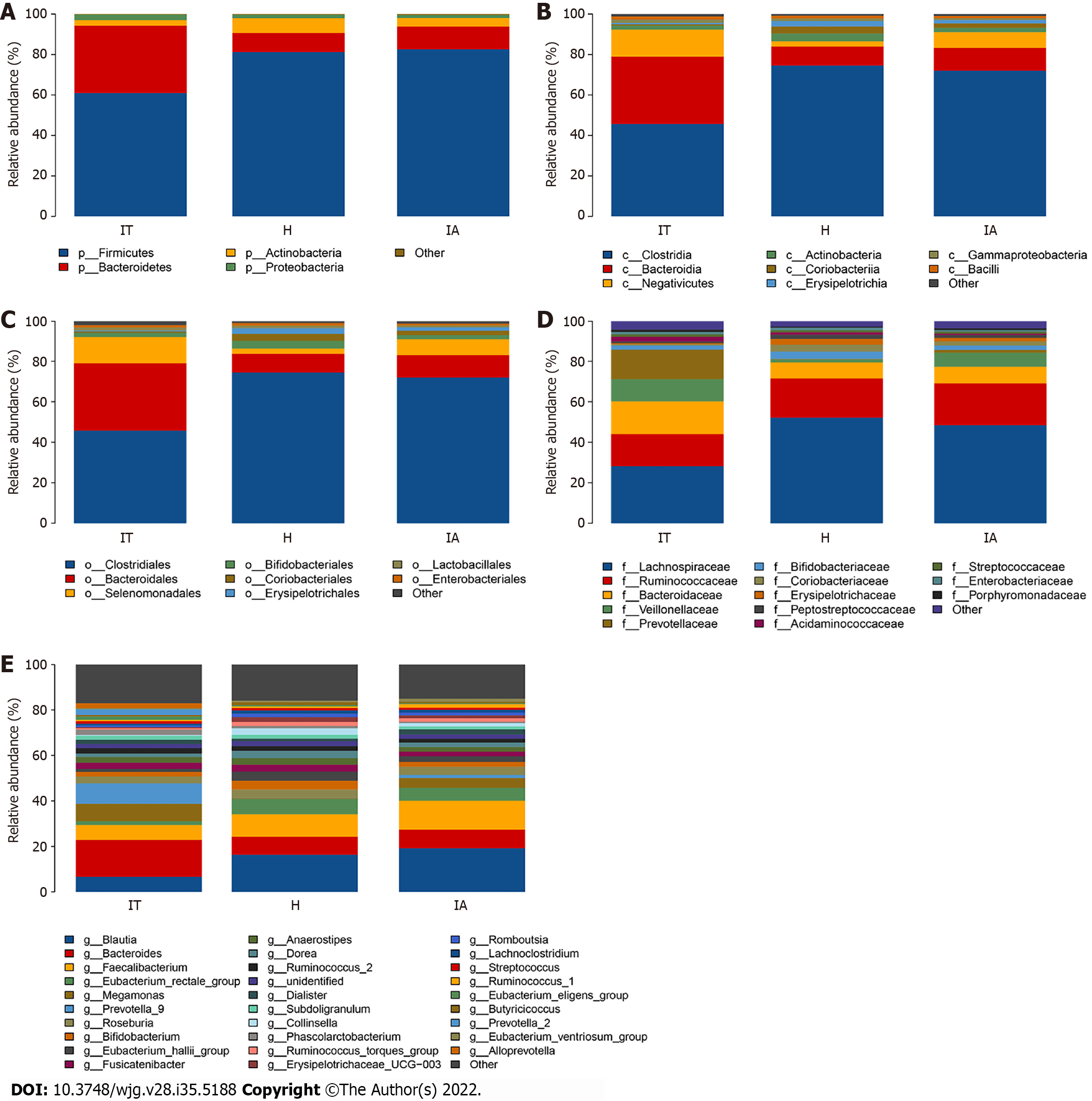
Figure 3 Distribution of the predominant bacteria at different taxonomic levels (phylum, class, order, family, and genus).
A-E: Stacked bars of the phylum, class, order, family, and genus level in healthy controls, immune-tolerant phase hepatitis B virus infection, and immune-active phase hepatitis B virus infection. IT: Immune-tolerant; IA: Immune-active; H: Healthy.
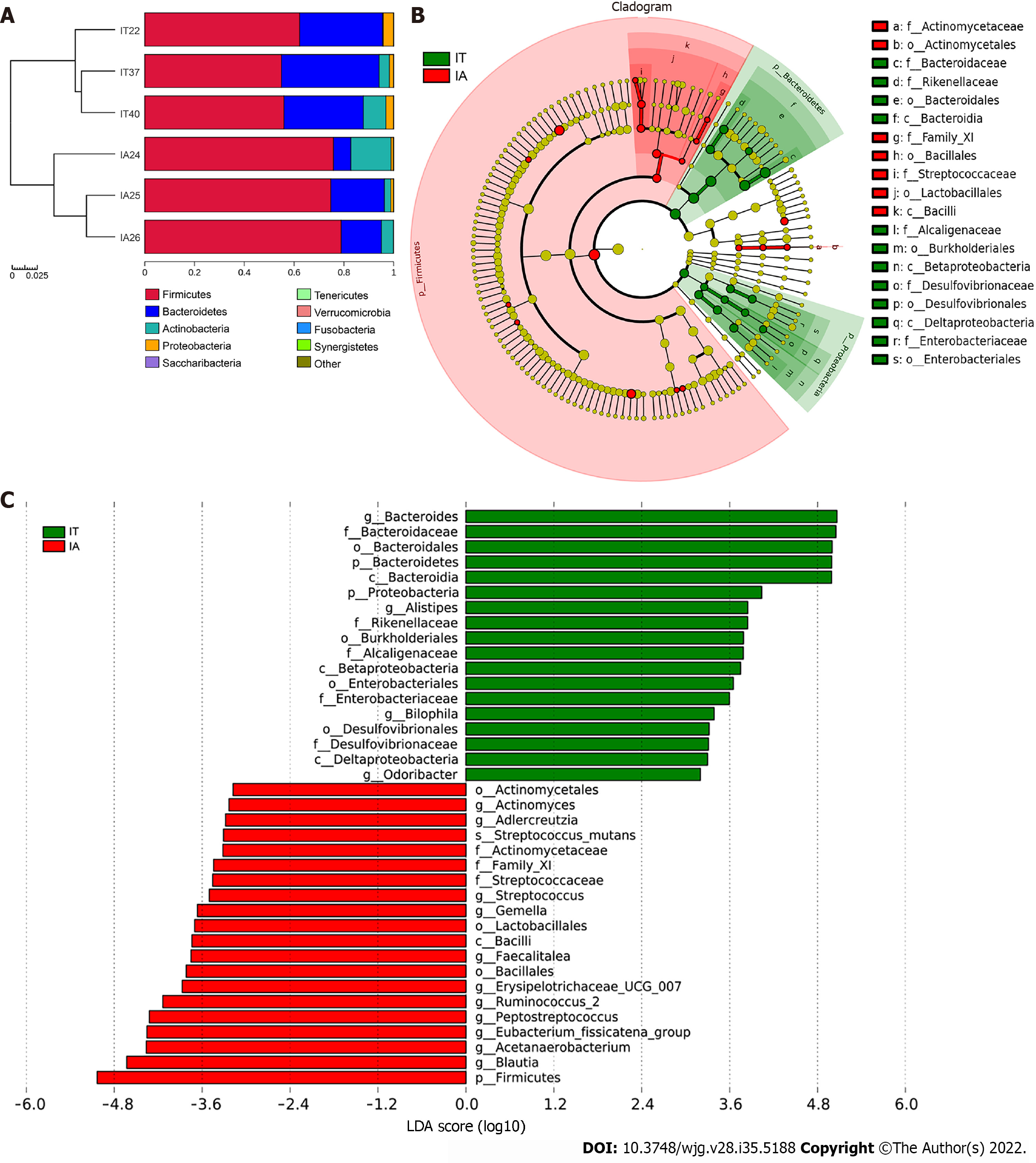
Figure 4 Comparison of variations in the microbiota of 3 patients before and after disease progression using the linear discriminant analysis effect size online tool.
A: Distribution of the predominant bacteria at the phylum level in the 3 patients in the immune-tolerant and immune-active phases; B: The taxonomic cladogram obtained from linear discriminant analysis effect size analysis of 16S sequences and taxonomic representation of statistically significant differences between the two groups. The diameter of each circle is proportional to the taxon abundance; C: Histogram of the linear discriminant analysis scores for differentially abundant genera between the two groups (a logarithmic linear discriminant analysis score > 3 indicated a higher relative abundance in the corresponding group compared to the other group). LDA: Linear discriminant analysis; IT: Immune-tolerant; IA: Immune-active.
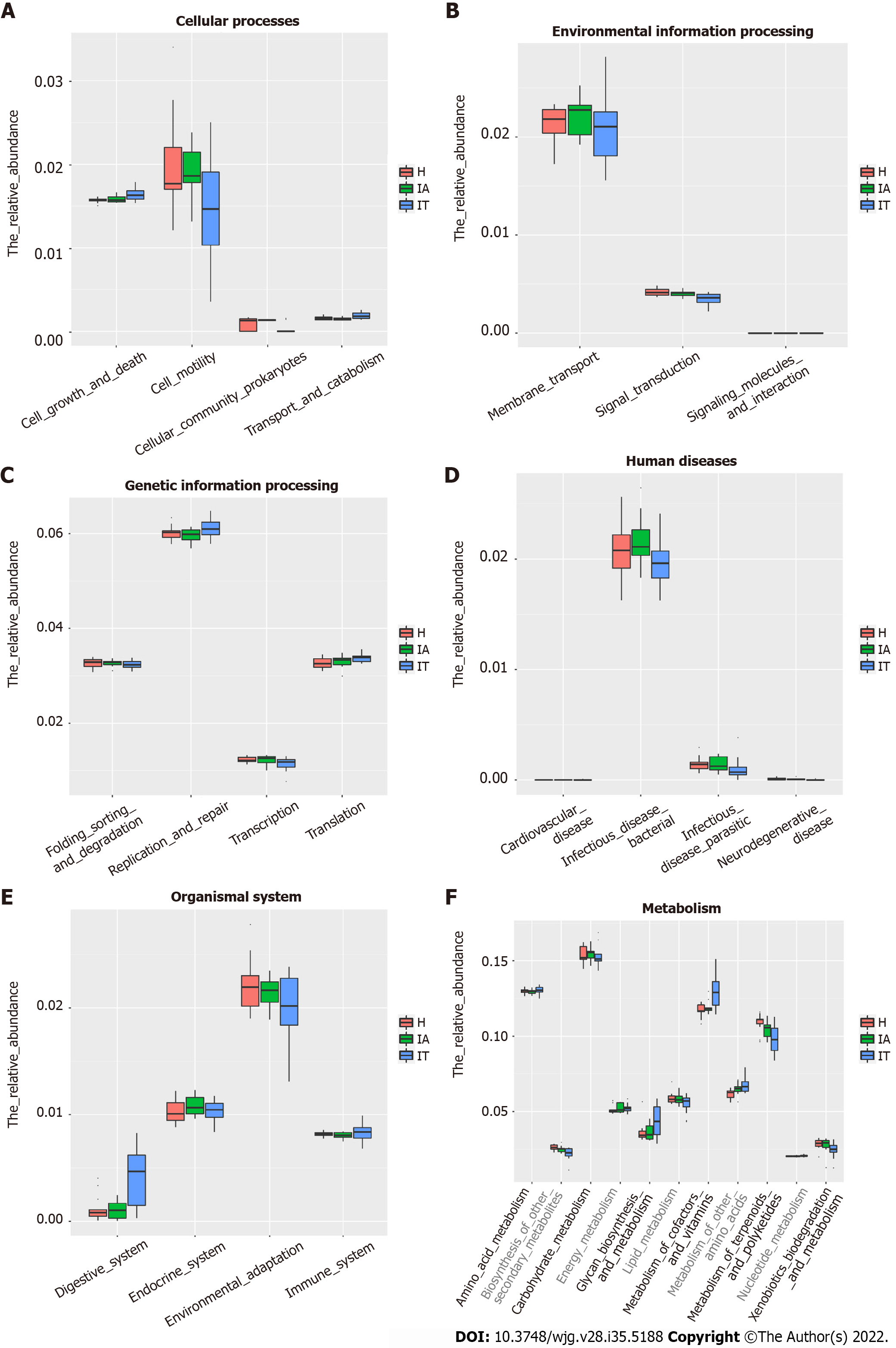
Figure 5 Alteration of the predicted microbial functional composition from the 16S rDNA sequencing data, analyzed using Phylogenetic Investigation of Communities by Reconstruction of Unobserved States.
A and B: Representing the differences at the cellular processes among healthy and patients in the immune-tolerant and immune-active phases; C and D: Representing the differences at the genetic information processing and human diseases among healthy and patients in the immune-tolerant and immune-active phases; E and F: Representing the differences at the organismal systems and metabolism among healthy and patients in the immune-tolerant and immune-active phases. IT: Immune-tolerant; IA: Immune-active; H: Healthy.
- Citation: Li YN, Kang NL, Jiang JJ, Zhu YY, Liu YR, Zeng DW, Wang F. Gut microbiota of hepatitis B virus-infected patients in the immune-tolerant and immune-active phases and their implications in metabolite changes. World J Gastroenterol 2022; 28(35): 5188-5202
- URL: https://www.wjgnet.com/1007-9327/full/v28/i35/5188.htm
- DOI: https://dx.doi.org/10.3748/wjg.v28.i35.5188