Copyright
©2010 Baishideng.
World J Gastroenterol. Apr 28, 2010; 16(16): 1953-1969
Published online Apr 28, 2010. doi: 10.3748/wjg.v16.i16.1953
Published online Apr 28, 2010. doi: 10.3748/wjg.v16.i16.1953
Figure 1 Summary of sequence changes detected over time within the hepatitis C virus (HCV) non-structural 3 (NS3) fragment of a single patient.
HCV RNA extracted from patient sera at months 12, 28 and 63 was amplified by reverse transcription polymerase chain reaction (RT-PCR). A panel of molecular clones spanning the NS3 fragment was aligned with the wild-type consensus sequence which is shown on top (underlined). From a total of 51 nucleotide substitutions found, 39 were nonsynonymous, causing amino acid alterations (capital letters). The amplified NS3 fragment encompasses epitopes NS3 1073-1081, 1131-1139 and 1169-1177. Dashed lines indicate identity to the consensus sequences, whereas positions of synonymous mutations not causing amino acid alterations are marked with the greek delta sign (Δ).
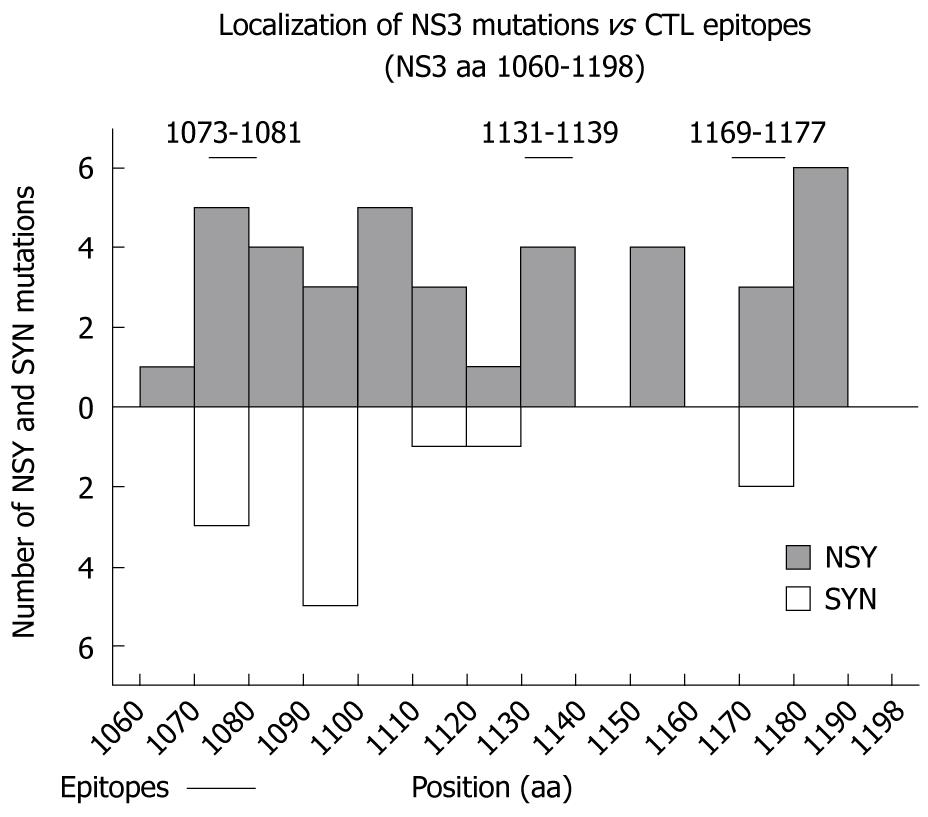
Figure 2 Localization of NS3 mutations in the proximity of three cytotoxic T lymphocyte (CTL) epitopes.
Sequence alterations within the NS3 region between amino acid (aa) 1060-1198 are graphically visualized. Nonsynonymous (top) and synonymous (bottom) mutations are shown separately. NS3 CTL epitope regions 1073-1081, 1131-1139 and 1169-1177 are indicated.
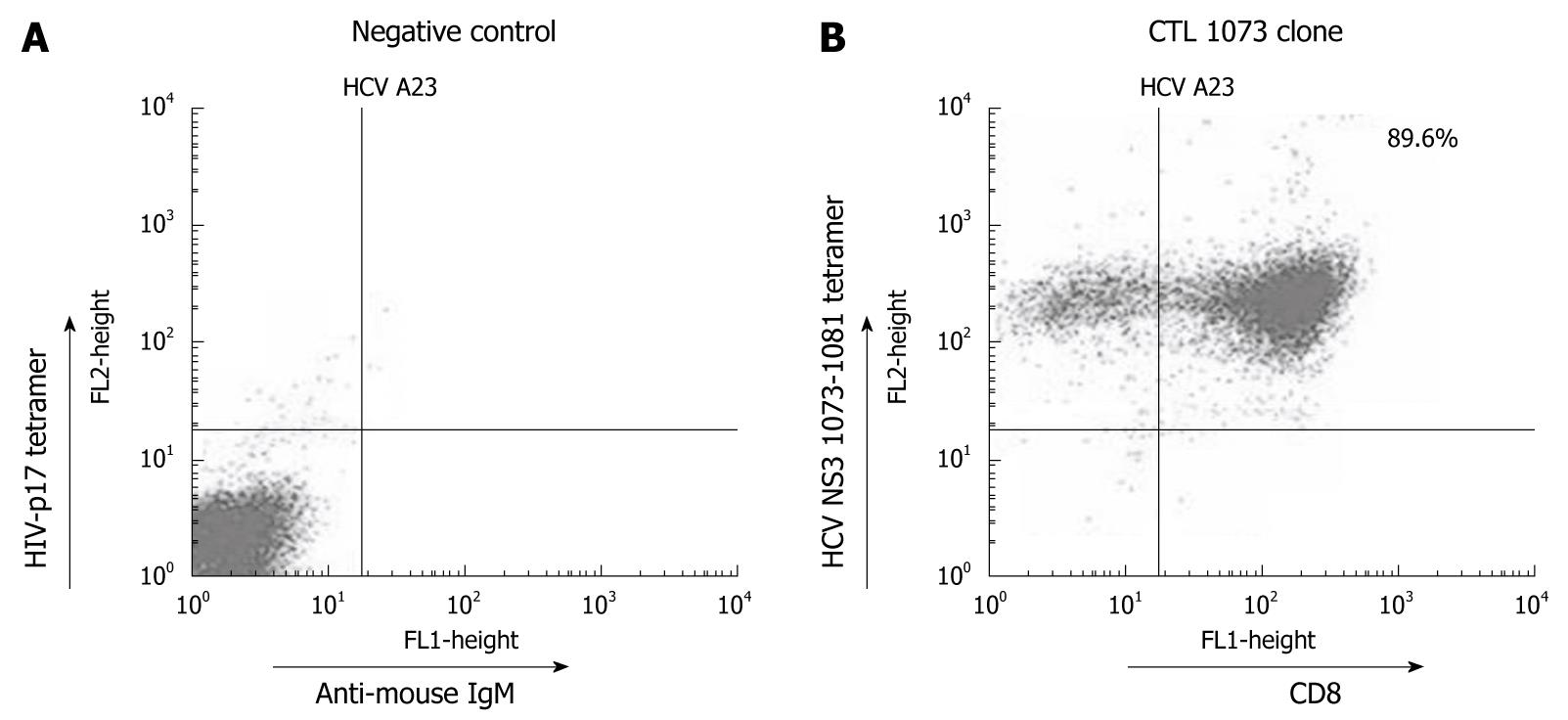
Figure 3 HCV NS3 1073-1081 tetramer staining using a CTL clone specific for wt1073.
A: As a negative control, CTL clone 1073 was stained with the HIV-p17 tetramer reagent and anti-mouse IgM FITC. As expected, no cell shift was observed; B: The same CTL clone was stained with the HCV NS3 1073-1081 tetramer and anti-CD8+-FITC. A positive HCV NS3 1073-1081 tetramer staining is shown with tetramer/CD8+ cells located in the upper right quadrant. The origin of cells seen in the upper left quadrant could not be identified and need further investigation. All staining experiments were repeated three times with similar results. Identical results were obtained with HCV NS3 1073-1081 tetramer staining using other CTL clones for wt1073 (data not shown).
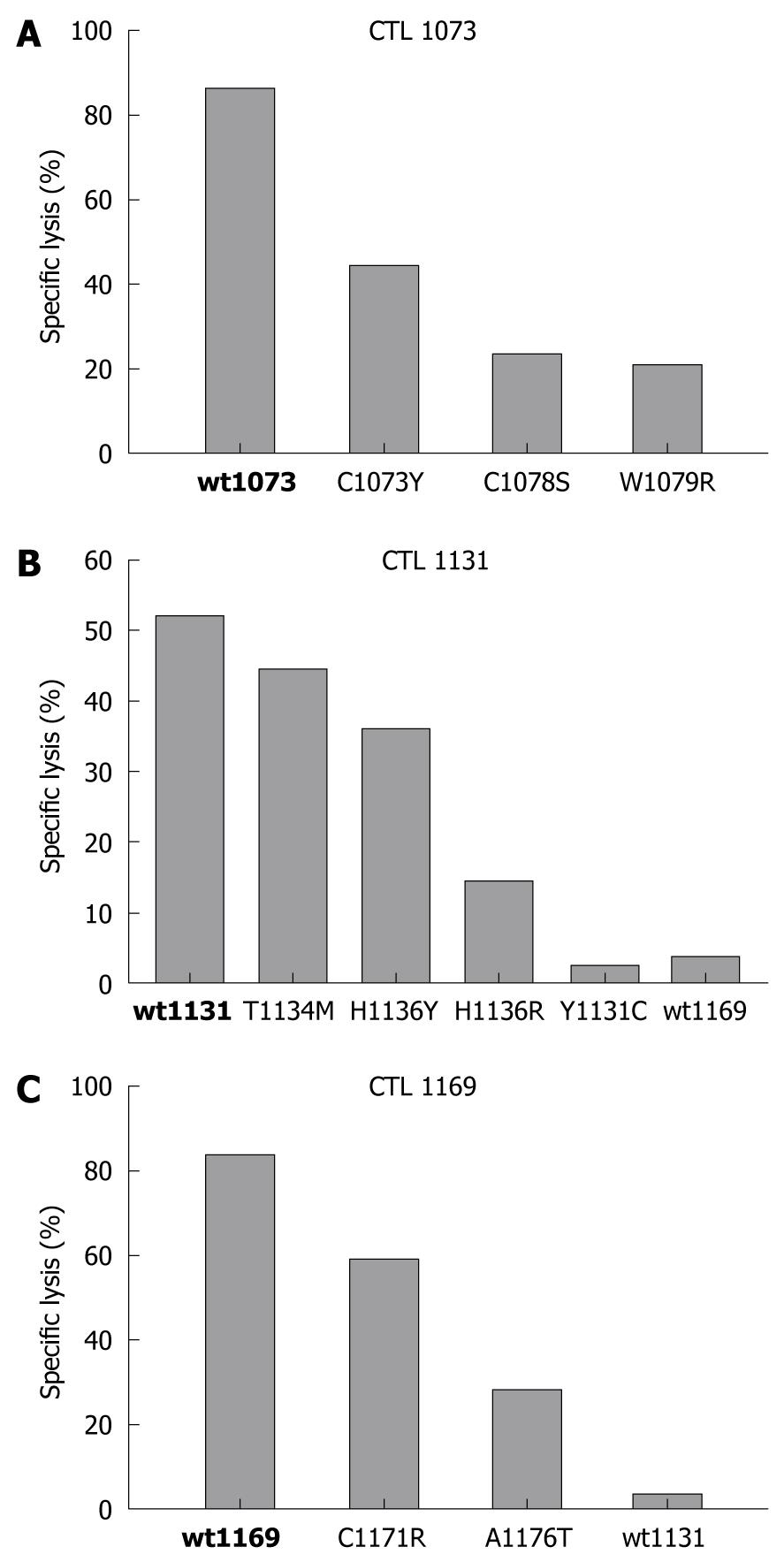
Figure 4 Effect of amino acid changes on epitope-specific CD8+ CTL responses.
The relative ability of synthetic peptides corresponding to the indicated wild type (bold) and variant epitope sequences to sensitize autologous target cell lines for lysis by CTL clone 1073 specific for wt1073 (A), clone 1131 specific for wt1131 (B) and clone 1169 specific for wt1169 peptide (C) was determined using in vitro cytotoxicity assays. The results shown are the specific lysis (%) of target cells incubated with each wild type (bold) and variant peptide at a concentration of 10 nmol/L. As expected, the highest cytotoxicity was found against wild-type peptides followed by various degrees of lysis for epitope variants. Target cells used in these assays were prepared from the B cell line (L.B3019) with an effector to target ratio of 20:1.
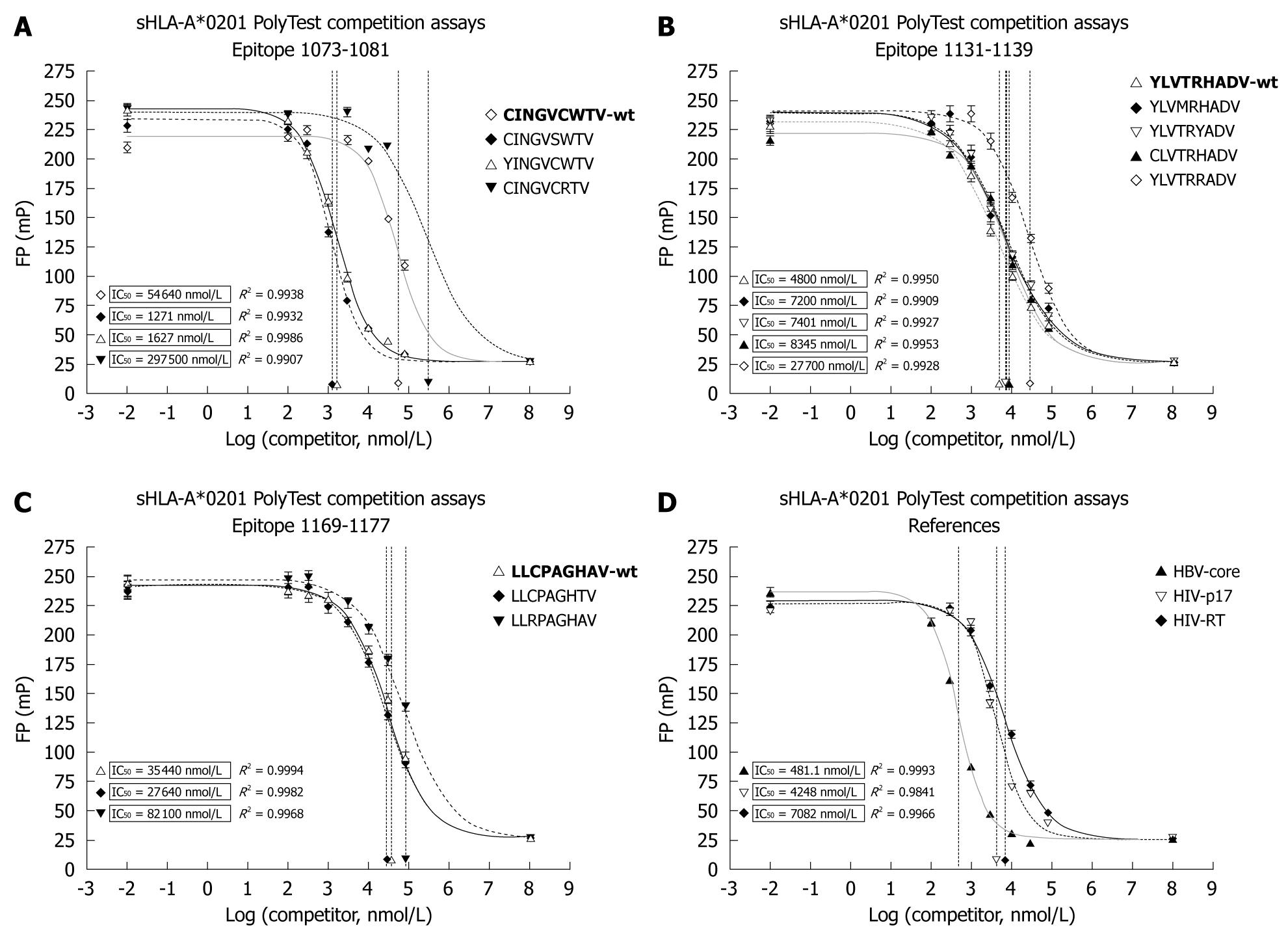
Figure 5 Soluble human leukocyte antigen (HLA)-A*0201 competition assays determining the binding capacity (IC50) of natural HCV epitope variants.
The affinity of three wild type HCV peptide epitopes (bold) wt1073 (A), wt1131 (B), wt1169 (C) derived from three locations within the NS3 region and their variants C1073Y, C1078S and W1079R (A), Y1131C, T1134M, H 1136Y and H1136R (B), C1171R and A1176T (C) was determined. Control peptides HBV-Core, HIV-p17 and HIV-reverse transcriptase (RT) were included to provide reference values to other viral systems. Their affinities are presented in graph (D). For each experiment, 8 serial dilutions (100 nmol/L-80 μmol/L) of the unlabeled viral peptide were used to compete against a FITC-labeled tracer peptide. After reaching equilibrium, IC50 values for all peptides were determined by fitting the data to a dose-response model using the software Prism. R2 values indicate the goodness of fit.
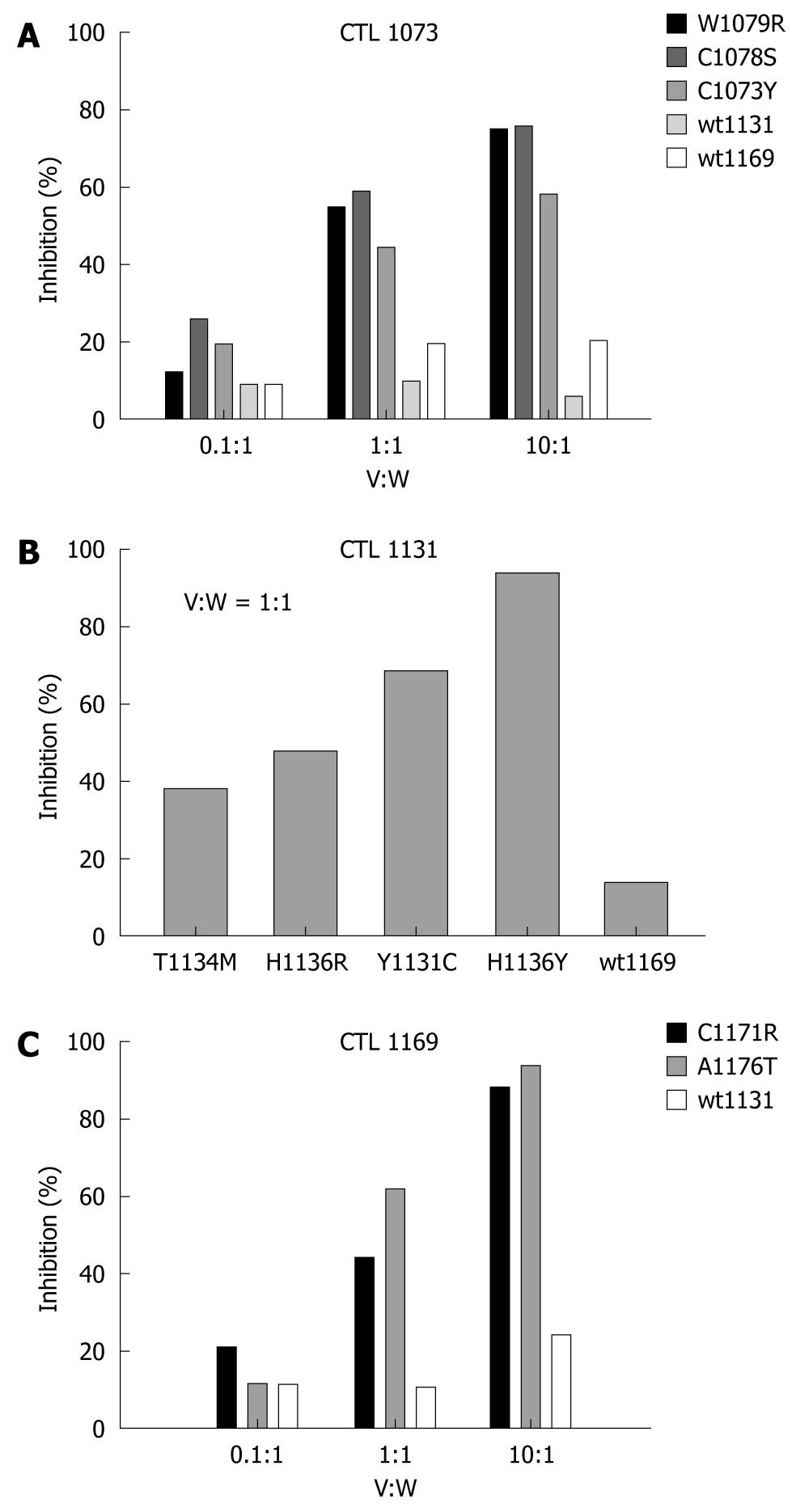
Figure 6 Antagonistic effect of NS3 peptide variants inhibiting specific cytolytic activity of specific CTL clones.
A detailed inhibition profile is shown for CTL clones 1073 (A) and 1169 (C) in which target cells were pre-pulsed with 10 nmol/L of wild-type peptide, and then incubated at indicated ratios of variant to wild-type peptide concentrations (V:W). Wild-type peptides wt1131 and wt1169 were used as negative controls for experiments using CTL clone 1073, where only a single control (wt1131) was used for tests involving CTL clone 1169. Both data sets were determined at effector to target ratios of 20:1. Because of lack of material, a more simplified profile is shown for CTL clone 1131 (B) testing only a V:W ratio of 1:1 with wt1169 as negative control. All experiments were performed in duplicates.
- Citation: Wang S, Buchli R, Schiller J, Gao J, VanGundy RS, Hildebrand WH, Eckels DD. Natural epitope variants of the hepatitis C virus impair cytotoxic T lymphocyte activity. World J Gastroenterol 2010; 16(16): 1953-1969
- URL: https://www.wjgnet.com/1007-9327/full/v16/i16/1953.htm
- DOI: https://dx.doi.org/10.3748/wjg.v16.i16.1953