Copyright
©2009 The WJG Press and Baishideng.
World J Gastroenterol. Nov 21, 2009; 15(43): 5377-5396
Published online Nov 21, 2009. doi: 10.3748/wjg.15.5377
Published online Nov 21, 2009. doi: 10.3748/wjg.15.5377
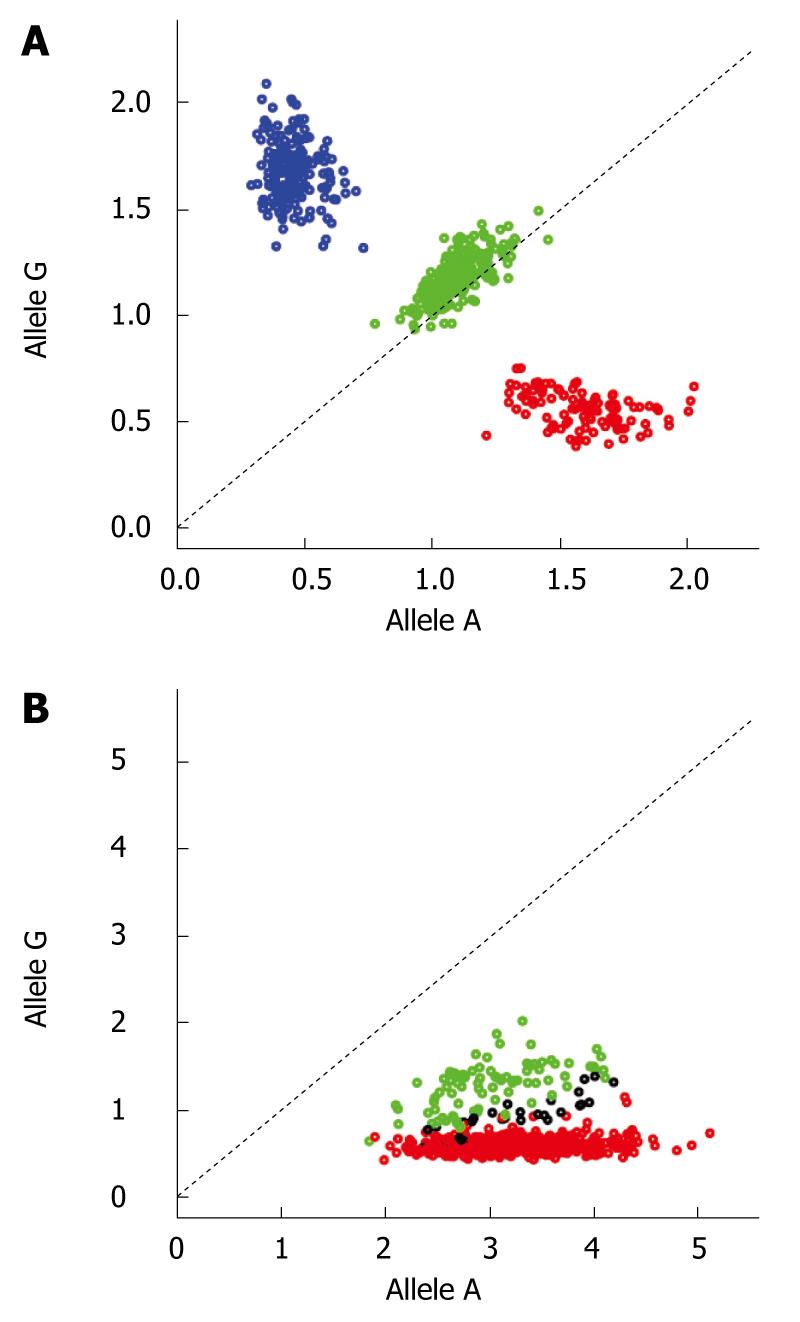
Figure 1 Example of cluster plots for two SNPs.
Plot A shows the plotting of the normalized intensity values for a SNP with good clustering. Each color represents the respective genotype (blue for GG, green for AG and red for AA). Plot B demonstrates the cluster plot for a SNP with bad clustering (same color coding as in A). Disease-associated SNPs demonstrating cluster plots as in B should be discarded as the significant associations at such SNPs are most likely technical artifacts.
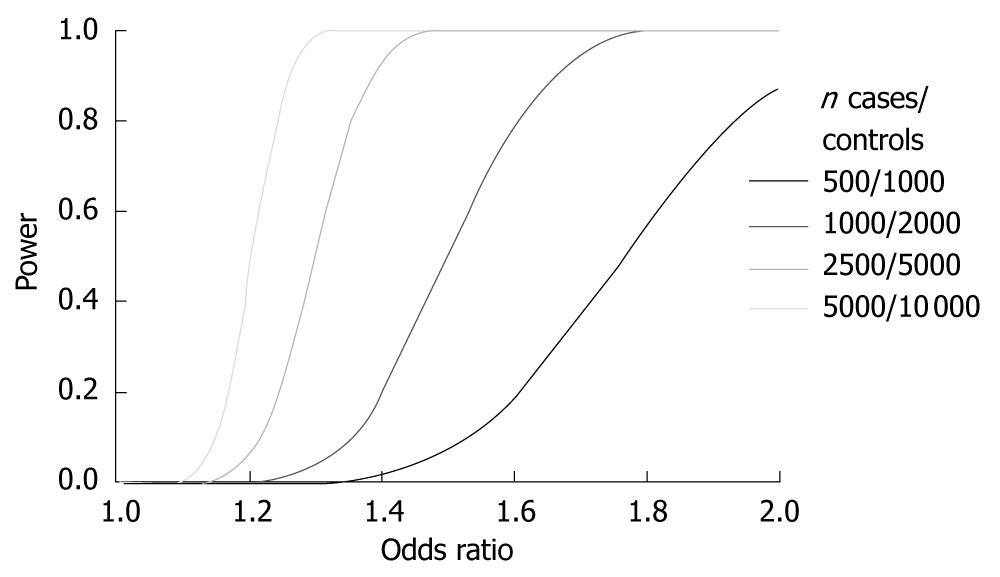
Figure 2 Power calculations for different case-control study panel sizes.
Power calculations for different case-control study panel sizes using an allelic based association test and a P-value of P < 5 × 10-7. All calculations assume a minor allele frequency of 30% and 1:2 ratio of cases vs controls.
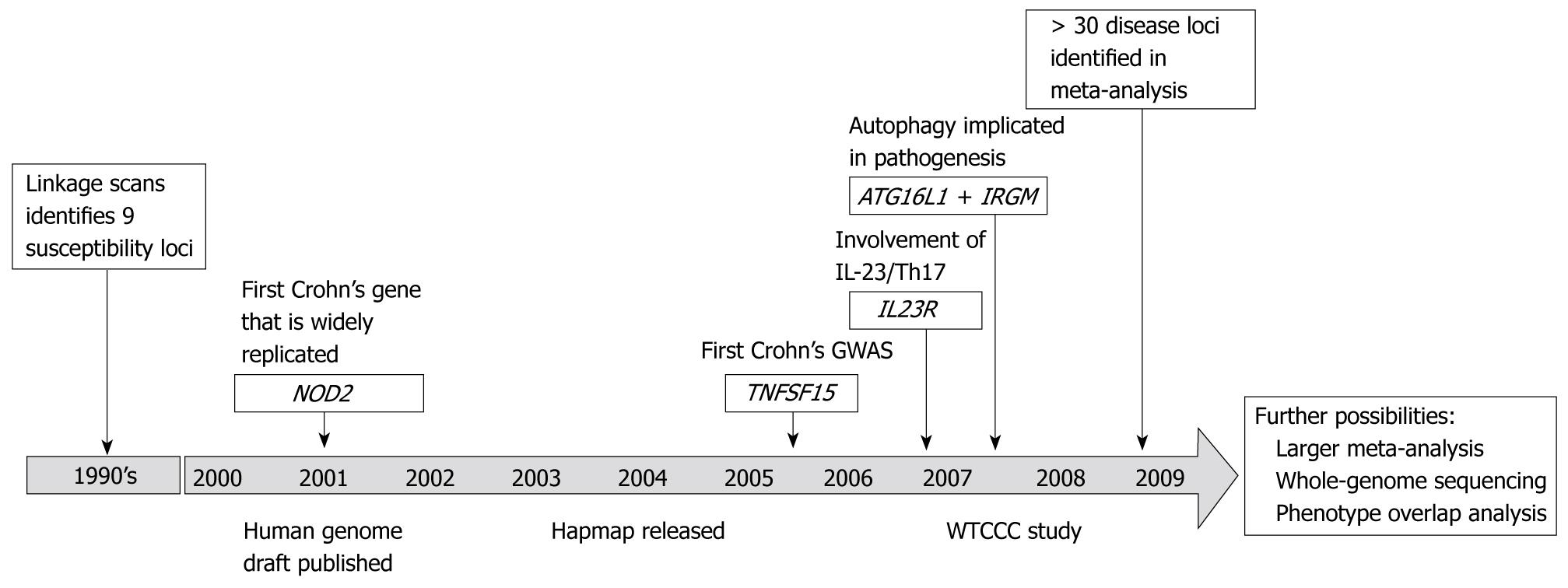
Figure 3 Historical milestones in Crohn’s disease genetics.
Developments in the knowledge of the genetics of Crohn’s disease. Only milestone gene discoveries are shown. With the publication of the Crohn’s disease meta-analysis in 2009 the number of replicated loci is now greater than 30.
- Citation: Melum E, Franke A, Karlsen TH. Genome-wide association studies - A summary for the clinical gastroenterologist. World J Gastroenterol 2009; 15(43): 5377-5396
- URL: https://www.wjgnet.com/1007-9327/full/v15/i43/5377.htm
- DOI: https://dx.doi.org/10.3748/wjg.15.5377