Published online Jun 28, 2022. doi: 10.37126/aige.v3.i3.31
Peer-review started: January 17, 2022
First decision: March 8, 2022
Revised: March 24, 2022
Accepted: June 20, 2022
Article in press: June 20, 2022
Published online: June 28, 2022
Processing time: 161 Days and 22.9 Hours
Colorectal cancer (CRC) is a heterogeneous illness characterized by various epigenetic and microenvironmental changes and is the third-highest cause of cancer-related death in the US. Artificial intelligence (AI) with its ability to allow automatic learning and improvement from experiences using statistical methods and Deep learning has made a distinctive contribution to the diagnosis and treatment of several cancer types. This review discusses the uses and application of AI in CRC screening using automated polyp detection assistance technologies to the development of computer-assisted diagnostic algorithms capable of accurately detecting polyps during colonoscopy and classifying them. Furth
Core Tip: Artificial intelligence (AI) and its potential in diagnosing colorectal cancer have been the subject of various reviews in the literature. However, this review reports the most recent discoveries and studies on artificial and machine learning in colorectal cancer screening, diagnosis, and treatment, as well as the future roles that AI applications may play in assisting in the treatment of colorectal cancer. Furthermore, this review talks about prospects and constraints for the use of AI systems, as well as the need for large-scale randomized clinical trials to examine AI algorithms before they can be implemented.
- Citation: Awidi M, Bagga A. Artificial intelligence and machine learning in colorectal cancer. Artif Intell Gastrointest Endosc 2022; 3(3): 31-43
- URL: https://www.wjgnet.com/2689-7164/full/v3/i3/31.htm
- DOI: https://dx.doi.org/10.37126/aige.v3.i3.31
In the United States, the third leading cause of cancer-related deaths is colorectal cancer (CRC)[1]. Since 1980, the number of people diagnosed with colon or rectal cancer has decreased due to improved screening guidelines and lifestyle-related risk factors modification. In addition, treatments for colorectal cancer have improved over the last few decades[2]. CRC is a diverse group of diseases with differences in epidemiology, histology, genomics, and host immune responses[3,4]. Recognizing the diversity of the disease, and the importance of personalized medicine, machine learning models have been utilized to improve detection rates, diagnosis, and treatment of CRC.
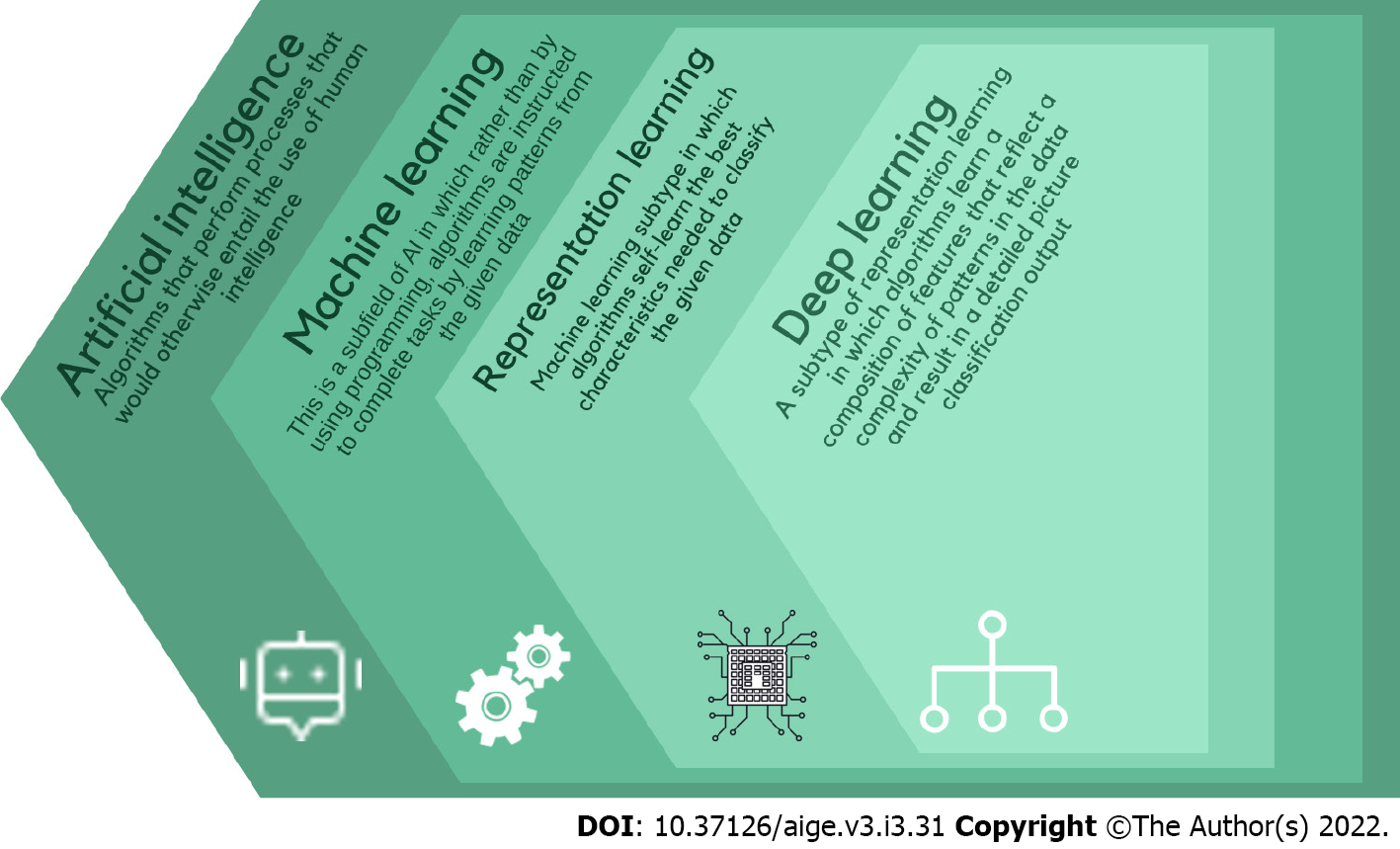
Artificial intelligence (AI) is a computer science field dedicated to developing systems capable of performing tasks that typically require human-level intelligence[5]. It is a broad term used to encompass Machine learning (ML), a subset of AI algorithms that allows automatic learning and improvement from experiences using statistical methods and deep learning which imitates higher level human data processing by using multi-layered neural networks for extractions and self-training algorithms[6] (Figure 1).
The increased utilization of this novel technology has made a distinctive contribution to the diagnosis and treatment of several cancer types. From AI models to reduce rates of missed adenomas to novel computer assisted drug delivery techniques and robotic surgery colorectal carcinoma treatment entered a new area rapidly moving towards precision and personalized medicine[7,8].
Our review aims to analyze the AI uses and application in CRC screening, diagnosis, and treatment. In addition, we will discuss potential future directions and limitations for the use of AI systems.
Colorectal screening remains the gold standard for improving patient clinical outcomes, such as avoiding treatment delays and lowering CRC morbidity and mortality[9]. CRC patients are diagnosed at advanced stages of the disease in 60%–70% of cases[9].
It is thought that the alterations from the normal mucosa to malignant state lesion take almost 10 to 20 years[10]. Colonoscopy, flexible sigmoidoscopy, and less invasive capsule endoscopy, computed tomography chorography, blood in stool tests, fecal immune-chemical testing, and multi-target cell DNA testing are just a few of the screening options available for CRC[11,12]. Colonoscopy is the gold standard screening test, though it is not without flaws[13]. It has been reported that around 9% of cases of CRC occurred within three years following a negative colonoscopy[14]. Adenoma detection rates are very variable with reported detection rates of 7% to 50%[15]. The wide range of detection rates is due to different factors, including endoscopic procedural experience, pre-procedure bowel preparation, time of procedure termination, use of sedation, flexure visualization, image enhanced endoscopy, and the presence of flat or diminished polyps[16,17].
The growing interest of AI in CRC yielded automated polyp detection assisted technology to aid in the detection and diagnosis of polyps during colonoscopy[5]. In addition, technologies that use deep learning techniques to improve detection rates and localize premalignant lesions are available and being applied[18].
A recent randomized controlled trial studied the effect of computer aided detection deep learning models on polyps and adenoma detection rates. The trial randomized 1058 patients to either conventional colonoscopy (n = 536) or colonoscopy with computer aided detection system (n = 522). In the computer aided detection system group there was an increase in both the adenoma detection rates, 29.1% vs 20.3%, P < 0.001, in addition to the mean number of identified adenomas per patient, 0.53 vs 0.31, P < 0.001, in comparison to the group assigned standard colonoscopy. This trial, however, did not reveal a significant statistical difference for the detection of large adenomas between the groups (77 vs 58, P = 0.075). Interestingly, the computer aided detection system arm had more hyperplastic adenomas (114 vs 52, P < 0.001) and diminutive polyps (185 vs 102, P < 0.001) identified. This study demonstrates the impact of AI-assisted colonoscopy technologies on the detection of small polyps that even highly trained endoscopists may miss[19].
Karkanis et al[20] used color and texture analysis of mucosal surfaces based on color wave covariance features were used to develop a computer-assisted diagnostic algorithm for automatic polyp identification. Rather than a real-time recognition system, the system was able to identify precancerous lesions in static endoscopic images. It accomplished that by examining frame images extracted from 60 colonoscopy video sequences containing small polyps with a sensitivity and specificity of 99.3% and 93.6% respectively.
In a study to evaluate deep learning algorithms for automated polyp detection during colonoscopy using colonoscopy images, colonoscopy videos obtained from four different datasets resulted a significant improvement in real-time colonoscopy video analysis byprocessing at least 25 frames per second with a latency of 76.8 milliseconds[65].
A recent systematic review and meta-analysis that included 48 studies showed a significant increase in both polyp detection rates [odds ratio (OR) 1.75, 95%CI 1.56-1.96; P < 0.001] as well as adenoma detection rates (OR 1.53, 95%CI 1.32-1.77; P < 0.001) patients who had a colonoscopy with AI compared to those who did not[21].
Recognizing that colonoscopy is a highly operator-dependent procedure, challenges such as light conditions, morphology of colorectal polyps during colonoscopy, and size could be overcome by AI computer assisted diagnostic systems as they serve as an “extra pair of eyes” and improve adenoma detection rates.
Several alternative screening tools to conventional colonoscopy have been developed. A modified computed tomography (CT) examination known as virtual colonoscopy or computed tomographic colonography (CTC) was first described in 1994[22]. Its ability to evaluate the entire colorectum, rapid acquisition of imaging, and lack of sedation makes it a valuable alternative for certain patients. The effectiveness of CTC in detecting asymptomatic colorectal lesions is still a point of contention. Several studies reported identification of 90 percent of patients with asymptomatic adenomas or cancers (≥ 10 mm in diameter) using CT colonography[23,24]. AI-based algorithm concepts have been used to obtain optimal diagnostics standards and image qualities to aid in CRC detection and diagnosis using CTC. Grosu et al[25] developed a machine learning method that had an area under the curve (AUC) of 0.91, a sensitivity of 82%, a specificity of 85% in differentiating between benign and precancerous lesions in average risk asymptomatic patients using CTC. In another study, Song et al[26] developed a virtual pathological model to see if image high-order differentiations (curvature and gradient) could be used to distinguish colorectal lesions (neoplastic and non-neoplastic). The results revealed an improvement of receiver operating characteristic (ROC) curve (AUC) from 0.74 (Using image intensity alone) to 0.85 (Using texture features from high-order differentiations).
In cases of incomplete colonoscopy or when evaluating the small intestines, capsule endoscopy (CE) is used as a minimally invasive technique. It acquires images as it passes through the gastrointestinal tract[27]. Hence, CE can be affected by laxative use. In addition, it requires manual interpretation and analysis of acquired images which is particularly time consuming[28,29]. AI-based systems are being used to automate the reading and examination of the results to reduce the time and the human error inherently present when reading images thereby improving adenoma detection rates[30,31]. Novel algorithms were developed to match CE and colonoscopy-identified polyps based on their size, morphology and location as well as utilizing deep convolutional neural networks for automatic colorectal polyp detection. When compared to the manual process of polyp detection, localization had a high sensitivity (97.1%), accuracy (96.4%), and specificity (93.3%) for identifying polyps[30].
Blood-based screening approaches have been developed to detect CRC at early stages. Demographic characteristics and blood test results such as complete blood count (CBC), which may indicate iron deficiency, microcytic anemia, or elevated red cell distribution width are frequently used to evaluate the risk of developing CRC[32-34]. An AI-assisted prediction model (MeScore®, Calgary, Alberta, Canada) was designed to identify people at high risk for CRC using parameters such as age, sex, and CBC data collected 3 to 6 mo prior to cancer diagnosis. A study using this AI-assisted prediction model revealed a 2.1-fold increase in cancer detection rates when the model is used in combination with FOBT[35]. Furthermore, a study using CellMax (CMx®) platform to detect and isolate circulating tumor cells in peripheral blood samples resulted in a sensitivity and specificity of 80%[36]. Table 1 highlights studies focusing on screening.
| Ref. | Objective | Results |
| Wang et al[19], 2019 | Effect of computer aided detection deep learning models on polyps and adenoma detection rates | Increase in adenoma detection rates [29.1% vs 20.3%, P < 0.001] and mean number of identified adenomas per patient [0.53 vs 0.31, P <0.001]; More hyperplastic adenomas (114 vs 52, P < 0.001) and diminutive polyps (185 vs 102, P < 0.001) identified |
| Nazarian et al[20], 2021 | Detection rates of polyp and adenoma with AI vs without AI | Increase in both polyp detection rates (odds ratio [OR] 1.75, 95%CI 1.56-1.96; P < 0.001) as well as adenoma detection rates (OR 1.53, 95%CI 1.32-1.77; P < 0.001) |
| Johnson et al[23], 2008; Pickhardt et al[24], 2003 | Degree to which CTC is effective in detecting asymptomatic colorectal lesions | Reported identification of 90% of patients with asymptomatic adenomas or cancers (≥ 10 mm in diameter) using CT colonography |
| Grosu et al[25], 2021 | Development of machine learning method differentiating between benign and precancerous lesions in average risk asymptomatic patients using CTC | Sensitivity of 82%, specificity of 85% and AUC of 0.91 |
| Song et al[26], 2015 | Development of virtual pathological model to assess the suitability of using image high-order differentiations to distinguish colorectal lesions | Improvement of ROC curve (AUC) from 0.74 to 0.85 |
| Blanes-Vidal et al[30], 2019 | Algorithms developed to match CE and colonoscopy-identified polyps based on their estimated size, morphology and location as well as utilizing deep convolutional neural networks for automatic colorectal polyp detection | Localization resulted in high sensitivity (97.1%), specificity (93.3%), and accuracy (96.4%) for identifying polyps when compared to the manual process of polyp detection |
| Kinar et al[35], 2017 | AI-assisted prediction model (MeScore®, Calgary, Alberta, Canada) was designed to identify people at high risk for CRC | Revealed a 2.1-fold increase in cancer detection rates when the model is used in combination with FOBT |
| Gupta et al[36], 2019 | Using CellMax (CMx®) platform to detect and isolate circulating tumor cells in peripheral blood samples | A sensitivity and specificity of 80% |
A machine learning algorithm can be trained to identify or differentiate polyps in real time in the field of endoscopy. Techniques for analyzing non-magnified endoscopic images and techniques for cellular imaging at a microscopic level have both been investigated (i.e., optical biopsy). The theory behind these methods is that they will improve polyp detection rates, reduce missed adenomas, and thus lower the risk of CRC. However, the increase in polyp detection rates will lead to an increase in financial burdens on health systems, specifically histopathological departments involved in the analysis of resected tissue. Current research initiatives are geared towards building a computer assisted diagnostic algorithm capable of reliably detecting polyps while also characterizing them as hyperplastic or adenomatous during colonoscopy[37].
The Preservation and Incorporation of Valuable endoscopic Innovations (PIVI) an American Society of Gastrointestinal Endoscopy program set a threshold of negative predictive value (NPV) > 90% for the development of new endoscopic technologies, such as the optical diagnosis of small colorectal polyps[38].
Many AI applications have been developed to assist endoscopist with the aim of adopting a “diagnose and leave” strategy for hyperplastic polyps and a “resect and discard” strategy for diminutive adenomas[39]. In one study a system was designed to predict the histology of colorectal polyps (adenomatous vs non-adenomatous) by analyzing linked color imaging demonstrated an 83.3% sensitivity, 70.1% specificity, 82.6% positive predictive value (PPV), 71.2% NPV and an accuracy of 78.4% when compared to expert endoscopists[40].
Magnification Endoscopy with Narrow-Band Imaging (NBI), Endocytoscopy, Magnifying Chromoendoscopy, Confocal Laser Endomicroscopy, Laser-Induced Fluorescence Spectroscopy, Autofluorescence Endoscopy, and White Light Endoscopy are example of advanced endoscopic techniques currently used to aid in the detection and diagnosis of polyps.
Magnification Endoscopy with NBI is a imaging system that allows observation of mucosal surfaces and microvascular patterns[41]. It improves the diagnostic accuracy of benign from premalignant lesions by evaluating depth of submucosal lesions[42-44]. Gross et al[45] developed a computer-assisted model for polyp classification by analyzing 9 vessel features, including perimeter and brightness from patients who underwent magnifying endoscopy with NBI. The model had a higher sensitivity (95% vs 86%), specificity (90.3% vs 87.8%) and accuracy (93.1% vs 86.8%) when compared to novice endoscopists however, they are comparable to those of experienced endoscopists (sensitivity, specificity, and accuracy of 93.4%, 91.8% and 92.7%, respectively).
In addition, Chen et al[46] used magnifying NBI images with 284 diminutive colorectal polyps extracted to create a deep learning model to classify diminutive colorectal polyps When compared to expert endoscopists, the algorithm was able to distinguish between neoplastic and hyperplastic lesions in less time (0.45 vs 1.54 s). It had a sensitivity, specificity, accuracy, PPV, and NPV of 96.3%, 78.1%, 90.1%, 89.6%, and 91.5% respectively.
Endocytoscopy is an endoscopic imaging modality, that allows in vivo microscopic imaging and real-time diagnosis of cellular structures at high magnifications (400× magnification power in endoscope-based to 1400× magnification in probe-based endocytoscopy) during colonoscopy[47]. A computer-aided algorithm was designed to histologically differentiate colorectal lesions in vivo using endocytoscopy[48]. Initially, this model used nuclear features (area, standard deviation of area, circularity, circularity of the 20 largest nuclei, shortest and longest diameter) after nuclear segmentation from the endocytoscopic images with a 92% sensitivity and 89.2% accuracy in establishing a histological diagnosis. This model was later improved by extracting features from texture analysis and utilizing SVM to classify benign, adenomatous lesions or invasive carcinoma[49,50]. Another model looked at the role of a computer-aided endocytoscopy system in the diagnosis of invasive colorectal carcinoma, and found that it had 89.4% sensitivity, 98.9% specificity, 98.8% positive predictive value, 90.1 percent negative predictive value, and 94.1 percent accuracy[51].
Magnifying Chromoendoscopy is a technique that uses dye to inspect and analyze the pit patterns of the polyp surfaces resulting in high diagnostic performance (97.8% sensitivity, 91.4% specificity and 97.1% accuracy) when performed by expert endoscopists[52]. Takemura et al[53] created a software model to automatically quantify and classify pit patterns. They used texture and quantitative analysis (area, perimeter, and circularity) to classify pit patterns. Using this model type I and II pit patterns were in complete agreement with the endoscopic diagnosis on discriminant analysis. Type III was found in 29 of the 30 cases (96.7%), while type IV was found in one. Type IV pit pattern was found in 29 of the 30 cases (96.7%). The computerized recognition system's overall accuracy was 132 out of 134 (98.5%).
Confocal Laser Endomicroscopy is a microscopic imaging modality that allows in vivo examination of cellular and subcellular structures at 1000× magnification power[54]. Andréet al[55] used an automated polyp characterization system to distinguish between benign and malignant lesions using the k-nearest neighbor classification with an accuracy of 89.6%. A neural network analysis algorithm had an accuracy of 84.5% in differentiating advanced colorectal adenocarcinomas from normal mucosa[56]. Algorithms using Confocal Laser Endomicroscopy are yet to be validated in randomized clinical trials.
Autofluorescence imaging endoscope characterizes colorectal polyps by analyzing different color emissions of tissue after exposure to a light source. It has shown promising results in differentiating non-neoplastic from neoplastic lesions during colonoscopy[57,58].
White light endoscopy and laser-induced fluorescence spectroscopy technologies have been tested as potential models to discriminate between neoplastic and non-neoplastic lesions with results that were inferior to NBI or chromoendoscopy with or without magnification[59,60]. Table 2 summarized relevant diagnostic research.
| Ref. | Objective | Results |
| Min et al[40], 2019 | System designed to predict the histology of colorectal polyps by analyzing linked color imaging | 83.3% sensitivity, 70.1% specificity, 82.6% PPV, 71.2% NPV and an accuracy of 78.4% when compared to expert endoscopists |
| Gross et al[45], 2011 | Development of computer-assisted model for polyp classification by analyzing 9 vessel features, from patients who underwent magnifying endoscopy with NBI | Higher sensitivity (95% vs 86%), specificity (90.3% vs 87.8%) and accuracy (93.1% vs 86.8%) when compared to novice endoscopists but comparable to those of expert endoscopists (sensitivity, specificity, and accuracy of 93.4%, 91.8% and 92.7%, respectively) |
| Chen et al[46], 2018 | Designed a deep learning model to classify diminutive colorectal polyps using magnifying NBI images with 284 diminutive colorectal polyps extracted | Able to distinguish between neoplastic and hyperplastic lesions in a shorter period compared to expert endoscopists (0.45 vs 1.54 seconds) and had a sensitivity, specificity, accuracy, PPV, and NPV of 96.3%, 78.1%, 90.1%, 89.6% and 91.5% respectively |
| Mori et al[48], 2015 | Computer-aided algorithm designed to histologically differentiate colorectal lesions in vivo using endocytoscopy | 92% sensitivity and 89.2% accuracy in establishing a histological diagnosis. |
| Takeda et al[51], 2017 | Model investigated the role of a computer-aided endocytoscopy system on the diagnosis of invasive colorectal carcinoma | 89.4% sensitivity, 98.9% specificity, 98.8% PPV, 90.1% NPV and 94.1% accuracy |
| Takemura et al[53], 2010 | Software model to automatically quantify and classify pit patterns. Used texture and quantitative analysis to classify pit patterns | Type I and II pit patterns were in complete agreement with the endoscopic diagnosis on discriminant analysis. Type III was diagnosed in 29 of 30 cases (96.7%) and type IV was diagnosed in one case. Twenty-nine of 30 cases (96.7%) were diagnosed as type IV pit pattern. The overall accuracy of the computerized recognition system was 132 of 134 (98.5%) |
| André et al[55], 2012 | Automated polyp characterization system to distinguish between benign and malignant lesions using the k-nearest neighbor classification | Accuracy of 89.6% |
| Ştefănescu et al[56], 2016 | A neural network analysis algorithm differentiating advanced colorectal adenocarcinomas from the normal mucosa | Accuracy of 84.5% |
Colorectal cancer is a heterogenic disease with numerous epigenetic and microenvironment alterations that affects drug response, aggressiveness, and prognosis[61,62]. The shift to a more personalized and tailored treatment tactic considering the various alternations is evolving to improve disease outcomes[63].
AI is being integrated in treatment selection to provide a true individualized treatment strategy. A MATCH system was developed to integrate clinical and genetic sequence data using data from hospitals, pharmaceutical laboratories, and research centers. The MATCH system aided in correlating between medical features and genetic data, giving the oncologist the opportunity to understand patient’s individual situation[64].
Machine learning techniques are also being used to predict protein-protein interactions of a potential therapeutic target protein (S100A9) with different drugs[65]. Several other models are being developed to identify molecular biomarkers and targets by integrating transcriptomics, proteomics data, and RNA-sequencing data[66,67].
Chemotherapy, neoadjuvant chemoradiotherapy (nCRT) and other approaches are treatment options for CRC. Studies have applied AI technology to CRC treatment to help clinicians choose the appropriate treatment option and improve efficacy and limit potential toxicities.
In a study based on an unsupervised machine learning algorithm comparing pharmacological response relationships between cancer therapies, distinct intrinsic subpopulation sensitivity to one drug but resistance to others was identified. They also identified genetic alterations that could be used as biomarkers for those subpopulations[68].
In another study, artificial neural network K-nearest neighbors, support vector machine, naïve Bayesian classifier, mixed logistic regression models were used to predict response demonstrated an accuracy of 0.88, AUC of 0.86 and sensitivity of 0.94[69].
Ferrari et al[70] used AI models to assess response to therapy in locally advanced rectal cancer. The AI model was able to identify patients who will have complete response at the end of the treatment and those who will not respond to therapy at an early stage of the treatment with an AUC of 0.83.
Shayesteh et al[71] used MRI based ensemble learning methods to predict the response to nCRT with AUC of 95% and accuracy of 90%.
Other algorithms to identify pathological complete responders (CR) and non-responders (NR) patients after neoadjuvant chemoradiotherapy (CRT) in locally advanced rectal cancer showed an AUC of 0.86 and 0.83 for pathological CRs and NRs respectively by analyzing textural features of T2-weighted magnetic resonance images[70]. Shi et al[72] created a model to predict the neoadjuvant CRT response by using pre-treatment and early-treatment MRI imaging. They reported that using deep learning achieved a higher accuracy of prediction.
Oyaga-Iriarte et al[73] used algorithms in metastatic CRC patients to predict Irinotecan toxicity with an accuracy of 76%, 75%, and 91% for predicting leukopenia, neutropenia, and diarrhea respectively. Abraham et al[74] used machine learning to predict the efficacy of bevacizumab combined with oxaliplatin based chemotherapies in patients with metastatic colorectal cancers.
AI technology is also being incorporated in drug research. Drug delivery models using nanoparticles are being developed[75,76]. Cruz et al[77] created a model using molecular and nuclear magnetic resonance to detect the half-maximal inhibitory concentration of a drug against HCT116 cell line with predicted accuracy of over 63% for both training and test sets.
Traditional mathematical and statistical analysis does not provide accurate predictions on patient’s progress. However, AI can process and analyze many features based on previous data to potentially predict prognosis.
Weiser et al[78], developed a nomogram to predict recurrence of CRC after curative resection to identify patients who may benefit from adjuvant therapy and early follow-up.
In addition, long term prediction models using independent prognostic factors such as tumor size, high mitotic count, non-gastric location, and sex are established and accurately predict patients who may be cured by surgery alone[79].
The prognosis in CRC is highly dependent on pathology. Kather et al[80] used CNN to automatically extract prognostic factors from HE-stained CRC tissues. They used 420 digitalized HE-stained samples to predict the 5-year survival with an AUC of 0.69 consistent with “expect level” accuracy.
Sailer et al[81] compared ten data mining algorithm’s to predict the 5-year survival based on seven attributes and reported an accuracy of 67.7% compared to clinical judgment of 59%. Table 3 summarizes relevant treatment, toxicity, and prognosis studies.
| Ref. | Objective | Results |
| Huang et al[69], 2020 | Artificial neural network K-nearest neighbors, support vector machine, naïve Bayesian classifier, mixed logistic regression models were used to predict response | Accuracy of 0.88, AUC of 0.86 and sensitivity of 0.94 |
| Ferrari et al[70], 2019 | AI models to assess response to therapy in locally advanced rectal cancer | Able to identify patients who will have complete response at the end of the treatment and those who will not respond to therapy at an early stage of the treatment with an AUC of 0.83 |
| Shayesteh et al[71], 2019 | MRI based ensemble learning methods to predict the response to nCRT | AUC of 95% and accuracy of 90% |
| Ferrari et al[71], 2019 | Algorithms to identify pathological CR and NR patients after neoadjuvant chemoradiotherapy (CRT) in locally advanced rectal cancer | AUC of 0.86 and 0.83 for pathological CRs and NRs |
| Oyaga-Iriarte et al[73], 2019 | Algorithms in metastatic CRC patients to predict Irinotecan toxicity | Accuracy of 76%, 75%, and 91% for predicting leukopenia, neutropenia, and diarrhea respectively |
| Sailer et al[81], 2015 | Compared ten data mining algorithms to predict the 5-yr survival based on seven attributes | Accuracy of 67.7% compared to clinical judgment of 59% |
Artificial intelligence and deep learning algorithms assist physicians in detecting and diagnosing CRC. They are also used to develop and identify treatment strategies to personalize CRC treatment. Until now, AI tools have been able to detect and diagnose CRC in a manner that is comparable to, if not superior to, that of humans (Figure 2).
Despite the significant advance in AI applications, AI-based technologies have several limitations. Machine training is a complex task and requires integrating the technology into clinical practice to provide high quality large volume training data to train the AI systems and obtain the best results. This process requires robust computational infrastructure.
The variability between patients’ clinical presentation could lead to a deviation from the training model environment which could result in the unpredictable performance of an algorithm[82]. Furthermore, the input and output data of an algorithm is known, there is limited information on the exact working and process in-between, frequently referred to as the “black box” problem in machine learning. As a result of this limited visibility, factors used by a deep learning algorithm to reach a particular decision could be missed potentially leading to significant confounders in output data[82].
Additionally, there is a lack of evidence-based standards in AI development. The data used to train algorithms vary in size, number, and quality. This results in inconsistencies in validating machine learning systems deterring their implementation on a wide scale clinical setting. Limited research on the application of AI in CRC treatment is currently present. Most of the existing studies assessed AI algorithm’s ability to predict response after nCRT and chemotherapy. However, they have small sample sizes and therefore lack generalization[83]. In addition, current AI algorithms linking clinical features to prognostic status are promising. However, there is a significant difference between sensitivities, specificities, and accuracies of different AI applications.
Machine learning systems can unintentionally exacerbate health disparities by magnifying existing biases used in their training datasets[84].
Machine learning and artificial intelligence is evolving, though the medical community remains highly optimistic about the future of AI, wide scale randomized clinical trials are needed to evaluate and validate AI algorithms prior to wide scale clinical implementation. Additionally, these systems should provide a high-quality standard with robust ethical and legal frameworks prior to integration in health systems.
With the rapid expansion in AI research and technology we believe that AI algorithms will improve and personalize patient care.
Initially, AI algorithms integrate clinical data such as age, health status, disease history and other comorbidities to stratify patients. Though the current gold standard for CRC screening and diagnosis is endoscopy and pathological biopsy[12], it carries a significant risk in a subset of patients. We believe that future research directives will focus on less invasive technologies in certain patient groups for diagnosis instead on colonoscopy. Any model must maintain or even exceed the diagnostic accuracy offered by conventional diagnostic modalities. Furthermore, incorporating AI in screen colonoscopy may improve the diagnosis of precancerous lesions.
Moreover, AI technologies could assist in a establishing a more accurate staging system that incorporates not only the classical TNM stages but also proteomics, metabolomics, and genetic data to account for the heterogeneous presentation of CRC. This algorithm would potentially identify patients who would benefit from neoadjuvant therapy.
As more datasets are made available, a sufficiently large dataset could support the prediction of the prognosis of AI technology. This can help identify factors with the greatest impact on prognosis and establish future prognostic and intervention research.
Artificial intelligence and deep learning are becoming an integral part of modern-day medicine. Though the research advances in the field is an exciting new venture, it currently remains in the infant stage. Colorectal cancer screening, diagnosis and treatment will be distinctly enhanced by the incorporation of artificial intelligence technologies. AI has showed promise in therapeutic recommendations and prediction of treatment toxicity and responses this will hopefully result in a better and more personalized treatments for those in need.
Provenance and peer review: Invited article; Externally peer reviewed.
Peer-review model: Single blind
Specialty type: Oncology
Country/Territory of origin: United States
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): B
Grade C (Good): C, C
Grade D (Fair): 0
Grade E (Poor): E
P-Reviewer: Li C, China; Nazari N, Iran; Yakar M, Turkey S-Editor: Liu JH L-Editor: A P-Editor: Liu JH
| 1. | Cancer Facts & Figures 2021. Atlanta, Ga: American Cancer Society, 2021. |
| 2. | Howlader N NA, Krapcho M, Miller D, Brest A, Yu M, Ruhl J, Tatalovich Z, Mariotto A, Lewis DR, Chen HS, Feuer EJ, Cronin KA (eds). SEER Cancer Statistics Review, 1975-2016. National Cancer Institute, 2019. [RCA] [DOI] [Full Text] [Cited by in Crossref: 10] [Cited by in RCA: 10] [Article Influence: 1.1] [Reference Citation Analysis (0)] |
| 3. | Guinney J, Dienstmann R, Wang X, de Reyniès A, Schlicker A, Soneson C, Marisa L, Roepman P, Nyamundanda G, Angelino P, Bot BM, Morris JS, Simon IM, Gerster S, Fessler E, De Sousa E Melo F, Missiaglia E, Ramay H, Barras D, Homicsko K, Maru D, Manyam GC, Broom B, Boige V, Perez-Villamil B, Laderas T, Salazar R, Gray JW, Hanahan D, Tabernero J, Bernards R, Friend SH, Laurent-Puig P, Medema JP, Sadanandam A, Wessels L, Delorenzi M, Kopetz S, Vermeulen L, Tejpar S. The consensus molecular subtypes of colorectal cancer. Nat Med. 2015;21:1350-1356. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 3408] [Cited by in RCA: 3558] [Article Influence: 355.8] [Reference Citation Analysis (0)] |
| 4. | Becht E, de Reyniès A, Giraldo NA, Pilati C, Buttard B, Lacroix L, Selves J, Sautès-Fridman C, Laurent-Puig P, Fridman WH. Immune and Stromal Classification of Colorectal Cancer Is Associated with Molecular Subtypes and Relevant for Precision Immunotherapy. Clin Cancer Res. 2016;22:4057-4066. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 325] [Cited by in RCA: 409] [Article Influence: 45.4] [Reference Citation Analysis (0)] |
| 5. | Shalev-Shwartz S, Ben-David S. Understanding machine learning: From theory to algorithms: Cambridge university press, 2014. [DOI] [Full Text] |
| 6. | Hamet P, Tremblay J. Artificial intelligence in medicine. Metabolism. 2017;69S:S36-S40. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 728] [Cited by in RCA: 832] [Article Influence: 104.0] [Reference Citation Analysis (0)] |
| 7. | Mori Y, Kudo SE, Berzin TM, Misawa M, Takeda K. Computer-aided diagnosis for colonoscopy. Endoscopy. 2017;49:813-819. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 90] [Cited by in RCA: 97] [Article Influence: 12.1] [Reference Citation Analysis (0)] |
| 8. | Jiménez-Rodríguez RM, Rubio-Dorado-Manzanares M, Díaz-Pavón JM, Reyes-Díaz ML, Vazquez-Monchul JM, Garcia-Cabrera AM, Padillo J, De la Portilla F. Learning curve in robotic rectal cancer surgery: current state of affairs. Int J Colorectal Dis. 2016;31:1807-1815. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 102] [Cited by in RCA: 87] [Article Influence: 9.7] [Reference Citation Analysis (0)] |
| 9. | Maida M, Macaluso FS, Ianiro G, Mangiola F, Sinagra E, Hold G, Maida C, Cammarota G, Gasbarrini A, Scarpulla G. Screening of colorectal cancer: present and future. Expert Rev Anticancer Ther. 2017;17:1131-1146. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 143] [Cited by in RCA: 126] [Article Influence: 15.8] [Reference Citation Analysis (0)] |
| 10. | Morson B. President's address. The polyp-cancer sequence in the large bowel. Proc R Soc Med. 1974;67:451-457. [PubMed] |
| 11. | Smith RA, Andrews KS, Brooks D, Fedewa SA, Manassaram-Baptiste D, Saslow D, Wender RC. Cancer screening in the United States, 2019: A review of current American Cancer Society guidelines and current issues in cancer screening. CA Cancer J Clin. 2019;69:184-210. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 295] [Cited by in RCA: 422] [Article Influence: 70.3] [Reference Citation Analysis (0)] |
| 12. | Issa IA, Noureddine M. Colorectal cancer screening: An updated review of the available options. World J Gastroenterol. 2017;23:5086-5096. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 434] [Cited by in RCA: 392] [Article Influence: 49.0] [Reference Citation Analysis (11)] |
| 13. | Bressler B, Paszat LF, Chen Z, Rothwell DM, Vinden C, Rabeneck L. Rates of new or missed colorectal cancers after colonoscopy and their risk factors: a population-based analysis. Gastroenterology. 2007;132:96-102. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 442] [Cited by in RCA: 449] [Article Influence: 24.9] [Reference Citation Analysis (0)] |
| 14. | Morris EJ, Rutter MD, Finan PJ, Thomas JD, Valori R. Post-colonoscopy colorectal cancer (PCCRC) rates vary considerably depending on the method used to calculate them: a retrospective observational population-based study of PCCRC in the English National Health Service. Gut. 2015;64:1248-1256. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 81] [Cited by in RCA: 109] [Article Influence: 10.9] [Reference Citation Analysis (0)] |
| 15. | Corley DA, Jensen CD, Marks AR, Zhao WK, Lee JK, Doubeni CA, Zauber AG, de Boer J, Fireman BH, Schottinger JE, Quinn VP, Ghai NR, Levin TR, Quesenberry CP. Adenoma detection rate and risk of colorectal cancer and death. N Engl J Med. 2014;370:1298-1306. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1251] [Cited by in RCA: 1561] [Article Influence: 141.9] [Reference Citation Analysis (0)] |
| 16. | Kaminski MF, Thomas-Gibson S, Bugajski M, Bretthauer M, Rees CJ, Dekker E, Hoff G, Jover R, Suchanek S, Ferlitsch M, Anderson J, Roesch T, Hultcranz R, Racz I, Kuipers EJ, Garborg K, East JE, Rupinski M, Seip B, Bennett C, Senore C, Minozzi S, Bisschops R, Domagk D, Valori R, Spada C, Hassan C, Dinis-Ribeiro M, Rutter MD. Performance measures for lower gastrointestinal endoscopy: a European Society of Gastrointestinal Endoscopy (ESGE) quality improvement initiative. United European Gastroenterol J. 2017;5:309-334. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 142] [Cited by in RCA: 177] [Article Influence: 22.1] [Reference Citation Analysis (0)] |
| 17. | Spadaccini M, Frazzoni L, Vanella G, East J, Radaelli F, Spada C, Fuccio L, Benamouzig R, Bisschops R, Bretthauer M, Dekker E, Dinis-Ribeiro M, Ferlitsch M, Gralnek I, Jover R, Kaminski MF, Pellisé M, Triantafyllou K, Van Hooft JE, Dumonceau JM, Marmo C, Alfieri S, Chandrasekar VT, Sharma P, Rex DK, Repici A, Hassan C. Efficacy and Tolerability of High- vs Low-Volume Split-Dose Bowel Cleansing Regimens for Colonoscopy: A Systematic Review and Meta-analysis. Clin Gastroenterol Hepatol. 2020;18:1454-1465.e14. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 63] [Cited by in RCA: 65] [Article Influence: 13.0] [Reference Citation Analysis (0)] |
| 18. | Nogueira-Rodríguez A, Domínguez-Carbajales R, López-Fernández H, Iglesias Á, Cubiella J, Fdez-Riverola F, Reboiro-Jato M, Glez-Peña D. Deep Neural Networks approaches for detecting and classifying colorectal polyps. Neurocomputing. 2021;423:721-734. [RCA] [DOI] [Full Text] [Cited by in Crossref: 33] [Cited by in RCA: 35] [Article Influence: 8.8] [Reference Citation Analysis (0)] |
| 19. | Wang P, Berzin TM, Glissen Brown JR, Bharadwaj S, Becq A, Xiao X, Liu P, Li L, Song Y, Zhang D, Li Y, Xu G, Tu M, Liu X. Real-time automatic detection system increases colonoscopic polyp and adenoma detection rates: a prospective randomised controlled study. Gut. 2019;68:1813-1819. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 398] [Cited by in RCA: 548] [Article Influence: 91.3] [Reference Citation Analysis (0)] |
| 20. | Karkanis SA, Iakovidis DK, Maroulis DE, Karras DA, Tzivras M. Computer-aided tumor detection in endoscopic video using color wavelet features. IEEE Trans Inf Technol Biomed. 2003;7:141-152. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 319] [Cited by in RCA: 207] [Article Influence: 9.4] [Reference Citation Analysis (0)] |
| 21. | Nazarian S, Glover B, Ashrafian H, Darzi A, Teare J. Diagnostic Accuracy of Artificial Intelligence and Computer-Aided Diagnosis for the Detection and Characterization of Colorectal Polyps: Systematic Review and Meta-analysis. J Med Internet Res. 2021;23:e27370. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 36] [Cited by in RCA: 46] [Article Influence: 11.5] [Reference Citation Analysis (0)] |
| 22. | Vining D, Gelfand DW, Bechtold RE. Technical feasibility of colon imaging with helical CT and virtual reality. [DOI] [Full Text] |
| 23. | Johnson CD, Chen MH, Toledano AY, Heiken JP, Dachman A, Kuo MD, Menias CO, Siewert B, Cheema JI, Obregon RG, Fidler JL, Zimmerman P, Horton KM, Coakley K, Iyer RB, Hara AK, Halvorsen RA Jr, Casola G, Yee J, Herman BA, Burgart LJ, Limburg PJ. Accuracy of CT colonography for detection of large adenomas and cancers. N Engl J Med. 2008;359:1207-1217. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 848] [Cited by in RCA: 706] [Article Influence: 41.5] [Reference Citation Analysis (0)] |
| 24. | Pickhardt PJ, Choi JR, Hwang I, Butler JA, Puckett ML, Hildebrandt HA, Wong RK, Nugent PA, Mysliwiec PA, Schindler WR. Computed tomographic virtual colonoscopy to screen for colorectal neoplasia in asymptomatic adults. N Engl J Med. 2003;349:2191-2200. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1495] [Cited by in RCA: 1284] [Article Influence: 58.4] [Reference Citation Analysis (0)] |
| 25. | Grosu S, Wesp P, Graser A, Maurus S, Schulz C, Knösel T, Cyran CC, Ricke J, Ingrisch M, Kazmierczak PM. Machine Learning-based Differentiation of Benign and Premalignant Colorectal Polyps Detected with CT Colonography in an Asymptomatic Screening Population: A Proof-of-Concept Study. Radiology. 2021;299:326-335. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 11] [Cited by in RCA: 34] [Article Influence: 8.5] [Reference Citation Analysis (0)] |
| 26. | Song B, Zhang G, Lu H, Wang H, Zhu W, J Pickhardt P, Liang Z. Volumetric texture features from higher-order images for diagnosis of colon lesions via CT colonography. Int J Comput Assist Radiol Surg. 2014;9:1021-1031. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 58] [Cited by in RCA: 50] [Article Influence: 4.5] [Reference Citation Analysis (0)] |
| 27. | Nakamura T, Terano A. Capsule endoscopy: past, present, and future. J Gastroenterol. 2008;43:93-99. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 119] [Cited by in RCA: 83] [Article Influence: 4.9] [Reference Citation Analysis (0)] |
| 28. | Eliakim R, Fireman Z, Gralnek IM, Yassin K, Waterman M, Kopelman Y, Lachter J, Koslowsky B, Adler SN. Evaluation of the PillCam Colon capsule in the detection of colonic pathology: results of the first multicenter, prospective, comparative study. Endoscopy. 2006;38:963-970. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 239] [Cited by in RCA: 222] [Article Influence: 11.7] [Reference Citation Analysis (0)] |
| 29. | Van Gossum A, Munoz-Navas M, Fernandez-Urien I, Carretero C, Gay G, Delvaux M, Lapalus MG, Ponchon T, Neuhaus H, Philipper M, Costamagna G, Riccioni ME, Spada C, Petruzziello L, Fraser C, Postgate A, Fitzpatrick A, Hagenmuller F, Keuchel M, Schoofs N, Devière J. Capsule endoscopy versus colonoscopy for the detection of polyps and cancer. N Engl J Med. 2009;361:264-270. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 267] [Cited by in RCA: 225] [Article Influence: 14.1] [Reference Citation Analysis (0)] |
| 30. | Blanes-Vidal V, Baatrup G, Nadimi ES. Addressing priority challenges in the detection and assessment of colorectal polyps from capsule endoscopy and colonoscopy in colorectal cancer screening using machine learning. Acta Oncol. 2019;58:S29-S36. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 38] [Cited by in RCA: 62] [Article Influence: 10.3] [Reference Citation Analysis (0)] |
| 31. | Hosoe N, Limpias Kamiya KJL, Hayashi Y, Sujino T, Ogata H, Kanai T. Current status of colon capsule endoscopy. Dig Endosc. 2021;33:529-537. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 30] [Cited by in RCA: 23] [Article Influence: 5.8] [Reference Citation Analysis (0)] |
| 32. | Spell DW, Jones DV Jr, Harper WF, David Bessman J. The value of a complete blood count in predicting cancer of the colon. Cancer Detect Prev. 2004;28:37-42. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 88] [Cited by in RCA: 105] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 33. | Kinar Y, Kalkstein N, Akiva P, Levin B, Half EE, Goldshtein I, Chodick G, Shalev V. Development and validation of a predictive model for detection of colorectal cancer in primary care by analysis of complete blood counts: a binational retrospective study. J Am Med Inform Assoc. 2016;23:879-890. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 81] [Cited by in RCA: 80] [Article Influence: 8.9] [Reference Citation Analysis (0)] |
| 34. | Hornbrook MC, Goshen R, Choman E, O'Keeffe-Rosetti M, Kinar Y, Liles EG, Rust KC. Early Colorectal Cancer Detected by Machine Learning Model Using Gender, Age, and Complete Blood Count Data. Dig Dis Sci. 2017;62:2719-2727. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 58] [Cited by in RCA: 77] [Article Influence: 9.6] [Reference Citation Analysis (0)] |
| 35. | Kinar Y, Akiva P, Choman E, Kariv R, Shalev V, Levin B, Narod SA, Goshen R. Performance analysis of a machine learning flagging system used to identify a group of individuals at a high risk for colorectal cancer. PLoS One. 2017;12:e0171759. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 31] [Cited by in RCA: 44] [Article Influence: 5.5] [Reference Citation Analysis (0)] |
| 36. | Gupta P, Gulzar Z, Hsieh B, Lim A, Watson D, Mei R. Analytical validation of the CellMax platform for early detection of cancer by enumeration of rare circulating tumor cells. J Circ Biomark. 2019;8:1849454419899214. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 8] [Cited by in RCA: 13] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 37. | Mori Y, Kudo SE, Misawa M, Mori K. Simultaneous detection and characterization of diminutive polyps with the use of artificial intelligence during colonoscopy. VideoGIE. 2019;4:7-10. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 44] [Cited by in RCA: 41] [Article Influence: 6.8] [Reference Citation Analysis (0)] |
| 38. | Lui TKL, Guo CG, Leung WK. Accuracy of artificial intelligence on histology prediction and detection of colorectal polyps: a systematic review and meta-analysis. Gastrointest Endosc. 2020;92:11-22.e6. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 55] [Cited by in RCA: 80] [Article Influence: 16.0] [Reference Citation Analysis (0)] |
| 39. | Mori Y, Kudo SE, East JE, Rastogi A, Bretthauer M, Misawa M, Sekiguchi M, Matsuda T, Saito Y, Ikematsu H, Hotta K, Ohtsuka K, Kudo T, Mori K. Cost savings in colonoscopy with artificial intelligence-aided polyp diagnosis: an add-on analysis of a clinical trial (with video). Gastrointest Endosc. 2020;92:905-911.e1. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 65] [Cited by in RCA: 100] [Article Influence: 20.0] [Reference Citation Analysis (0)] |
| 40. | Min M, Su S, He W, Bi Y, Ma Z, Liu Y. Computer-aided diagnosis of colorectal polyps using linked color imaging colonoscopy to predict histology. Sci Rep. 2019;9:2881. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 34] [Cited by in RCA: 38] [Article Influence: 6.3] [Reference Citation Analysis (0)] |
| 41. | Barbeiro S, Libânio D, Castro R, Dinis-Ribeiro M, Pimentel-Nunes P. Narrow-Band Imaging: Clinical Application in Gastrointestinal Endoscopy. GE Port J Gastroenterol. 2018;26:40-53. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 61] [Cited by in RCA: 53] [Article Influence: 7.6] [Reference Citation Analysis (0)] |
| 42. | Takemura Y, Yoshida S, Tanaka S, Kawase R, Onji K, Oka S, Tamaki T, Raytchev B, Kaneda K, Yoshihara M, Chayama K. Computer-aided system for predicting the histology of colorectal tumors by using narrow-band imaging magnifying colonoscopy (with video). Gastrointest Endosc. 2012;75:179-185. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 81] [Cited by in RCA: 84] [Article Influence: 6.5] [Reference Citation Analysis (0)] |
| 43. | Hirata M, Tanaka S, Oka S, Kaneko I, Yoshida S, Yoshihara M, Chayama K. Evaluation of microvessels in colorectal tumors by narrow band imaging magnification. Gastrointest Endosc. 2007;66:945-952. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 117] [Cited by in RCA: 119] [Article Influence: 6.6] [Reference Citation Analysis (0)] |
| 44. | Maeyama Y, Mitsuyama K, Noda T, Nagata S, Nagata T, Yoshioka S, Yoshida H, Mukasa M, Sumie H, Kawano H, Akiba J, Araki Y, Kakuma T, Tsuruta O, Torimura T. Prediction of colorectal tumor grade and invasion depth through narrow-band imaging scoring. World J Gastroenterol. 2018;24:4809-4820. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 3] [Cited by in RCA: 7] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 45. | Gross S, Trautwein C, Behrens A, Winograd R, Palm S, Lutz HH, Schirin-Sokhan R, Hecker H, Aach T, Tischendorf JJ. Computer-based classification of small colorectal polyps by using narrow-band imaging with optical magnification. Gastrointest Endosc. 2011;74:1354-1359. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 92] [Cited by in RCA: 97] [Article Influence: 6.9] [Reference Citation Analysis (0)] |
| 46. | Chen PJ, Lin MC, Lai MJ, Lin JC, Lu HH, Tseng VS. Accurate Classification of Diminutive Colorectal Polyps Using Computer-Aided Analysis. Gastroenterology. 2018;154:568-575. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 250] [Cited by in RCA: 276] [Article Influence: 39.4] [Reference Citation Analysis (0)] |
| 47. | Neumann H, Fuchs FS, Vieth M, Atreya R, Siebler J, Kiesslich R, Neurath MF. Review article: in vivo imaging by endocytoscopy. Aliment Pharmacol Ther. 2011;33:1183-1193. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 78] [Cited by in RCA: 69] [Article Influence: 4.9] [Reference Citation Analysis (0)] |
| 48. | Mori Y, Kudo SE, Wakamura K, Misawa M, Ogawa Y, Kutsukawa M, Kudo T, Hayashi T, Miyachi H, Ishida F, Inoue H. Novel computer-aided diagnostic system for colorectal lesions by using endocytoscopy (with videos). Gastrointest Endosc. 2015;81:621-629. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 103] [Cited by in RCA: 104] [Article Influence: 10.4] [Reference Citation Analysis (0)] |
| 49. | Mori Y, Kudo SE, Chiu PW, Singh R, Misawa M, Wakamura K, Kudo T, Hayashi T, Katagiri A, Miyachi H, Ishida F, Maeda Y, Inoue H, Nimura Y, Oda M, Mori K. Impact of an automated system for endocytoscopic diagnosis of small colorectal lesions: an international web-based study. Endoscopy. 2016;48:1110-1118. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 81] [Cited by in RCA: 74] [Article Influence: 8.2] [Reference Citation Analysis (0)] |
| 50. | Mori Y, Kudo SE, Mori K. Potential of artificial intelligence-assisted colonoscopy using an endocytoscope (with video). Dig Endosc. 2018;30 Suppl 1:52-53. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 20] [Cited by in RCA: 15] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 51. | Takeda K, Kudo SE, Mori Y, Misawa M, Kudo T, Wakamura K, Katagiri A, Baba T, Hidaka E, Ishida F, Inoue H, Oda M, Mori K. Accuracy of diagnosing invasive colorectal cancer using computer-aided endocytoscopy. Endoscopy. 2017;49:798-802. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 80] [Cited by in RCA: 101] [Article Influence: 12.6] [Reference Citation Analysis (0)] |
| 52. | Kudo SE, Mori Y, Wakamura K, Ikehara N, Ichimasa K, Wada Y, Kutsukawa M, Misawa M, Kudo T, Hayashi T, Miyachi H, Inoue H, Hamatani S. Endocytoscopy can provide additional diagnostic ability to magnifying chromoendoscopy for colorectal neoplasms. J Gastroenterol Hepatol. 2014;29:83-90. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 39] [Cited by in RCA: 29] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 53. | Takemura Y, Yoshida S, Tanaka S, Onji K, Oka S, Tamaki T, Kaneda K, Yoshihara M, Chayama K. Quantitative analysis and development of a computer-aided system for identification of regular pit patterns of colorectal lesions. Gastrointest Endosc. 2010;72:1047-1051. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 49] [Cited by in RCA: 47] [Article Influence: 3.1] [Reference Citation Analysis (1)] |
| 54. | Neumann H, Kiesslich R, Wallace MB, Neurath MF. Confocal laser endomicroscopy: technical advances and clinical applications. Gastroenterology. 2010;139:388-392, 392.e1. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 183] [Cited by in RCA: 176] [Article Influence: 11.7] [Reference Citation Analysis (0)] |
| 55. | André B, Vercauteren T, Buchner AM, Krishna M, Ayache N, Wallace MB. Software for automated classification of probe-based confocal laser endomicroscopy videos of colorectal polyps. World J Gastroenterol. 2012;18:5560-5569. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 64] [Cited by in RCA: 59] [Article Influence: 4.5] [Reference Citation Analysis (0)] |
| 56. | Ştefănescu D, Streba C, Cârţână ET, Săftoiu A, Gruionu G, Gruionu LG. Computer Aided Diagnosis for Confocal Laser Endomicroscopy in Advanced Colorectal Adenocarcinoma. PLoS One. 2016;11:e0154863. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 49] [Cited by in RCA: 47] [Article Influence: 5.2] [Reference Citation Analysis (0)] |
| 57. | Aihara H, Saito S, Inomata H, Ide D, Tamai N, Ohya TR, Kato T, Amitani S, Tajiri H. Computer-aided diagnosis of neoplastic colorectal lesions using 'real-time' numerical color analysis during autofluorescence endoscopy. Eur J Gastroenterol Hepatol. 2013;25:488-494. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 42] [Cited by in RCA: 48] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 58. | Inomata H, Tamai N, Aihara H, Sumiyama K, Saito S, Kato T, Tajiri H. Efficacy of a novel auto-fluorescence imaging system with computer-assisted color analysis for assessment of colorectal lesions. World J Gastroenterol. 2013;19:7146-7153. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 32] [Cited by in RCA: 33] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 59. | Komeda Y, Handa H, Watanabe T, Nomura T, Kitahashi M, Sakurai T, Okamoto A, Minami T, Kono M, Arizumi T, Takenaka M, Hagiwara S, Matsui S, Nishida N, Kashida H, Kudo M. Computer-Aided Diagnosis Based on Convolutional Neural Network System for Colorectal Polyp Classification: Preliminary Experience. Oncology. 2017;93 Suppl 1:30-34. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 174] [Cited by in RCA: 129] [Article Influence: 16.1] [Reference Citation Analysis (0)] |
| 60. | Kuiper T, Alderlieste YA, Tytgat KM, Vlug MS, Nabuurs JA, Bastiaansen BA, Löwenberg M, Fockens P, Dekker E. Automatic optical diagnosis of small colorectal lesions by laser-induced autofluorescence. Endoscopy. 2015;47:56-62. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 13] [Cited by in RCA: 21] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 61. | Linnekamp JF, Wang X, Medema JP, Vermeulen L. Colorectal cancer heterogeneity and targeted therapy: a case for molecular disease subtypes. Cancer Res. 2015;75:245-249. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 124] [Cited by in RCA: 151] [Article Influence: 15.1] [Reference Citation Analysis (0)] |
| 62. | Awidi M, Ababneh N, Shomaf M, Al Fararjeh F, Owaidi L, AlKhatib M, Al Tarawneh B, Awidi A. KRAS and NRAS mutational gene profile of metastatic colorectal cancer patients in Jordan. PLoS One. 2019;14:e0226473. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 10] [Cited by in RCA: 6] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 63. | Buikhuisen JY, Torang A, Medema JP. Exploring and modelling colon cancer inter-tumour heterogeneity: opportunities and challenges. Oncogenesis. 2020;9:66. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 29] [Cited by in RCA: 66] [Article Influence: 13.2] [Reference Citation Analysis (0)] |
| 64. | Siddiqi J, Akhgar B, Gruzdz A, Zaefarian G, Ihnatowicz A. Automated Diagnosis System to Support Colon Cancer Treatment: MATCH. Proceedings of the Fifth International Conference on Information Technology: New Generations (itng 2008); 2008; 201-205. [DOI] [Full Text] |
| 65. | Lee J, Kumar S, Lee SY, Park SJ, Kim MH. Development of Predictive Models for Identifying Potential S100A9 Inhibitors Based on Machine Learning Methods. Front Chem. 2019;7:779. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 20] [Cited by in RCA: 21] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 66. | Ding D, Han S, Zhang H, He Y, Li Y. Predictive biomarkers of colorectal cancer. Comput Biol Chem. 2019;83:107106. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 20] [Cited by in RCA: 38] [Article Influence: 6.3] [Reference Citation Analysis (0)] |
| 67. | Pacheco MP, Bintener T, Ternes D, Kulms D, Haan S, Letellier E, Sauter T. Identifying and targeting cancer-specific metabolism with network-based drug target prediction. EBioMedicine. 2019;43:98-106. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 34] [Cited by in RCA: 59] [Article Influence: 9.8] [Reference Citation Analysis (0)] |
| 68. | Keshava N, Toh TS, Yuan H, Yang B, Menden MP, Wang D. Defining subpopulations of differential drug response to reveal novel target populations. NPJ Syst Biol Appl. 2019;5:36. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 17] [Cited by in RCA: 18] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 69. | Huang CM, Huang MY, Huang CW, Tsai HL, Su WC, Chang WC, Wang JY, Shi HY. Machine learning for predicting pathological complete response in patients with locally advanced rectal cancer after neoadjuvant chemoradiotherapy. Sci Rep. 2020;10:12555. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 30] [Cited by in RCA: 31] [Article Influence: 6.2] [Reference Citation Analysis (0)] |
| 70. | Ferrari R, Mancini-Terracciano C, Voena C, Rengo M, Zerunian M, Ciardiello A, Grasso S, Mare' V, Paramatti R, Russomando A, Santacesaria R, Satta A, Solfaroli Camillocci E, Faccini R, Laghi A. MR-based artificial intelligence model to assess response to therapy in locally advanced rectal cancer. Eur J Radiol. 2019;118:1-9. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 42] [Cited by in RCA: 61] [Article Influence: 10.2] [Reference Citation Analysis (0)] |
| 71. | Shayesteh SP, Alikhassi A, Fard Esfahani A, Miraie M, Geramifar P, Bitarafan-Rajabi A, Haddad P. Neo-adjuvant chemoradiotherapy response prediction using MRI based ensemble learning method in rectal cancer patients. Phys Med. 2019;62:111-119. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 24] [Cited by in RCA: 36] [Article Influence: 6.0] [Reference Citation Analysis (0)] |
| 72. | Shi L, Zhang Y, Nie K, Sun X, Niu T, Yue N, Kwong T, Chang P, Chow D, Chen JH, Su MY. Machine learning for prediction of chemoradiation therapy response in rectal cancer using pre-treatment and mid-radiation multi-parametric MRI. Magn Reson Imaging. 2019;61:33-40. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 59] [Cited by in RCA: 82] [Article Influence: 13.7] [Reference Citation Analysis (0)] |
| 73. | Oyaga-Iriarte E, Insausti A, Sayar O, Aldaz A. Prediction of irinotecan toxicity in metastatic colorectal cancer patients based on machine learning models with pharmacokinetic parameters. J Pharmacol Sci. 2019;140:20-25. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 19] [Cited by in RCA: 32] [Article Influence: 5.3] [Reference Citation Analysis (0)] |
| 74. | Abraham JP, Magee D, Cremolini C, Antoniotti C, Halbert DD, Xiu J, Stafford P, Berry DA, Oberley MJ, Shields AF, Marshall JL, Salem ME, Falcone A, Grothey A, Hall MJ, Venook AP, Lenz HJ, Helmstetter A, Korn WM, Spetzler DB. Clinical Validation of a Machine-learning-derived Signature Predictive of Outcomes from First-line Oxaliplatin-based Chemotherapy in Advanced Colorectal Cancer. Clin Cancer Res. 2021;27:1174-1183. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 12] [Cited by in RCA: 42] [Article Influence: 8.4] [Reference Citation Analysis (0)] |
| 75. | Torchilin VP. Passive and Active Drug Targeting: Drug Delivery to Tumors as an Example. In: Schäfer-Korting M, editor Drug Delivery. Berlin, Heidelberg: Springer Berlin Heidelberg, 2010: 3-53. [RCA] [DOI] [Full Text] [Cited by in Crossref: 313] [Cited by in RCA: 314] [Article Influence: 20.9] [Reference Citation Analysis (0)] |
| 76. | Martel S, Mohammadi M. Switching between Magnetotactic and Aerotactic Displacement Controls to Enhance the Efficacy of MC-1 Magneto-Aerotactic Bacteria as Cancer-Fighting Nanorobots. Micromachines (Basel). 2016;7. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 10] [Cited by in RCA: 16] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 77. | Cruz S, Gomes SE, Borralho PM, Rodrigues CMP, Gaudêncio SP, Pereira F. In Silico HCT116 Human Colon Cancer Cell-Based Models En Route to the Discovery of Lead-Like Anticancer Drugs. Biomolecules. 2018;8. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 18] [Cited by in RCA: 29] [Article Influence: 4.1] [Reference Citation Analysis (0)] |
| 78. | Weiser MR, Landmann RG, Kattan MW, Gonen M, Shia J, Chou J, Paty PB, Guillem JG, Temple LK, Schrag D, Saltz LB, Wong WD. Individualized prediction of colon cancer recurrence using a nomogram. J Clin Oncol. 2008;26:380-385. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 190] [Cited by in RCA: 225] [Article Influence: 13.2] [Reference Citation Analysis (0)] |
| 79. | Joensuu H, Vehtari A, Riihimäki J, Nishida T, Steigen SE, Brabec P, Plank L, Nilsson B, Cirilli C, Braconi C, Bordoni A, Magnusson MK, Linke Z, Sufliarsky J, Federico M, Jonasson JG, Dei Tos AP, Rutkowski P. Risk of recurrence of gastrointestinal stromal tumour after surgery: an analysis of pooled population-based cohorts. Lancet Oncol. 2012;13:265-274. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 576] [Cited by in RCA: 672] [Article Influence: 48.0] [Reference Citation Analysis (0)] |
| 80. | Kather JN, Krisam J, Charoentong P, Luedde T, Herpel E, Weis CA, Gaiser T, Marx A, Valous NA, Ferber D, Jansen L, Reyes-Aldasoro CC, Zörnig I, Jäger D, Brenner H, Chang-Claude J, Hoffmeister M, Halama N. Predicting survival from colorectal cancer histology slides using deep learning: A retrospective multicenter study. PLoS Med. 2019;16:e1002730. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 555] [Cited by in RCA: 492] [Article Influence: 82.0] [Reference Citation Analysis (0)] |
| 81. | Sailer F, Pobiruchin M, Bochum S, Martens UM, Schramm W. Prediction of 5-Year Survival with Data Mining Algorithms. Stud Health Technol Inform. 2015;213:75-78. [PubMed] |
| 82. | Auger SD, Jacobs BM, Dobson R, Marshall CR, Noyce AJ. Big data, machine learning and artificial intelligence: a neurologist's guide. Pract Neurol. 2020;. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 9] [Cited by in RCA: 14] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 83. | Huang S, Yang J, Fong S, Zhao Q. Artificial intelligence in cancer diagnosis and prognosis: Opportunities and challenges. Cancer Lett. 2020;471:61-71. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 133] [Cited by in RCA: 296] [Article Influence: 49.3] [Reference Citation Analysis (1)] |
| 84. | Rigby MJ. Ethical Dimensions of Using Artificial Intelligence in Health Care. AMA J Ethics. 2019;21:121-124. [DOI] [Full Text] |