INTRODUCTION
Artificial intelligence (AI) has directly impacted the field of endoscopy by nurturing questions directed at the status quo and eventually opened up new paradigms that redefined the boundaries of our abilities as an endoscopist. AI is a broad term that encompasses the development and application of algorithms that can perform tasks that generally necessitate human intelligence[1]. Machine learning (ML), on the other hand, is a subset of AI which refers to a specific algorithm, capable of analyzing features in a dataset, based on raw data, in order to deliver a classification output[2,3]. One of the areas where ML has shown a lot of promise is in image discrimination and diagnosis, which has many applications in the field of gastro-intestinal (GI) endoscopy. The advent of advanced imaging techniques such as high-definition white light endoscopy (HD-WLE) and pre-processing techniques like optical chromo-endoscopy, have paved the way for AI to make a significant impact in diagnostic endoscopy. Currently, AI in GI endoscopy is witnessing a paradigm shift, from mere ‘identification’ to a more composite and clinically relevant ‘interpretation’ of the images[4]. This paradigm shift, in combination with rapid improvement in computing power, has enabled ML algorithms to occupy a central role in the world of endoscopy.
Machine learning has already demonstrated remarkable success in several areas of medicine, such as radiology and pathology[5-9]. More importantly, there has been a deluge of published literature on the utility and potential of ML within the domain of endoscopy in the past decade[10-18]. Deep learning has strengthened the reality that the use of ML in endoscopy is an eventuality that is here to stay[19]. However, we are still in the early stages of understanding its full potential in image differentiation and classification of endoscopic lesions, with many unanswered questions leading to poor acceptance of these technologies.
The relative novelty of ML in the field of endoscopy, coupled with the frequent use of technical terminology around machine learning, has been a major factor that has affected its widespread acceptance among clinicians. Moreover, understanding the progress made in this area and adopting this new tool for clinical practice necessitates a working knowledge of the technical basis and a familiarity of the terminology used. In this review, the common terminology as well as a brief technical basis of image interpretation by AI-based applications will be described. This will be followed by an update on the role of AI in the prevention of colorectal cancer (CRC) and the evaluation of specific pancreatic lesions using EUS.
TECHNICAL BASIS AND COMMON TERMINOLOGY USED
ML in healthcare is a convergence of two diverse and complex areas, namely data science engineering and medicine, each with its unique expertise and jargon, which often results in a relationship that is fraught with misinterpretation and ambiguity. This fosters a disconnect that can be one of the major barriers of progress in this field. In this section, we define the relevant terminology and, in the process, also briefly describe the technical basis of the use of ML in endoscopy.
AI and ML
‘Artificial intelligence’ is a popular term that is commonly used interchangeably with ‘machine learning’. In essence however, AI encompasses a broader field that includes path finding, logic representation and reasoning[4]. While ML is used to accomplish specific tasks, AI attempts to provide a more generic path for autonomous learning. The field of ML involves the use of existing data to build mathematical models that can predict expected outcomes on new data. There are two broad subtypes of ML models, namely, supervised and unsupervised learning. Supervised learning is achieved on a model with labelled data points (e.g.: Benign vs malignant), following which, the algorithm attempts to predict the labels upon a test set of unseen datapoints. On the other hand, unsupervised learning is used only to find the underlying structure, or a pattern within an unlabelled dataset; in other words, there is access to data but the outcome is not labelled (malignant or benign). Common examples of ML algorithms include deep neural networks (deep learning), support vector machines (SVM), gradient boosted trees and K-nearest neighbours.
Feature extraction
Before the generation of a predictive model, the data needs to be transformed into a numerical representation that can be fed to the ML algorithms. This process is called feature extraction and generally requires the input of medical experts in the field. Alternatively, modern ML techniques have automated this process and enabled extraction of features automatically from vision, language and sound datasets.
Deep learning
Deep learning (DL) is a type of ML algorithm originally known as Artificial Neural Networks (ANN’s). ANN’s are loosely inspired by the biological process found in a brain. They are comprised of mathematical neurons which “fire” if they are activated, and each neuron is connected to other neurons with “weights”. This connection of neurons and weights makes up what is known as “layers” in the neural network. Deep learning is when you have many layers (10’s to 100’s) connected, with millions of neurons and weights all interconnected. Deep learning models are very promising because they achieve extremely high rates of success in the fields of computer vision, natural language processing, machine translation, and speech recognition. This success is possible because of the enormous amount of data available, modern computing architectures and improved optimization algorithms. The attractiveness of deep learning is that it requires little expert domain knowledge in the form of feature extraction. The algorithm learns directly from the raw data (pixels, sound waves, text) and will automatically learn the correct “weights” which produce the most accurate results.
It has been shown that the lower layers of a deep learning model learn more abstract concepts such as “edges, shapes, lines” and the higher layers of the network learn more specific representations such as “nose, hair, eyes”.
Computer aided detection and computer aided diagnosis
ML algorithms that are applied to assist in the interpretation of medical images/videos are referred to as computer-aided detection (CADe) and computer-aided diagnosis (CADx). Distinction between CADe and CADx algorithms is important as the former is mainly used to ‘detect’ pathology, while the latter is able to ‘classify’ the pathology. For example, CADe will be used to identify a colonic polyp in a study, while CADx will enable characterization of the polyp as adenomatous or non-adenomatous. This has profound implications in the management of patients undergoing colonoscopy. Therefore, it necessitates a high degree of accuracy, reliability and external validity. Apart from this, ML algorithms can also be applied to guide interventions and is usually referred to as ‘image-guided interventions’; like the use of ML to guide the necessity and site of biopsy using EUS imaging.
ROLE OF AI IN SCREENING COLONOSCOPY FOR CRC
CRC is a leading cause of death with a rising incidence especially in younger age-groups, both in western countries as well as many Asian countries in the recent past[20,21]. Most CRC develops from pre-existing adenomas which are pre-cancerous lesions[22]. Resection of adenomas during a screening colonoscopy has been shown to be instrumental in lowering the risk of CRC[23]. Thus, adenoma detection rate (ADR) in particular, apart from withdrawal time, clean colon and caecal intubation rate, is considered to be a vital quality indicator of CRC screening programs. For every 1% increase in adenoma detection rate, there is an associated 3% decrease in interval incidence of colon cancer[23]. Non-visualization is a major factor that can lower ADR in most cases. This can mainly be attributed to polyps hidden in poorly accessible areas like the left colon, or behind mucosal folds. Besides hidden polyps, those that are technically in the visual field may still be missed if they are subtle, diminutive, transiently visible, partially obscured by debris, or seen on the edge of the screen[24]. High quality bowel preparation, strict adherence to globally accepted standards for withdrawal time, meticulous mucosal inspection techniques and the use of endoscopes with wider viewing angles can, to a certain extent, address these issues[25]. However, even with the currently performed, careful colonoscopy, rates of missed adenomas can be as high as 26% for adenomatous polyps less than 5 mm in size[26]. Even in the case of advanced adenomas, adenoma missed rates (AMR) has been reported to be as high as 5.4%[27]. An intutitive approach to this problem would be to employ measures that can supplement our capacity for visualisation. To that end, recent studies using full-spectrum colonoscopy (FUSE), which provides 330 angle of view, have been described to access previously hidden areas during a colonoscopy. However, results have been sub-optimal with a persistent AMR ranging between 7% to 20.5%[28,29]. Another option explored was the use of second observers (nurse observers/trainees). However, even this approach was not effective in bridging the gap and reducing AMR during screening colonoscopy[30-32]. This indicates that extending the field of vision or supplementing the limits of visualization with additional human eyes, may not fully overcome the inherent deficiencies of human attention and visualization, especially in the context of subtle colonic lesions.
In this context, the recent innovation of AI plays a pivotal role in CADe and CADx systems for polyp detection and characterization respectively. They have been pegged as a potentially disruptive technology that can herald a new era in CRC prevention strategies. The success and practical utility of these systems hinges on a low false positive rate and low latency time defined as the time from the first appearance of the polyp to detection in real time[33]. In other words, these systems have to show high accuracy, fidelity, consistency and enable real-time detection (low latency time) of polyps that are otherwise missed[34]. In this section, we will summarise the current status of ML systems in this area and discuss the future of this technology in the CRC prevention programs.
Evolution of AI in polyp detection
Initial application of AI in gastroenterology was limited to ‘edge detection’ by identifying sharp changes in image brightness and ‘region growing’ by a group of pixels of similar properties. This was essentially useful in lesions when edges were undetectable in standard endoscopic images[35]. The first polyp detection software CoLD (colorectal lesions detector), was developed in 2003 with an accuracy of 93%[36]. With the advancement of endoscopic imaging quality, subsequent DNN systems could make use of additional features like color, temporal factors and texture of the polyps with a high level of precision[37]. Subsequently, novel deep learning techniques were applied that could take advantage of image processing and vast datasets, to enable complex functions like polyp classification leading to a shift in our approach. Since, then, multiple systems have been developed that have shown improved results and accuracy[16,17,38,39]. Moreover, robust image databases and the use of video-based algorithms have provided an effective training as well as testing platform. This has led to an array of CADe and CADx systems that have become commercially available in the last 5 years[16,38,40].
Real time use of CADe systems for polyp detection
CADe systems have been well-validated in real-time colonoscopic examinations. They have demonstrated high accuracy for polyp detection, especially for polyps less than 5mm and those between 5-9 mm. These systems have enabled the identification of lesions that are subtle, obscured by debris, poorly visualised due to specular reflections or lesions at the edge of the screen[41]. Different CADe techniques have demonstrated promising results in polyp detection, especially when combining different DL methodologies. Not surprisingly, larger datasets appear to improve overall measures of performance[17]. Among these, a CADe system developed by Wang et al[34] was the first one to be validated in a large multi-centric trial. The system was developed on a large dataset of over 1200 patients and was independently validated on two separate datasets, including over 27000 images and nearly 200 colonoscopy videos, generating 100% specificity and a latency of 76.8 ms. Patients were then randomized to undergo routine diagnostic colonoscopy (n = 536) or real-time CADe assisted colonoscopy (n = 522). The CADe system significantly increased ADR (29.1% vs 20.3%; P < 0.001), mean number of adenomas per patient (0.53 vs 0.31; P < 0.001), and overall polyp detection rate (45% vs 29%, P < 0.001). Not only did the CADe system increase polyp and adenoma detection rates, it identified significantly more flat and sessile polyps, as well as diminutive polyps. There were however, a few false positives in this study (0.075 per colonoscopy) which were attributed to air bubbles, mucosal inflammation and retained fecal matter. The same study group then performed another study of their CADe system to assess its efficacy in reducing AMR among patients undergoing screening colonoscopy. In this study tandem colonoscopies were performed for each participant by the same blinded endoscopist, wherein, patients were randomly assigned to groups that received either CADe assisted colonoscopy or routine colonoscopy first, followed immediately by the other procedure. They found that AMR was significantly lower with CADe assisted colonoscopy (13.89%) than with routine colonoscopy (40%)[24].
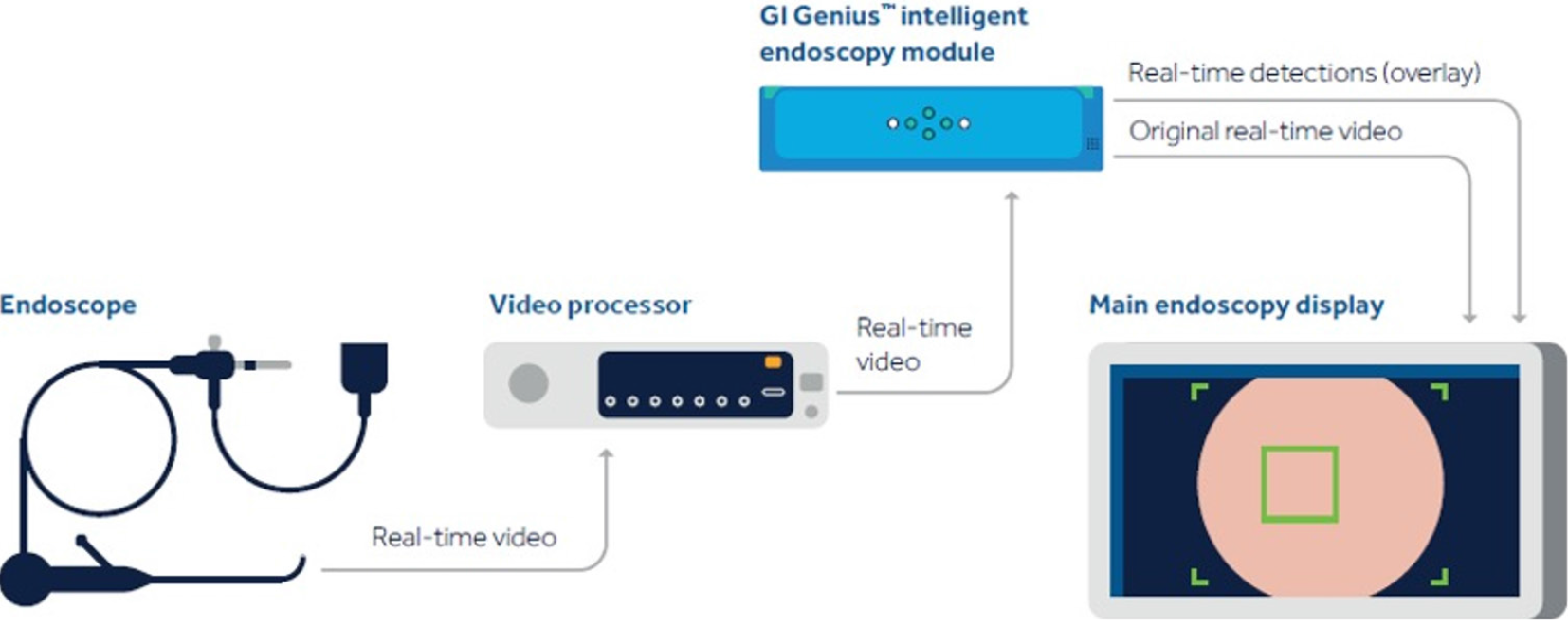
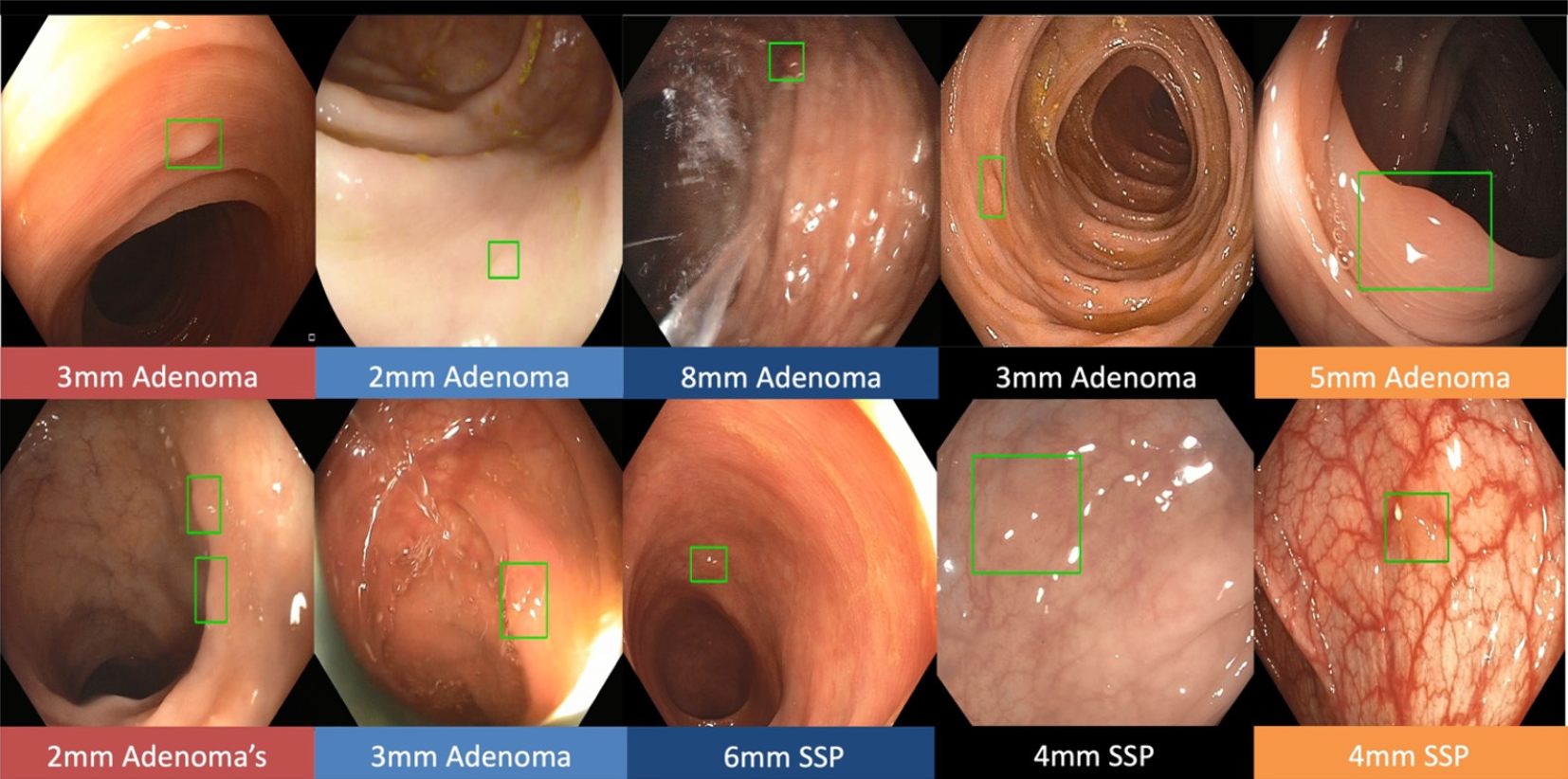
Real-time CADe during screening colonoscopy, tested on several hours of colonoscopy videos, were also found to have a high accuracy of almost 97%[15,38]. In a study by authors Urban et al[15], deep neural networks (DNN) to detect polyps was developed using a diverse and representative set of 8641 hand labeled images from screening colonoscopies collected from over 2000 patients. This was tested on 20 colonoscopy videos. Gold standards were developed with the help of experts who were asked to identify all polyps in de-identified videos. They found that their CADe system had an accuracy of 96.5% and can detect and localize polyps well within real-time constraints. In a recent publication, Repici et al[42] evaluated the AI system developed by Medtronic based on a convolutional neural network, called GI-GeniusTM (Figure 1). In this randomized, controlled study, GI-GeniusTM detected significantly more adenomas with an adenoma detection rate of 54.8%, irrespective of withdrawal time[31] (Figure 2). Adenomas detected per colonoscopy were also higher in the GI-GeniusTM group (mean 1.07 ± 1.54) than in the control group (mean 0.71 ± 1.20) (incidence rate ratio 1.46; 95%CI, 1.15-1.86). This improved ADR was mainly seen in polyps < 5 mm and polyps with 5-9 mm diameter. These findings indicate that CADe systems are clearly an effective strategy to increase ADR and could prove to be indispensable in the future[42]. The imperative question however, is not whether it can merely ‘detect’ what was missed by the human eye, but whether it can provide additional information by identifying patterns that are otherwise invisible to the human eye?
Figure 1 Gastrointestinal GeniusTM Intelligent endoscopy module by Medtronic.
©2020 Medtronic. All rights reserved. Used with the permission of Medtronic.
Figure 2 The green boxes indicate examples of challenging polyps detected by Gastrointestinal GeniusTM Intelligent endoscopy module by Medtronic, including diminutive polyps, flat polyps, or polyps obscured by light reflection.
©2020 Medtronic. All rights reserved. Used with the permission of Medtronic.
The leap from polyp detection to histological characterization
The leap from merely detecting a polyp to accurate histological characterization has opened up a new paradigm of screening colonoscopy for CRC prevention. Two alternate strategies have been proposed for the management of diminutive polyps that may have far-reaching consequences in clinical practice and healthcare economics. These two approaches are ‘Resect and discard’ and ‘leave-in-situ’ strategies[43,44]. The advanced imaging capabilities achieved through CADx make the above choices a welcome reality. Thus, when an adenomatous dimunitive polyp is diagnosed by a CADx system, ‘resect and discard’ approach can be safely undertaken. At the same time, a non-neoplastic diminutive polyp found on colonoscopy can be safely managed with ‘leave-in-situ strategy. These alternate strategies have important advantages like cost reduction, avoiding adverse events related to polypectomy with its resultant shorter procedure time[45]. Both these strategies are highly dependent on advanced imaging systems that provides a precise, real-time identification of the polyp. However, both strategies have not found good penetration outside of expert centres as current imaging systems do not meet the appropriate thresholds for accuracy[44,46]. CADx systems could be the answer in these situations by improving the diagnostic accuracy of existing imaging systems[47].
Initial experience with CADx systems showed that they were able to discriminate adenomatous from hyperplastic polyps when using magnification chromoendoscopy or magnification narrow-band imaging (NBI)[18,48,49]. However, these used traditional AI techniques which limited its real-time application as it required manual segmentation of polyp margins and captured images that required magnification technologies that were not widely available. With the development of DNN techniques, newer CADx systems addressed these issues and have shown a lot of promise in preliminary real-time polyp classification. In a prospective single-operator trial of 41 patients, diagnostic accuracy of 93.2% was shown for a real-time CADx system on 118 colorectal lesions evaluated with magnifying NBI before resection. Among the subset of patients with diminutive polyps, exceeding the Preservation and Incorporation of Valuable Endoscopic Innovations (PIVI) initiative threshold of ≥ 90% for the “resect and discard” strategy, 92.7% showed concordance between the CADx diagnosis and the pathological findings[50]. This highlights the massive impact that CADx systems can potentially have in reducing costs associated with CRC screening programs.
Advanced imaging techniques such as NBI have come into routine use and supplemented our ability to better characterize colonic polyps. Moreover, emerging techniques of incorporating NBI images, with and without magnification, to create datasets for CADx systems, especially with larger image and video banks, have yielded highly sensitive systems with high negative predictive values[16,48,51]. The level of performance of these CADx systems in conjunction with NBI imaging have been shown to meet the minimum threshold for a ‘diagnose and leave-in-situ’ strategy (90% NPV) as proposed by the American Society for Gastrointestinal Endoscopy PIVI initiative[43]. In a very interesting study by Jin et al[14], CADx improved the overall accuracy of optical polyp diagnosis from 82.5% to 88.5% (P < 0.05). In particular, CADx assistance was most beneficial to novices with limited training in using enhanced imaging techniques for polyp characterization, where accuracy jumped from 73.8% to 85.6% which was comparable to the endoscopy experts. This finding has significant implications on the feasibility of implementation of CADx systems in routine practice.
Endocytoscopy
Endocytoscopy is another evolving technology that involves ultramagnification that can detect microscopic changes at the level of the nuclei (abnormal spindle shaped nucleus, loss of polarity)[52]. It is conceivable that innovation in endoscytoscopy with CADx systems may one day, replace conventional histopathological examination through tissue acquisition, fixation, staining and microscopic examination. In a study of 791 consecutive patients who underwent colonoscopy with endocytoscopes, CADx was able to characterize diminutive rectosigmoid polyps in real time with an accuracy of 94% and an NPV of 96%, which supports the use of “diagnose and leave in situ strategy” for nonneoplastic polyps[11].
Limitations of AI in screening colonoscopy
Although automatic polyp detection has shown promising results, it is yet to live up to expectations. A number of factors can affect the performance of AI-based systems including camera motion, strong light reflection, poor focus, polyp morphology, presence of bubbles and retained fecal material. When it comes to CADx systems, accuracy of tissue characterisation can be affected by inadequate staining and surface cleaning and inability to obtain a cross sectional view[53]. Nevertheless, the advent of AI system through improved detection and histological characterisation could lead to increased ADR and reduce missed adenomas, leading to lowered incidence of interval CRC.
Future of AI in screening colonoscopy
CADx systems, once validated in real-time use for polyp characterization, could enable the implementation of ‘Resect and discard’ and ‘leave-in-situ’ startegies. These strategies have been shown to reduce the cost of care dramatically. In a study by Mori et al[54], the use of CADx system for polyp characterisation in order to implement ‘leave in situ’ strategy resulted in a significant cost saving of 10.9%. In addition, these strategies could potentially reduce procedure time and reduce adverse events related to unnecessary polypectomies.
Recent findings have shown promising results with the use of video analysis and its potential advantages. Video-based algorithms have several advantages over image-based algorithms. Since a video is basically a series of images over time, it provides vital spatiotemporal information as in real life, that is not available in still images. When such spatiotemporal information is combined with CAD system, its performance can be significantly improved. This is especially true for colonic polyps since there is marked difference between the polyp and the surrounding mucosa which is easily picked up on a video analysis[55]. However, video-based algorithms need further validation in controlled settings.
Another aspect where AI could potentially improve colonoscopy performance, in general and screening colonoscopy in particular, is its role in quality control and monitoring[56,57]. These algorithms can potentially monitor endoscopic quality, by which it can indicate colonic surface missed during withdrawal, need for a slower speed of withdrawal, areas of poor bowel preparation necessitating adequate cleansing before moving on. Although this area has not been investigated thoroughly, an argument can be made that this might have an equal, if not bigger, impact on clinical outcomes of CRC screening programs than a specific lesion detection tool for a specific pathology.
Several questions remain to be answered in order to fine-tune the role of AI in polyp detection. However, with the advent of advanced systems that combine multiple functions, the time seems appropriate to embrace this technology and troubleshoot issues along the way, rather than delay the adoption of AI in our daily practice in the hope of achieving perfection.
ROLE OF AI IN THE EVALUATION OF PANCREATIC DUCTAL ADENOCARCINOMA USING EUS
Pancreatic ductal adenocarcinoma (PDAC) has a dismal prognosis with a five-year survival rate of approximately 6%[58]. PDAC is also associated with significant morbidity and accounts for 3.9% Disability Adjusted Life Years(DALY) related to cancers. Moreover, future estimates indicate that the PDAC burden is likely to double within the next four decades[59]. The incidence of PDAC in the United States is increasing by 0.5% to 1.0% per year, and is expected to be the second-leading cause of cancer-related mortality by 2030[60].
Most patients with PDAC are unresectable at the time of diagnosis owing to locally advanced (30%-35%) or metastatic disease (50%-55%) at presentation[60]. Surgical resection is possible only in around 20% of patients[61]. Despite curative resection, most of these patients will eventually have a recurrence, with a 5 year survival of around 25%[62]. However, cancers < 1 cm in size at the time of diagnosis, have been shown to have an excellent response following resection with a survival rate as high as 84.4%[63]. This highlights the paramount importance of screening and early detection for PDAC. Unfortunately, well-defined pre-malignant conditions and proper guidelines are lacking for pancreatic cancer, as compared to CRC. Moreover, current modalities of screening are inadequate and merit further evaluation before recommending routine clinical use.
Diagnosis of PDAC relies on accurate identification of the tumor by various imaging modalities, followed by a reliable method of tissue acquisition to confirm the histological characteristics of malignancy. Currently available modalities for imaging include transabdominal ultrasonography, computed tomography (CT), magnetic resonance imaging, EUS, and endoscopic retrograde cholangiopancreatography. Of these imaging modalities, EUS enables real-time observation of the pancreas with high spatial resolution, and the sensitivity of detection of PDAC using EUS has been reported to be as high as 94%[64]. Numerous studies indicate that EUS is a highly sensitive modality for the detection of pancreatic tumours and its application is especially useful for lesions less than 2 cm in size which may be missed on contrast enhanced CT studies[65]. Although the sensitivity for tumour detection is high, it is also important to note that it has a very high negative predictive value (NPV) in the background of a normal pancreas[66].
The major drawback of EUS is the fact that it is highly operator dependent and the learning curve to perfect the techniques of EUS imaging can be quite long. The American Society for Gastrointestinal Endoscopy recommends that a trainee should undergo at least two years of standard GI fellowship followed by one year of pancreatic EUS training prior to independently performing EUS[67]. ASGE also recommends that an endosonographer should perform a minimum of 150 supervised EUS procedures, including 75 pancreaticobiliary cases and 50 EUS-guided fine needle aspiration (EUS-FNA) procedures, to achieve competence in this area. In addition, specialised EUS training centres are usually inaccessible hampering the widespread application of standardised protocols for EUS screening of the pancreas[68].
Another major challenge that is faced by endosonographers is inability to correctly identify PDAC in patients with chronic pancreatitis (CP). Several studies have shown that the diagnostic yield of EUS and EUS-guided fine needle aspiration (FNA) are markedly decreased in the presence of CP[69,70]. This can be attributed to the fact that neoplastic lesions and inflammatory masses usually have a similar sono-morphology with very subtle differentiating characteristics. Studies by Fritscher-Ravens et al[71] and Varadarajulu et al[70] found EUS sensitivity to range from 54% to 73.4% respectively, in patients with CP[70,71].
AI could potentially address both these issues. In this section, a brief account of the progress made by AI-based CAD systems in image differentiation among patients with chronic pancreatitis will be presented; followed by the recent developments in the field of AI assisted EUS training systems.
Evolution of AI in endosonography
Similar to screening colonoscopy, AI is being actively investigated in the early diagnosis of PDAC. However, its application in this area is still in its infancy with no commercially available CAD systems yet. Initial reports focus on integrating AI with EUS imaging to identify PDAC in the background of CP. Several sonographic features of CP such as calcification and the presence of pseudotumors with intense desmoplasia pose significant challenges to making an accurate diagnosis of PDAC in these patients[72]. The first report of the use of an AI based system for the diagnosis of PDAC was by Norton et al[73] in 2001. In this study, 35 patients were included, of which 21 patients were histologically proven to have PDAC, while 14 patients had focal CP. Representative images with the region of interest were fed into a CAD system which was then trained to identify subtle differences in the gray scale and overall brightness within the images. These features were then assessed to differentiate between PDAC and focal CP. This early CAD system was found to have an overall diagnostic sensitivity of nearly 89%. In an effort to reduce the chances of missed malignancy, the authors found that even when the sensitivity for malignancy was set to 100%, the overall diagnostic accuracy was still around 80%. This was remarkably close to the 85% accuracy that was observed among blinded, trained endosonographers[73]. Although the technology used in this study was primitive to say the least, it was the first study that demonstrated the feasibility of integrating AI into diagnostic studies using EUS, and formed the foundation to the studies that followed. Since then, many attempts at applying conventional CAD using ANNs or SVMs have been tested, both with traditional grayscale texture features on B-mode imaging as well as on elastography images. The Area under Receiver operating characteristic curve (AUROC) in these studies ranged from 0.8 to 0.94[74-78]. Though these studies showed promising results, the accuracy in the background of CP was still far from ideal.
One of the promises of AI in the field of endoscopy, is the ability of the machine to make a diagnosis in real time imaging and assist the endoscopist in planning the next step in the management of the patient during the procedure itself. However, the multiple intricate post-processing steps that were needed in the studies that assessed the role of CAD system in EUS precluded their use during real time imaging. This was one of the main reasons for the technology remaining dormant for years after the initial proof of concept in 2001. However, encouraged by the benefits of CADe and CADx systems in screening colonoscopy, there has been renewed interest, in recent years, on the application of AI systems in EUS. A sudden surge of publications that have employed novel CAD systems for pancreatic lesions combining EUS elastography and contrast enhanced EUS studies has opened up new avenues for the role of AI based technology in this area.
AI and EUS elastography
EUS elastography (EUS-E) can transform the tissue properties based on elastic coefficients, into visible images composed of color pixels. This can provide vital information regarding the pathological state of the tissue under study and has been shown to be useful in the evaluation of pancreatic lesions. In a seminal study by Săftoiu et al[79] real-time EUS-E avoided motion artifacts and color perception errors that arose from individual selection, manipulation bias and static image analysis. Following this, a large multicentric trial was conducted in Europe in which, 744 EUS-E images from 258 patients with pancreatic lesions were studied. A detailed analysis of the color hue histogram data from the dynamic sequence of EUS-E was performed using a novel neural network, in order to distinguish benign from malignant patterns. An overall sensitivity of 87.6%, specificity of 82.9%, and positive predictive value (PPV) of 96.3% indicated that the combination of EUS-E with AI based software, could be beneficial in the real-time evaluation of pancreatic lesions[80].
Role of AI in contrast EUS and fine needle biopsy
EUS guided fine needle biopsy (EUS-FNB) has enabled reliable tissue acquisition and accurate histological diagnosis in patients with PDAC. In fact, it is considered to be the cornerstone of management of pancreatic lesions < 3 cms[81]. Multiple studies have documented a high diagnostic accuracy of EUS-FNB for PDAC with a pooled sensitivity of 87% and specificity of 96%[82]. However, these results have been negatively impacted by the presence of chronic pancreatitis. Intense desmoplasia, fibrosis and calcifications seen among patients with CP can decrease diagnostic yield of EUS-FNB because of the higher tissue impedance, poor visibility and inaccessibility of the lesion due to various factors[69,83]. Moreover, Rapid On site examination of the cytology obtained from EUS-FNA which has been shown to be a major factor that impacts diagnostic yield, is not feasible in many centers[84]. ML based algorithms have shown promise in this area by augmenting visual inspection of the histopathology slides. In a study by Inoue et al[85], an ML-based automated visual inspection system could reliably highlight areas of abnormal cellularity on the stained smears obtained after an EUS-FNB from solid pancreatic lesions.
Contrast harmonic EUS (C-EUS) uses the enhancement properties of the solid lesions and categorizes them into different patterns[86]. Multiple studies have shown C-EUS to have a pooled sensitivity of around 93% and specificity ranging between 80%-89% for pancreatic lesions[87-89]. Its ability to highlight areas of increased vascularity and to outline areas of reduced vascularity due to necrosis and fibrosis have been used during EUS-FNA, to increase the diagnostic yield[90-94].
In an elegant study by Saftiou and colleagues, a time intensity curve was made for patients with pancreatic lesions, using dynamic C-EUS examinations. Using a set of 7 features that were extracted from the data using a convolutional neural networks (CNN), sensitivity, specificity, NPV and PPV were 94.6%, 94.4%, 89.4% and 97.2%,respectively, was reported[95]. Since then, multiple studies are underway that highlight a significant ancillary role played by AI-based systems in improving the diagnostic yield of EUS-FNB with C-EUS.
Future of AI in the field of endosonography
The immediate clinical application of the results of studies using AI based systems in the field of endosonography are unfortunately limited, to say the least. This is in part due to the necessity of pre-analysis image preparation and post-processing steps that preclude real-time application[96].
A major factor in the development of machine learning models for EUS is the sheer volume of labelled images required to improve accuracy. ImageNet is one of the most popular datasets used in machine learning models. This dataset contains as many as 14 million labelled images, which is used by a majority of image recognition software. This essentially means that it takes millions of labelled images to train a machine to accurately interpret an image or video. To add to the problem, the concept of, "Garbage in and garbage out", is another cause for concern. This means that if we feed the machine poor quality/poorly labelled images, the output will be inaccurate. So, apart from the quantity of labelled images, quality is equally, if not more important.
With regard to EUS, trained endosonographers are not widely available. The time and resources required to have trained endosonographers read, label and edit an adequate number of high quality videos is impractical to implement. This is why, there has been a recent change in the paradigm of ML in EUS. Instead of depending on endosonographers alone to edit videos, investigators have begun training the machine to detect stations which can result in shortened videos focussed on the regions of interest. This would significantly reduce the time and resources required to create a high quality dataset of EUS images.
In a study by Zhang and colleagues, a novel CNN was evaluated for the accurate recognition of the EUS station as well as segment the pancreas for more detailed evaluation. Compared with EUS experts, the models achieved 90.0% accuracy in classification, which is comparable with that of experts[97]. In 2019, Kuwahara et al[98] evaluated the use of DL based CAD with CNN to achieve two objectives – accurately determine the station of the EUS probe as well as differentiate between malignant vs benign intraductal papillary mucinous neoplasms (IPMN) of the pancreas. The area under ROC curve for CAD systems to diagnose malignant IPMNs was found to be was as high as 0.98 (P < 0.001). The sensitivity, specificity, and accuracy was found to be 95.7%, 92.6%, and 94.0%, respectively; which was significantly higher as compared to expert endoscopists in the study.
In addition to accurate classification of lesions, AI based systems could potentially be beneficial by supplementing EUS training programs. This can eventually result in a uniform, high quality EUS examinations which are more amenable to the application of CAD systems that can identify and diagnose pancreatic lesions in real-time. In the study by Zhang et al[97], the developed CAD system was subsequently validated on trainees, where they found that diagnostic accuracy improved from 67.2% to a significant 78.4% for the evaluation of solid pancreatic lesions.
In the most recent study by Tonozuka et al[99], a complex CNN based DL method was employed for the detection of PDAC. They found improved performance of this automated system with AUROC of 0.924 and 0.940 in the validation and test setting, respectively. However, there have been very few head-to-head comparison studies that have compared the efficacy of CADe systems for the diagnosis of PDAC and its role in image differentiation merits further clarity.
There are several potential benefits likely to arise from the use of AI based CAD systems in the field of EUS. Firstly, AI can augment EUS expertise especially by shortening the learning curve. Although, there is very little data to support this statement, initial results are extremely encouraging and it would be reasonable to surmise a significant role played by AI-based automated systems in EUS training programs. Secondly, the recent innovations using CNN based DL algorithms have the potential to significantly augment the diagnostic accuracy of EUS and could, conceivably overcome the inherent deficiencies of human error, visualisation, inattention and fatigue. Finally, our rudimentary foray into this area, coupled with the encouraging results seen in the case of endocytoscopy-based CADx systems for colonic polyps; could pave the way for optical diagnosis of pancreatic lesions in the future. This could theoretically, expand the role of EUS in the context of solid pancreatic lesions, by enabling the accurate diagnosis of lesions which are poorly accessible, failed EUS-FNA (high tissue impedance, intervening vessels) or poor visualisation due to calcifications and fibrosis secondary to CP.
However, the current systems possess major drawbacks that hamper the uniform application of AI -based CAD systems for EUS in clinical practice. One of the major drawbacks is the "black box phenomenon" where the basis of a decision taken by the machine is not clearly understood by the programmers and developers. This makes it difficult to course-correct the system in case of sub-optimal accuracy. Another important drawback is the fact that real-time video and the tactile understanding of the location of the scope, plays a major role in decisions with regard to EUS-FNA. These data inputs are currently not factored into the DL algorithms and could significantly hamper its clinical applicability.
CONCLUSION
The tremendous progress witnessed in the field of artificial intelligence and machine learning has enabled the development of novel and innovative algorithms that can perform specific functions in the field of endoscopy. Although AI based systems have shown immense promise in the prevention of CRC by detecting and characterising colonic polyps, the systematic incorporation of these systems in our everyday practice is still lacking. While it is intuitive to engage our efforts on the implementation of these systems in our endoscopy practice, there needs to be a clear agreement and consensus as to the specific gaps that can be addressed by AI based systems. This could improve efficiency of implementation and efficacy, thereby enabling the translation from mere ‘promise’ to measurable ‘impact’ on global screening programs. There are encouraging steps taken in that regard, where novel approaches like ‘Leave-in-situ’ and ‘Resect and Discard’, can potentially change the landscape of CRC screening programs. Validated and reliable CADx systems can enable the adoption of these strategies. The most critical and exciting aspect is the potential to implement these strategies at the community level in emerging economies like India, where CRC prevalence have shown alarming upward trends in the past decade, owing to a higher prevalence of metabolic risk factors and changing patterns of diet and lifestyle practices. These strategies can reduce the cost of screening programs significantly by obviating the need for histopathological evaluation of small diminutive polyps. In addition, the reduced requirement of specialised man-power, logistical issues and equipment installation at primary care centres in the community can make CRC screening programs economically viable and a welcome addition to global efforts to reduce the burden of CRC.
AI in the field of EUS, however, is still in its infancy. Given the present lacunae in the diagnosis of early PDAC, there is significant scope for the application of AI-based CADe and CADx systems, which can augment our capabilities to manage patients with solid pancreatic lesions with/without CP in the future. However, there is an acute need to re-examine the available approaches to development of CADe and CADx systems in this area. The specific functions and questions that need the assistance of AI based systems needs to be clarified by expert consensus before we embark further on the development of newer systems.
In conclusion, there is an urgent need, now more than ever before, for future collaborative projects with the ever-expanding world of data science and artificial intelligence, which could pave the way for a brave new world, of man and machine, acting in concert to bring about the technological age of modern medicine.