Published online Aug 28, 2021. doi: 10.37126/aige.v2.i4.185
Peer-review started: June 11, 2021
First decision: June 24, 2021
Revised: June 25, 2021
Accepted: August 18, 2021
Article in press: August 18, 2021
Published online: August 28, 2021
Processing time: 86 Days and 19.6 Hours
Early gastrointestinal (GI) cancer has been the core of clinical endoscopic work. Its early detection and treatment are tightly associated with patients’ prognoses. As a novel technology, artificial intelligence has been improved and applied in the field of endoscopy. Studies on detection, diagnosis, risk, and prognosis evaluation of diseases in the GI tract have been in development, including precancerous lesions, adenoma, early GI cancers, and advanced GI cancers. In this review, research on esophagus, stomach, and colon was concluded, and associated with the process from precancerous lesions to early GI cancer, such as from Barrett’s esophagus to early esophageal cancer, from dysplasia to early gastric cancer, and from adenoma to early colonic cancer. A status quo of research on early GI cancers and artificial intelligence was provided.
Core Tip: Diagnosis and management of early gastrointestinal (GI) cancer is one of the cores of clinical practice. Endoscopy is the indispensable tool for standard surveillance and management. Artificial intelligence is a novel technology used in some fields of cancer including early GI cancer. Therefore, we provide an overview and introduce how artificial intelligence can be applied to endoscopy on early GI cancer mainly including esophagus, stomach, and colon from the point of view of the clinical diagnosis and management guidelines. Studies with quality control on the diagnosis and management of early GI cancer and their precancerous lesions have also been concluded.
- Citation: Yang H, Hu B. Early gastrointestinal cancer: The application of artificial intelligence. Artif Intell Gastrointest Endosc 2021; 2(4): 185-197
- URL: https://www.wjgnet.com/2689-7164/full/v2/i4/185.htm
- DOI: https://dx.doi.org/10.37126/aige.v2.i4.185
Artificial intelligence (AI) is essentially a process of learning human thinking and transferring human experience. Recognizing images based on artificial neural networks/convolutional neural networks (CNNs) is one of the novel and main fields of AI. Computer-aided diagnosis (CAD) systems are designed to interpret medical images using advances in AI from method learning to deep learning (DL) and includes mainly three groups (CADe, CADx, and CADm)[1].
AI has been widely involved in cancer[2]. In regard to digestive cancer, it has been utilized to find more intelligent ways to facilitate detection, diagnosis, risk evaluation, and prognosis. For instance, radiomics machine learning signature for diagnosing hepatocellular carcinoma in cirrhotic patients with indeterminate liver nodules was also validated in a multicenter retrospective cohort, which could enhance clinicians’ decisions[3].
In the aspect of pancreatic cancer, it continues to be one of the deadliest malignancies with less than 10% overall survival rate. Survival rates will increase if pancreatic cancer can be detected at an early stage[4]. Intraductal papillary mucinous neoplasms are precursor lesions of pancreatic adenocarcinoma. A DL model was shown to be a more accurate and objective method to diagnose malignancies of intraductal papillary mucinous neoplasms in comparison to human diagnosis and conventional endoscopic ultrasonography (EUS) images[5]. Pancreatic cystic lesions are also precursors of pancreatic cancer. Radiomics utilizing quantitative image analysis to extract features in conjunction with machine learning and AI methods helped differentiate benign pancreatic cystic lesions from malignant ones[6]. An artificial neural network was trained to help predict pancreatic ductal adenocarcinoma based on gene expression[7]. An AI-assisted CAD system using DL analysis of EUS images was efficient to help detect pancreatic ductal carcinoma[8]. The artificial neural network model could accurately predict the survival of pancreatic adenocarcinoma patients as a useful objective decision tool in complex treatment decisions[9].
In this review, we concluded the application and research of AI based on endoscopic examination related to early gastrointestinal (GI) cancer mainly including esophagus, stomach, and colon. The progression of carcinogenesis from Barrett’s esophagus (BE) to early esophageal cancer (EEC), from dysplasia to early gastric cancer (EGC), and from adenoma to early colonic cancer (ECC) were reviewed in detailed as well as related AI research on the histopathology and invasion depth detection of these GI cancer.
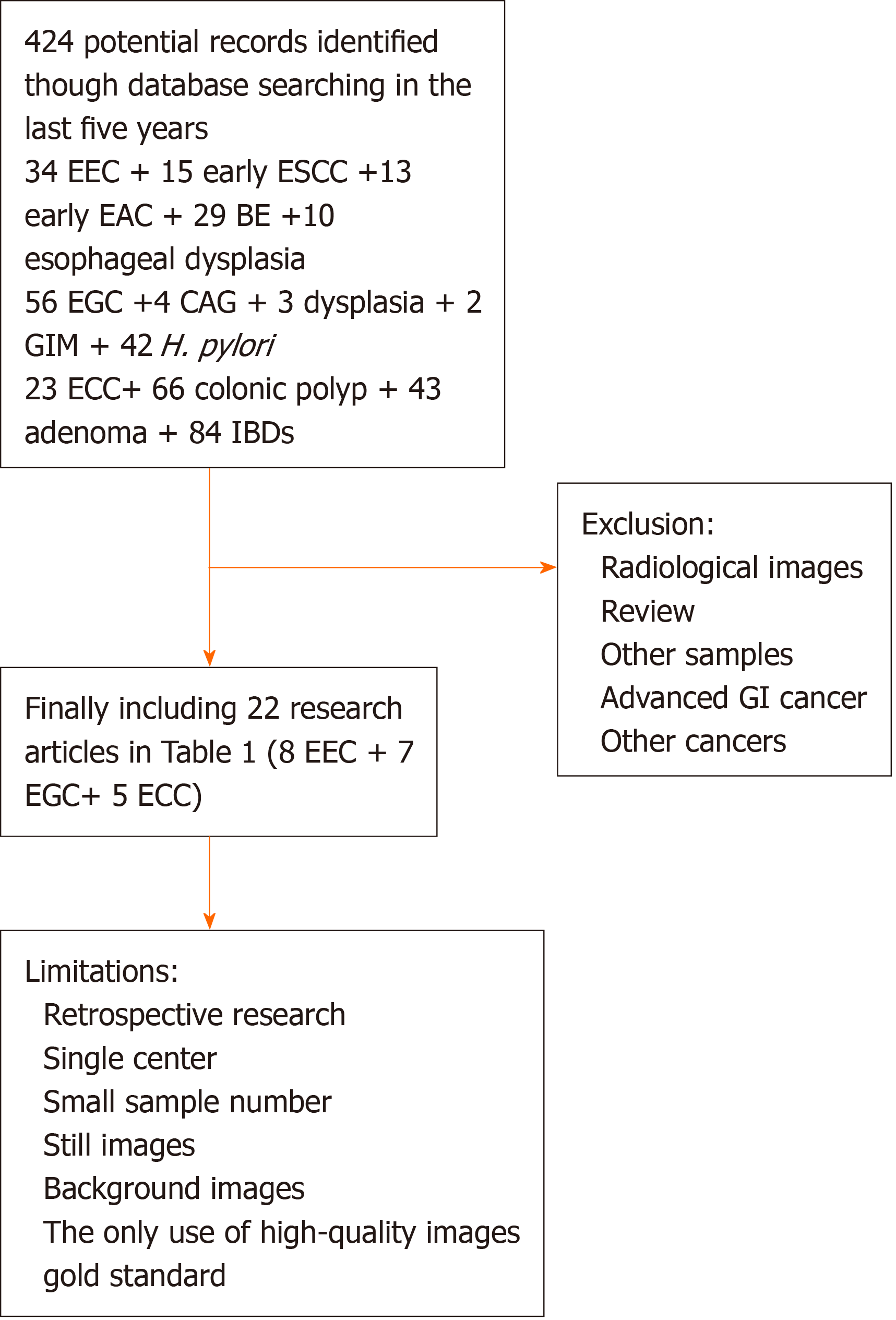
This review was aimed to make a qualitative only review of the application of AI on early GI cancer. We searched the PubMed database for articles that were published in the last 5 years using the term combinations of AI/DL and EEC, esophageal squamous cell carcinoma (ESCC), esophageal adenocarcinoma (EAC), EGC, and ECC for early GI cancer, and term combinations of AI/DL and precancerous lesions [BE/ dysplasia/chronic atrophic gastritis (CAG)/gastric intestinal metaplasia/Helicobacter pylori/adenoma/polyp/inflammatory bowel diseases] for precancerous lesions of early GI cancer. Endoscopic-related results were qualitatively concluded in Table 1.
| Ref. | Target disease | Prospective/ retrospective | AI | Endoscopy image | Training dataset | Validation dataset | Sensitivity | Specificity | Accuracy1/AUC | |
| [1] | Diagnosing ESCC and EAC | Retrospective | CNNs (SSD) | WLI and NBI | 8428 images | 1118 images | 98% | 95% | 98%1 | |
| [2] | Diagnosing ESCC | Retrospective | CAD (SegNet) | NBI/videos | 6473 images | 6671 images | 98.04% | 95.03% | 0.989 | |
| [3] | Detecting EEC and BE | Retrospective | CAD (ResNet-UNet) | WLI | 494364 images | 1704 images | 90% | 88% | 89%1 | |
| [4] | Detecting E/J cancers | Retrospective | CNNs (SSD) | WLI and NBI | 3443 images | 232 images | 94% | 42% | 66%1 | |
| [5] | Detecting ESCC | Retrospective | DCNNs-CAD | NBI | 2428 images | 187 images | 97.80% | 85.40% | 91.4%1 | |
| [6] | Diagnosing BE and EAC | Retrospective | CAD (ResNet) | WLI and NBI | 148/100 | Leave-one patient-out cross validation | 97%(WLI)/94%(NBI) | 88% (WLI)/80%(NBI) | ||
| [7] | Diagnosing ESCC | Retrospective | CAD (FCN) | ME-NBI | 3-fold cross-validation | |||||
| [8] | Detecting EAC | Retrospective | CNNs (SSD) | WLI | 100 images | 96% | 92% | |||
| [9] | Detecting EGC | Retrospective | CNNs | WLI | 348943 images | 9650 images | 80.00% | 94.80% | ||
| [10] | Diagnosing EGC | Retrospective | CNNs | WLI | 21217 images | 1091 images | 36.8 | 91.20% | ||
| [11] | Diagnosing EGC | Retrospective | CNNs (Inception-v3) | ME-NBI | 1702 images | 170 images | 91.18% | 90.64% | 90.91%1 | |
| [12] | Diagnosing EGC | Retrospective | CNNs (VGG16) | WLI | 896 t1a-EGC and 809 t1b-EGC | 5-fold cross-validation | Detection (0.981) | |||
| Depth prediction (0.851) | ||||||||||
| [13] | Detecting EGC | Retrospective | CNNs (VGG16 and ResNet-50) | WLI/NBI/BLI | 3170 images | 94.00% | 91.00% | 92.5%1 | ||
| [14] | Diagnosing EGC | Retrospective | CNNs (ResNet-50) | WLI | 790 images | 203 images | 76.47% | 95.56% | 89.16%1 | |
| [15] | Detecting EGC | Retrospective | CNNs (SSD) | WLI | 13584 images | 2940 images | 58.40% | 87.30% | 0.76 | |
| [16] | Classifying EGC | Retrospective | CNNs (Inception-ResNet-v2) | WLI | 5017 images | 5-fold cross-validation | 0.85 | |||
| [17] | Diagnosing EGC | Retrospective | CNNs (ResNet-50) | ME-NBI | 4460 images | 1114 images | 98% | 100% | 98.7%1 | |
| [18] | Detecting and localizing colonic adenoma | Representative | CNNs (VGG16,19, ResNet50) | WLI and NBI | 8641 images/9 videos, 11 videos | Cross-validation | ||||
| [19] | Detecting ECC | Representative | CNNs | WLI | 190 images | 3-fold cross-validation | 67.50% | 89.00% | 81.2%1/0.871 | |
| [20] | Classifying ECC | Representative | CNNs (ResNet-152) | WLI | 3-fold cross-validation | 95.40% | 30.10% | |||
| [21] | Detecting colonic adenoma | Prospective | Cade | 1058 patients | ADR (29.1% vs 20.3%) | |||||
| [22] | Detecting colonic adenoma | Prospective | Cade | 962 patients | ADR (34% vs 28%) | |||||
Initially, a total of 424 articles were identified. After manually screening and reading, 22 studies were tabulated in Table 1, and 2 prospective studies on detecting adenoma were also added in Table 1. Meanwhile, 13 studies on precancerous lesions of early GI cancer were showed in the review. The flowchart was presented in Figure 1.
Esophageal cancer is one of most common cancers related to a considerable decline in health-related quality of life and a reduction in survival rate. ESCC and EAC are two main histological types. Many patients with ESCC have a history of heavy tobacco and alcohol use[10] as well as other risk factors including polycyclic aromatic hydro
There is also study on AI being involved in preclinical stage. For instance, the diagnostic ability of AI using DL to detect esophageal cancer including superficial and advanced squamous cell carcinoma and adenocarcinoma was characterized as highly sensitive (98%) and efficient based on WLI images. Small cancer lesions less than 10 mm in size could be detected[15].
In terms of EAC, AI using DL to diagnose superficial esophagogastric junctional adenocarcinoma showed favorable sensitivity (94%) and acceptable specificity (42%) of WLI images compared with experts[16]. A CAD using DL (CAD-DL) model was trained by two datasets based on two different kinds of images (WLI and NBI images) used to detect early EAC. The diagnosis of EAC by CAD-DL reached sensiti
In terms of ESCC, the endocytoscopic system (ECS) helps in virtual realization of histology. The CNN method was applied to detect ESCC with an overall sensitivity of 92.6% based on ECS images aimed at replacing biopsy-based histology[19]. NBI is currently regarded as the standard modality for diagnosing ESCC. A CNN model was applied to detect ESCC based on NBI images and showed significantly higher sensitivity (91%), specificity (51%), and accuracy (63%) than those of endoscopic experts[20]. Besides NBI and ECS, AI was also applied in magnified endoscopy (ME). The accuracy, sensitivity, and specificity of AI based on ME images were 89%, 71%, and 95% for the AI system, respectively[21]. Accuracy, sensitivity, and specificity with WLI images were 87%, 50%, and 99%, respectively. Furthermore, as endoscopic resection (ER) is often used to treat ESCC when invasion depths are diagnosed as intraepithelial–submucosal layer (tumor invasion is within 0.5 mm of the muscularis mucosae). The invasion depth of superficial ESCC was also calculated by a CNN method based on WLI and NBI images, which demonstrated higher accuracy. The diagnosis accuracy of the CNN method was higher in the intraepithelial-lamina propria and muscularis mucosa groups (91.2% and 91.4%, respectively) than that in the submucosal layer group (67.8%)[22].
Recently, there have been some application and research of AI on precursor lesions of EEC including BE and dysplasia in squamous epithelium. For instance, AI could enhance the image of volumetric laser endomicroscopy to facilitate the surveillance BE[23]. The CNN method was developed to recognized early esophageal neoplasia in BE. It could correctly detect early neoplasia with the sensitivity of 96.4%, the specificity of 94.2%, and the accuracy of 95.4%. In addition, the object detection algorithm was able to draw a localization box around areas of dysplasia with a mean average accuracy of 75.33% and sensitivity of 95.60%[24]. Another similar research demonstrated that a CAD system used five independent endoscopy datasets to detect early neoplasia in patients with BE. In dataset 4, the CAD classified images as containing neoplasms or non-dysplastic BE with 89% accuracy, 90% sensitivity, and 88% specificity. The CAD also identified the optimal site for biopsy of detected neoplasia in 97% of cases in dataset 4[25].
Moreover, AI was also applied in esophageal histopathology; attention-based deep neural networks were used to detect cancerous and precancerous esophagus tissue on histopathological slides. Classification accuracies of the proposed model were 85% for the BE-no-dysplasia class, 89% for the BE-with-dysplasia class, and 88% for the adenocarcinoma class[26].
EGC is defined as a cancer confined to the mucosa or submucosa, regardless of lymph node metastasis (LNM). Standard WLI and image enhancement endoscopy, such as NBI and ME, have been widely used in screening and surveillance of EGC as well as EUS, which can enable the precise assessment of the risk of LNM of EGC[27]. Risk factors include Helicobacter pylori infection, age, high salt intake, diets low in fruit and vegetables, and genetic factors[28]. ER is a minimally invasive treatment for EGC with negligible risk of LNM[29]. Patients with CAG, intestinal metaplasia, or dysplasia are at risk for gastric adenocarcinoma and are recommended to accept the regular endoscopic surveillance. Virtual chromoendoscopy can guide biopsies for staging atrophic and metaplastic changes and can target neoplastic lesions[30]. The 5-year survival rate of EGC patients is significantly higher than that of advanced GC patients[31,32]. Early detection and treatment are always one of the top priorities.
In regard to the application of AI in EGC, there are some considerations both related on the promise such as the benefits for endoscopists and patients and limitations[33]. To detect and diagnose EGC via ME with NBI (ME-NBI) requires considerable experience; AI-assisted CNN CAD system based on ME-NBI images was constructed to diagnose EGC, and the overall accuracy, sensitivity, and specificity of the CNN were 98.7%, 98.0%, and 100%, respectively, in a short period of time[34]. Different deep CNN methods have been designed (such as VGG, Single-Shot Multibox Detector, and ResNet) based on different image types (such as WLI, NBI, and chromoendoscopy) and mucosal backgrounds (normal mucosa, superficial gastritis, and erosive mucosa) (shown in Table 1). There was also research on differentiating EGC from gastritis[35] and peptic ulcer[36] achieving reliable accuracy.
Moreover, training with video is considered to improve accuracy in a real clinical setting. A CNN model based on videos demonstrated a high detection rate (94.1%) with a high processing speed[37]. Furthermore, CNN-CAD was applied to diagnose the invasion depth of GC based on WLI images and distinguish EGC from advanced GC, with the sensitivity of 76.47%, specificity of 95.56%, and accuracy of 89.16%[38]. Another model was also involved in invasion depth. For instance, a CNN method (lesion-based VGG-16 model) was used to classify EGC with of sensitivity (91.0%), specificity (97.6%), and accuracy (98.1%), respectively. The prediction of invasion depth achieved sensitivity (79.2%), specificity (77.8%), and accuracy (85.1%), respectively, higher than results of non-lesion-based models, indicating a lesion-based CNN was an appropriate training method for AI in EGC[39].
In terms of histopathology, a CNN model trained with pixel-level annotated hematoxylin and eosin stained whole slide images achieved a sensitivity near 100% and an average specificity of 80.6% in diagnosing GC, aimed at alleviating the workload and increasing diagnostic accuracy[40]. Similarly, AI automatically classified GC in hematoxylin and eosin stained histopathological whole slide images from different groups and demonstrated favorable results[41,42]. Besides endoscopic images, machine learning based on radiographic-radiomic images could help predict adverse histopathological status of GC[43]. Dual-energy computed tomography based DL radiomics could improve LNM risk prediction for GC[44]
In the aspect of gastric precancerous conditions, the application of AI has also been focused. For example, atrophic gastritis, as a kind of precancerous condition was diagnosed by the pretrained CNN based on WLI images achieved an accuracy of 93% in an independent dataset, outperforming expert endoscopists[45]. The CNN method was trained by WLI images of gastric antrum in diagnosing CAG, and the diagnostic accuracy, sensitivity, and specificity were 94.2%, 94.5%, and 94.0%, respectively, which were higher than those of experts. The further detection rates of mild, moderate, and severe atrophic gastritis were 93%, 95%, and 99%, respectively[46]. Helicobacter pylori infection, as a dominant cause of CAG and GC, has also been detected via AI method based on endoscopic images, such as CNN (GoogLeNet) and CNN (ResNet-50 model), and achieved the higher accuracy and reliability in a considerably shorter time[47-49].
ECC has been defined as a carcinoma with invasion limited to the submucosa regardless of lymph node status and according to the Royal College of Pathologists as TNM stage T1NXM0[50]. If the dysplasia is restricted to the layer of epithelium, it is defined as low-grade or high-grade intraepithelial neoplasia. Mild or moderate dysplasia is the pathological character of low-grade intraepithelial neoplasia, and severe dysplasia is the pathological character of high-grade intraepithelial neoplasia or preinvasive carcinoma[51]. Colonic precancerous lesions include traditional serrated adenoma and sessile serrated adenoma/polyps[52,53]. The submucosal invasion in clinical practice is considered as the superficial depth of tumor invasion and further as a surrogate for nominal LNM risk. Meanwhile, it can be a general criterion to identify whether patients are eligible for local ER or surgery[54]. Curative ER is indicated for lesions confined to the mucosal layer or invading less than 1 mm into the submucosal layer[50]. Endoscopic screening is proven to decrease the risk of disease-specific morbidity and mortality[55]. Current guidelines recommend screening beginning at age 50 and continuing until age 75 with fecal immunochemical test every year, flexible sigmoidoscopy every 5 years, and/or colonoscopy every 10 years[56]. Early diagnosis and treatment are pivotal. When colon carcinoma is detected in a localized stage, the 5-year relative survival is 91.1%. However, the 5-year relative survival of colon carcinoma patients with regional metastasis or distant metastasis were 71.7% and 13.3%, respectively[57].
AI has been widely involved in the research of ECC on the aspect of detection, diagnosis, classification, invasion depth, and histopathology as well as inflammatory bowel diseases associated with inflammation-dysplasia-colon cancer pattern. Regarding the detection and diagnosis, a research trained Faster R-CNN with VGG16 based on WLI images and videos covering ECC (Tis or T1) and precursor lesions including hyperplastic polyps, sessile serrated adenoma/polyps, traditional serrated adenoma, low-grade intraepithelial neoplasia, high-grade intraepithelial neoplasia, and submucosal invasive cancer was conducted. It showed the sensitivity and specificity were 97.3% and 99.0%, respectively[58]. Another research used two CNN methods trained by WLI images. ResNet-152 showed a higher mean area under the curve for detecting tubular adenoma + lesions (0.818), and the mean area under the curve for detecting high-grade intraepithelial neoplasia + lesions reached 0.876 by ResNet-v2[59]. Regarding the invasion depth, for deeply invasive cT1 (SM) (hereafter, cT1b) or deeper colorectal cancer (CRC), there is a 10%–15% or higher risk of lymph node metastases. Further surgical resection including lymph node dissection is required[60]. For an accurate depth of invasion diagnosis, the CNN method was used to assist in cT1b diagnosis and demonstrated that cT1b sensitivity, specificity, and accuracy were 67.5%, 89.0%, and 81.2%, respectively[61].
In the research of AI application in precancerous lesions such as polyps, there has been some research of AI, especially retrospective research related to polyp detection and diagnosis with high accuracy[62,63]. For example, a local-feature-prioritized automatic CADe system could detect laterally spreading tumors and sessile serrated adenoma/polyps with high sensitivity from 85.71% to 100%[64]. Besides retrospective research, AI has been designed into some associated prospective research. For instance, a multicenter randomized trial used CAD to detect colorectal neoplasia. It showed a significant increase in adenoma detection rates and adenomas detected per colonoscopy without increasing withdrawal time (54.8% vs 40.4%). Additionally, the detection rate of adenomas 5 mm or smaller was significantly higher in the CAD group (33.7%) than in the control group[65]. Another randomized study used CAD to detect adenomas and achieved increased adenoma detection rates (29.1% vs 20.3%) and the mean number of adenomas per patient (0.53 vs 0.31). Similarly, a higher number of diminutive adenomas were found (185 vs 102)[66]. In addition, inflammatory bowel diseases including Crohn’s disease and ulcerative colitis are also associated precancerous lesions, and some AI methods aiding in scoring have been trained, such as DL model in grading endoscopic disease severity of patients with ulcerative colitis[67] and in predicting remission in patients with moderate to severe Crohn’s disease[68].
In the aspect of histopathology, AI has been used in ECC and precancerous lesions. A systematic review has concluded that AI use in CRC pathology image analysis included gland segmentation, tumor classification, tumor microenvironment characterization, and prognosis prediction[69]. A DL approach was developed to recognize four different stages of cancerous tissue development, including normal mucosa, early preneoplastic lesion, adenoma, and cancer and obtained an overall accuracy more than 95%[70]. Prediction of LNM for early CRC is critical for determining treatment strategies after ER. An LNM prediction algorithm for submucosal invasive (T1) CRC based on machine learning showed better LNM predictive ability than the conventional method on some datasets[71-82].
Endoscopy is usually the first choice in the diagnosis and management of early GI cancer. According to the Clinical Practice Guideline, ER is now a standard treatment for early GI cancers without regional LNM. Early GI cancers can completely be removed by en bloc fashion (resection of a tumor in one piece without visible residual tumor) via endoscopic mucosal resection and/or endoscopic submucosal dissection. High-definition white light endoscopy, chromoendoscopy, and image-enhanced endoscopy such as ME-NBI can be used to assess the edge and depth of early GI cancers for delineation of resection boundaries and prediction of the possibility of LNM before the decision of ER. Histopathological evaluation can confirm the depth of cancer invasion and lymphovascular invasion[83]. From this review, we can see AI as a novel technology has been penetrated in early GI cancer detection, diagnosis, boundaries, invasion depth, lymphovascular invasion, and prognosis prediction based on endoscopic images and videos and pathological tissue slides obtained after ER.
Both high-quality endoscopy and high-quality AI model construction research are crucial to ensure better health outcomes and benefits of patients. Some AI methods have been designed to identify and assure the quality of endoscopy to improve the detection rate of early GI cancer. In upper GI tract, missed EGC rates are an important measure of quality. A deep CNN model was built to monitor blind spots, time the procedure, and automatically generate photo-documentation during esophagogastroduodenoscopy[84]. Meanwhile, in colonoscopy, poorer adenoma detection rates are associated with poorer outcomes and higher rates of post-colonoscopy colonic cancer[85]. A deep CNN model was developed for timing withdrawal phase, supervising withdrawal stability, evaluating bowel preparation, and detecting colorectal polyps[86].
In the aspect of quality control of AI studies related to endoscopy, some limitations should be concerned. Different CNN models have demonstrated high accuracies or area under the curve and 7 out of 22 more than 90%/0.9 with high sensitivities and specificities in Table 1. These limitations were concentrated on the retrospective research, the single center, the small sample number, still images, background images, the only use of high-quality images, and not all images with lesions identified by gold standard such as pathology. They may discount the reliability of the results. As most endoscopic-related algorithms are trained in a supervised manner, labeling data is important. Meanwhile, videos and large, heterogenous, and prospectively collected data are less prone to biases[87].
AI has been widely used in medicine, although most studies have remained at the preclinical stage. In this review, we provided an overview of the associated application of AI in early GI cancer including EEC, EGC, and ECC as well as their precancerous lesions. Detection, diagnosis, classification, invasion depth, and histopathology have been involved. Indeed, AI will bring benefits to patients and doctors. It will provide useful support during endoscopies to achieve more precise diagnosis of early GI cancer after more intelligent detection and biopsy with high efficiency and reduce workload to fill the lack of clinical resources in the future.
Manuscript source: Unsolicited manuscript
Specialty type: Gastroenterology and hepatology
Country/Territory of origin: China
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): 0
Grade C (Good): C, C, C, C
Grade D (Fair): D, D
Grade E (Poor): 0
P-Reviewer: Balakrishnan DS, Lalmuanawma S, Tanabe S, Vijh S S-Editor: Liu M L-Editor: Filipodia P-Editor: Xing YX
| 1. | Ahmad OF, Soares AS, Mazomenos E, Brandao P, Vega R, Seward E, Stoyanov D, Chand M, Lovat LB. Artificial intelligence and computer-aided diagnosis in colonoscopy: current evidence and future directions. Lancet Gastroenterol Hepatol. 2019;4:71-80. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 102] [Cited by in RCA: 126] [Article Influence: 18.0] [Reference Citation Analysis (0)] |
| 2. | Huang S, Yang J, Fong S, Zhao Q. Artificial intelligence in cancer diagnosis and prognosis: Opportunities and challenges. Cancer Lett. 2020;471:61-71. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 133] [Cited by in RCA: 296] [Article Influence: 49.3] [Reference Citation Analysis (1)] |
| 3. | Mokrane FZ, Lu L, Vavasseur A, Otal P, Peron JM, Luk L, Yang H, Ammari S, Saenger Y, Rousseau H, Zhao B, Schwartz LH, Dercle L. Radiomics machine-learning signature for diagnosis of hepatocellular carcinoma in cirrhotic patients with indeterminate liver nodules. Eur Radiol. 2020;30:558-570. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 80] [Cited by in RCA: 125] [Article Influence: 25.0] [Reference Citation Analysis (0)] |
| 4. | Young MR, Abrams N, Ghosh S, Rinaudo JAS, Marquez G, Srivastava S. Prediagnostic Image Data, Artificial Intelligence, and Pancreatic Cancer: A Tell-Tale Sign to Early Detection. Pancreas. 2020;49:882-886. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 10] [Cited by in RCA: 9] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 5. | Kuwahara T, Hara K, Mizuno N, Okuno N, Matsumoto S, Obata M, Kurita Y, Koda H, Toriyama K, Onishi S, Ishihara M, Tanaka T, Tajika M, Niwa Y. Usefulness of Deep Learning Analysis for the Diagnosis of Malignancy in Intraductal Papillary Mucinous Neoplasms of the Pancreas. Clin Transl Gastroenterol. 2019;10:1-8. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 61] [Cited by in RCA: 127] [Article Influence: 25.4] [Reference Citation Analysis (0)] |
| 6. | Dalal V, Carmicheal J, Dhaliwal A, Jain M, Kaur S, Batra SK. Radiomics in stratification of pancreatic cystic lesions: Machine learning in action. Cancer Lett. 2020;469:228-237. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 37] [Cited by in RCA: 68] [Article Influence: 11.3] [Reference Citation Analysis (0)] |
| 7. | Almeida PP, Cardoso CP, de Freitas LM. PDAC-ANN: an artificial neural network to predict pancreatic ductal adenocarcinoma based on gene expression. BMC Cancer. 2020;20:82. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 38] [Cited by in RCA: 24] [Article Influence: 4.8] [Reference Citation Analysis (0)] |
| 8. | Tonozuka R, Itoi T, Nagata N, Kojima H, Sofuni A, Tsuchiya T, Ishii K, Tanaka R, Nagakawa Y, Mukai S. Deep learning analysis for the detection of pancreatic cancer on endosonographic images: a pilot study. J Hepatobiliary Pancreat Sci. 2021;28:95-104. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 36] [Cited by in RCA: 79] [Article Influence: 15.8] [Reference Citation Analysis (0)] |
| 9. | Walczak S, Velanovich V. An Evaluation of Artificial Neural Networks in Predicting Pancreatic Cancer Survival. J Gastrointest Surg. 2017;21:1606-1612. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 23] [Cited by in RCA: 26] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 10. | Prabhu A, Obi KO, Rubenstein JH. The synergistic effects of alcohol and tobacco consumption on the risk of esophageal squamous cell carcinoma: a meta-analysis. Am J Gastroenterol. 2014;109:822-827. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 208] [Cited by in RCA: 175] [Article Influence: 15.9] [Reference Citation Analysis (0)] |
| 11. | Abnet CC, Arnold M, Wei WQ. Epidemiology of Esophageal Squamous Cell Carcinoma. Gastroenterology. 2018;154:360-373. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 935] [Cited by in RCA: 1152] [Article Influence: 164.6] [Reference Citation Analysis (1)] |
| 12. | Coleman HG, Xie SH, Lagergren J. The Epidemiology of Esophageal Adenocarcinoma. Gastroenterology. 2018;154:390-405. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 425] [Cited by in RCA: 381] [Article Influence: 54.4] [Reference Citation Analysis (0)] |
| 13. | Spechler SJ, Souza RF. Barrett’s esophagus. N Engl J Med. 2014;371:836-845. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 360] [Cited by in RCA: 375] [Article Influence: 34.1] [Reference Citation Analysis (0)] |
| 14. | di Pietro M, Canto MI, Fitzgerald RC. Endoscopic Management of Early Adenocarcinoma and Squamous Cell Carcinoma of the Esophagus: Screening, Diagnosis, and Therapy. Gastroenterology. 2018;154:421-436. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 155] [Cited by in RCA: 173] [Article Influence: 24.7] [Reference Citation Analysis (0)] |
| 15. | Horie Y, Yoshio T, Aoyama K, Yoshimizu S, Horiuchi Y, Ishiyama A, Hirasawa T, Tsuchida T, Ozawa T, Ishihara S, Kumagai Y, Fujishiro M, Maetani I, Fujisaki J, Tada T. Diagnostic outcomes of esophageal cancer by artificial intelligence using convolutional neural networks. Gastrointest Endosc. 2019;89:25-32. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 240] [Cited by in RCA: 271] [Article Influence: 45.2] [Reference Citation Analysis (0)] |
| 16. | Iwagami H, Ishihara R, Aoyama K, Fukuda H, Shimamoto Y, Kono M, Nakahira H, Matsuura N, Shichijo S, Kanesaka T, Kanzaki H, Ishii T, Nakatani Y, Tada T. Artificial intelligence for the detection of esophageal and esophagogastric junctional adenocarcinoma. J Gastroenterol Hepatol. 2021;36:131-136. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 32] [Cited by in RCA: 27] [Article Influence: 6.8] [Reference Citation Analysis (0)] |
| 17. | Ebigbo A, Mendel R, Probst A, Manzeneder J, Souza LA Jr, Papa JP, Palm C, Messmann H. Computer-aided diagnosis using deep learning in the evaluation of early oesophageal adenocarcinoma. Gut. 2019;68:1143-1145. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 94] [Cited by in RCA: 103] [Article Influence: 17.2] [Reference Citation Analysis (0)] |
| 18. | Ghatwary N, Zolgharni M, Ye X. Early esophageal adenocarcinoma detection using deep learning methods. Int J Comput Assist Radiol Surg. 2019;14:611-621. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 38] [Cited by in RCA: 66] [Article Influence: 11.0] [Reference Citation Analysis (0)] |
| 19. | Kumagai Y, Takubo K, Kawada K, Aoyama K, Endo Y, Ozawa T, Hirasawa T, Yoshio T, Ishihara S, Fujishiro M, Tamaru JI, Mochiki E, Ishida H, Tada T. Diagnosis using deep-learning artificial intelligence based on the endocytoscopic observation of the esophagus. Esophagus. 2019;16:180-187. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 65] [Cited by in RCA: 65] [Article Influence: 10.8] [Reference Citation Analysis (0)] |
| 20. | Fukuda H, Ishihara R, Kato Y, Matsunaga T, Nishida T, Yamada T, Ogiyama H, Horie M, Kinoshita K, Tada T. Comparison of performances of artificial intelligence versus expert endoscopists for real-time assisted diagnosis of esophageal squamous cell carcinoma (with video). Gastrointest Endosc. 2020;92:848-855. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 73] [Cited by in RCA: 52] [Article Influence: 10.4] [Reference Citation Analysis (0)] |
| 21. | Shimamoto Y, Ishihara R, Kato Y, Shoji A, Inoue T, Matsueda K, Miyake M, Waki K, Kono M, Fukuda H, Matsuura N, Nagaike K, Aoi K, Yamamoto K, Nakahara M, Nishihara A, Tada T. Real-time assessment of video images for esophageal squamous cell carcinoma invasion depth using artificial intelligence. J Gastroenterol. 2020;55:1037-1045. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 24] [Cited by in RCA: 46] [Article Influence: 9.2] [Reference Citation Analysis (0)] |
| 22. | Tokai Y, Yoshio T, Aoyama K, Horie Y, Yoshimizu S, Horiuchi Y, Ishiyama A, Tsuchida T, Hirasawa T, Sakakibara Y, Yamada T, Yamaguchi S, Fujisaki J, Tada T. Application of artificial intelligence using convolutional neural networks in determining the invasion depth of esophageal squamous cell carcinoma. Esophagus. 2020;17:250-256. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 64] [Cited by in RCA: 83] [Article Influence: 16.6] [Reference Citation Analysis (0)] |
| 23. | Trindade AJ, McKinley MJ, Fan C, Leggett CL, Kahn A, Pleskow DK. Endoscopic Surveillance of Barrett’s Esophagus Using Volumetric Laser Endomicroscopy With Artificial Intelligence Image Enhancement. Gastroenterology. 2019;157:303-305. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 40] [Cited by in RCA: 42] [Article Influence: 7.0] [Reference Citation Analysis (0)] |
| 24. | Hashimoto R, Requa J, Dao T, Ninh A, Tran E, Mai D, Lugo M, El-Hage Chehade N, Chang KJ, Karnes WE, Samarasena JB. Artificial intelligence using convolutional neural networks for real-time detection of early esophageal neoplasia in Barrett’s esophagus (with video). Gastrointest Endosc. 2020;91:1264-1271.e1. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 172] [Cited by in RCA: 143] [Article Influence: 28.6] [Reference Citation Analysis (0)] |
| 25. | de Groof AJ, Struyvenberg MR, van der Putten J, van der Sommen F, Fockens KN, Curvers WL, Zinger S, Pouw RE, Coron E, Baldaque-Silva F, Pech O, Weusten B, Meining A, Neuhaus H, Bisschops R, Dent J, Schoon EJ, de With PH, Bergman JJ. Deep-Learning System Detects Neoplasia in Patients With Barrett’s Esophagus With Higher Accuracy Than Endoscopists in a Multistep Training and Validation Study With Benchmarking. Gastroenterology. 2020;158:915-929.e4. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 172] [Cited by in RCA: 222] [Article Influence: 44.4] [Reference Citation Analysis (0)] |
| 26. | Tomita N, Abdollahi B, Wei J, Ren B, Suriawinata A, Hassanpour S. Attention-Based Deep Neural Networks for Detection of Cancerous and Precancerous Esophagus Tissue on Histopathological Slides. JAMA Netw Open. 2019;2:e1914645. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 133] [Cited by in RCA: 100] [Article Influence: 16.7] [Reference Citation Analysis (0)] |
| 27. | Sumiyama K. Past and current trends in endoscopic diagnosis for early stage gastric cancer in Japan. Gastric Cancer. 2017;20:20-27. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 52] [Cited by in RCA: 65] [Article Influence: 8.1] [Reference Citation Analysis (0)] |
| 28. | Kumar S, Metz DC, Ellenberg S, Kaplan DE, Goldberg DS. Risk Factors and Incidence of Gastric Cancer After Detection of Helicobacter pylori Infection: A Large Cohort Study. Gastroenterology. 2020;158:527-536.e7. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 118] [Cited by in RCA: 189] [Article Influence: 37.8] [Reference Citation Analysis (0)] |
| 29. | Hatta W, Gotoda T, Koike T, Masamune A. History and future perspectives in Japanese guidelines for endoscopic resection of early gastric cancer. Dig Endosc. 2020;32:180-190. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 52] [Cited by in RCA: 77] [Article Influence: 15.4] [Reference Citation Analysis (0)] |
| 30. | Pimentel-Nunes P, Libânio D, Marcos-Pinto R, Areia M, Leja M, Esposito G, Garrido M, Kikuste I, Megraud F, Matysiak-Budnik T, Annibale B, Dumonceau JM, Barros R, Fléjou JF, Carneiro F, van Hooft JE, Kuipers EJ, Dinis-Ribeiro M. Management of epithelial precancerous conditions and lesions in the stomach (MAPS II): European Society of Gastrointestinal Endoscopy (ESGE), European Helicobacter and Microbiota Study Group (EHMSG), European Society of Pathology (ESP), and Sociedade Portuguesa de Endoscopia Digestiva (SPED) guideline update 2019. Endoscopy. 2019;51:365-388. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 712] [Cited by in RCA: 670] [Article Influence: 111.7] [Reference Citation Analysis (0)] |
| 31. | Suzuki H, Oda I, Abe S, Sekiguchi M, Mori G, Nonaka S, Yoshinaga S, Saito Y. High rate of 5-year survival among patients with early gastric cancer undergoing curative endoscopic submucosal dissection. Gastric Cancer. 2016;19:198-205. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 135] [Cited by in RCA: 188] [Article Influence: 20.9] [Reference Citation Analysis (0)] |
| 32. | Shi Y, Xu X, Zhao Y, Qian F, Tang B, Hao Y, Luo H, Chen J, Yu P. Long-term oncologic outcomes of a randomized controlled trial comparing laparoscopic versus open gastrectomy with D2 lymph node dissection for advanced gastric cancer. Surgery. 2019;165:1211-1216. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 26] [Cited by in RCA: 46] [Article Influence: 7.7] [Reference Citation Analysis (0)] |
| 33. | Mori Y, Berzin TM, Kudo SE. Artificial intelligence for early gastric cancer: early promise and the path ahead. Gastrointest Endosc. 2019;89:816-817. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 14] [Cited by in RCA: 10] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 34. | Ueyama H, Kato Y, Akazawa Y, Yatagai N, Komori H, Takeda T, Matsumoto K, Ueda K, Hojo M, Yao T, Nagahara A, Tada T. Application of artificial intelligence using a convolutional neural network for diagnosis of early gastric cancer based on magnifying endoscopy with narrow-band imaging. J Gastroenterol Hepatol. 2021;36:482-489. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 45] [Cited by in RCA: 89] [Article Influence: 22.3] [Reference Citation Analysis (0)] |
| 35. | Horiuchi Y, Aoyama K, Tokai Y, Hirasawa T, Yoshimizu S, Ishiyama A, Yoshio T, Tsuchida T, Fujisaki J, Tada T. Convolutional Neural Network for Differentiating Gastric Cancer from Gastritis Using Magnified Endoscopy with Narrow Band Imaging. Dig Dis Sci. 2020;65:1355-1363. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 70] [Cited by in RCA: 102] [Article Influence: 20.4] [Reference Citation Analysis (1)] |
| 36. | Namikawa K, Hirasawa T, Nakano K, Ikenoyama Y, Ishioka M, Shiroma S, Tokai Y, Yoshimizu S, Horiuchi Y, Ishiyama A, Yoshio T, Tsuchida T, Fujisaki J, Tada T. Artificial intelligence-based diagnostic system classifying gastric cancers and ulcers: comparison between the original and newly developed systems. Endoscopy. 2020;52:1077-1083. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 29] [Cited by in RCA: 31] [Article Influence: 6.2] [Reference Citation Analysis (0)] |
| 37. | Ishioka M, Hirasawa T, Tada T. Detecting gastric cancer from video images using convolutional neural networks. Dig Endosc. 2019;31:e34-e35. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 55] [Cited by in RCA: 55] [Article Influence: 9.2] [Reference Citation Analysis (0)] |
| 38. | Zhu Y, Wang QC, Xu MD, Zhang Z, Cheng J, Zhong YS, Zhang YQ, Chen WF, Yao LQ, Zhou PH, Li QL. Application of convolutional neural network in the diagnosis of the invasion depth of gastric cancer based on conventional endoscopy. Gastrointest Endosc. 2019;89:806-815.e1. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 201] [Cited by in RCA: 231] [Article Influence: 38.5] [Reference Citation Analysis (0)] |
| 39. | Yoon HJ, Kim S, Kim JH, Keum JS, Oh SI, Jo J, Chun J, Youn YH, Park H, Kwon IG, Choi SH, Noh SH. A Lesion-Based Convolutional Neural Network Improves Endoscopic Detection and Depth Prediction of Early Gastric Cancer. J Clin Med. 2019;8:1310. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 61] [Cited by in RCA: 91] [Article Influence: 15.2] [Reference Citation Analysis (0)] |
| 40. | Song Z, Zou S, Zhou W, Huang Y, Shao L, Yuan J, Gou X, Jin W, Wang Z, Chen X, Ding X, Liu J, Yu C, Ku C, Liu C, Sun Z, Xu G, Wang Y, Zhang X, Wang D, Wang S, Xu W, Davis RC, Shi H. Clinically applicable histopathological diagnosis system for gastric cancer detection using deep learning. Nat Commun. 2020;11:4294. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 152] [Cited by in RCA: 158] [Article Influence: 31.6] [Reference Citation Analysis (0)] |
| 41. | Yoshida H, Shimazu T, Kiyuna T, Marugame A, Yamashita Y, Cosatto E, Taniguchi H, Sekine S, Ochiai A. Automated histological classification of whole-slide images of gastric biopsy specimens. Gastric Cancer. 2018;21:249-257. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 63] [Cited by in RCA: 72] [Article Influence: 10.3] [Reference Citation Analysis (0)] |
| 42. | Sharma H, Zerbe N, Klempert I, Hellwich O, Hufnagl P. Deep convolutional neural networks for automatic classification of gastric carcinoma using whole slide images in digital histopathology. Comput Med Imaging Graph. 2017;61:2-13. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 176] [Cited by in RCA: 171] [Article Influence: 21.4] [Reference Citation Analysis (0)] |
| 43. | Li Q, Qi L, Feng QX, Liu C, Sun SW, Zhang J, Yang G, Ge YQ, Zhang YD, Liu XS. Machine Learning-Based Computational Models Derived From Large-Scale Radiographic-Radiomic Images Can Help Predict Adverse Histopathological Status of Gastric Cancer. Clin Transl Gastroenterol. 2019;10:e00079. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 11] [Cited by in RCA: 21] [Article Influence: 4.2] [Reference Citation Analysis (0)] |
| 44. | Li J, Dong D, Fang M, Wang R, Tian J, Li H, Gao J. Dual-energy CT-based deep learning radiomics can improve lymph node metastasis risk prediction for gastric cancer. Eur Radiol. 2020;30:2324-2333. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 44] [Cited by in RCA: 117] [Article Influence: 23.4] [Reference Citation Analysis (0)] |
| 45. | Guimarães P, Keller A, Fehlmann T, Lammert F, Casper M. Deep-learning based detection of gastric precancerous conditions. Gut. 2020;69:4-6. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 58] [Cited by in RCA: 80] [Article Influence: 16.0] [Reference Citation Analysis (0)] |
| 46. | Zhang Y, Li F, Yuan F, Zhang K, Huo L, Dong Z, Lang Y, Zhang Y, Wang M, Gao Z, Qin Z, Shen L. Diagnosing chronic atrophic gastritis by gastroscopy using artificial intelligence. Dig Liver Dis. 2020;52:566-572. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 45] [Cited by in RCA: 78] [Article Influence: 15.6] [Reference Citation Analysis (0)] |
| 47. | Shichijo S, Nomura S, Aoyama K, Nishikawa Y, Miura M, Shinagawa T, Takiyama H, Tanimoto T, Ishihara S, Matsuo K, Tada T. Application of Convolutional Neural Networks in the Diagnosis of Helicobacter pylori Infection Based on Endoscopic Images. EbioMedicine. 2017;25:106-111. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 235] [Cited by in RCA: 181] [Article Influence: 22.6] [Reference Citation Analysis (0)] |
| 48. | Zheng W, Zhang X, Kim JJ, Zhu X, Ye G, Ye B, Wang J, Luo S, Li J, Yu T, Liu J, Hu W, Si J. High Accuracy of Convolutional Neural Network for Evaluation of Helicobacter pylori Infection Based on Endoscopic Images: Preliminary Experience. Clin Transl Gastroenterol. 2019;10:e00109. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 64] [Cited by in RCA: 70] [Article Influence: 11.7] [Reference Citation Analysis (0)] |
| 49. | Bang CS, Lee JJ, Baik GH. Artificial Intelligence for the Prediction of Helicobacter Pylori Infection in Endoscopic Images: Systematic Review and Meta-Analysis Of Diagnostic Test Accuracy. J Med Internet Res. 2020;22:e21983. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 76] [Cited by in RCA: 63] [Article Influence: 12.6] [Reference Citation Analysis (0)] |
| 50. | Bianco F, Arezzo A, Agresta F, Coco C, Faletti R, Krivocapic Z, Rotondano G, Santoro GA, Vettoretto N, De Franciscis S, Belli A, Romano GM; Italian Society of Colorectal Surgery. Practice parameters for early colon cancer management: Italian Society of Colorectal Surgery (Società Italiana di Chirurgia Colo-Rettale; SICCR) guidelines. Tech Coloproctol. 2015;19:577-585. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 11] [Cited by in RCA: 19] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 51. | Dumoulin FL, Hildenbrand R. Endoscopic resection techniques for colorectal neoplasia: Current developments. World J Gastroenterol. 2019;25:300-307. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 47] [Cited by in RCA: 66] [Article Influence: 11.0] [Reference Citation Analysis (0)] |
| 52. | Murakami T, Sakamoto N, Nagahara A. Endoscopic diagnosis of sessile serrated adenoma/polyp with and without dysplasia/carcinoma. World J Gastroenterol. 2018;24:3250-3259. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 33] [Cited by in RCA: 18] [Article Influence: 2.6] [Reference Citation Analysis (5)] |
| 53. | Kawasaki K, Fujii M, Sugimoto S, Ishikawa K, Matano M, Ohta Y, Toshimitsu K, Takahashi S, Hosoe N, Sekine S, Kanai T, Sato T. Chromosome Engineering of Human Colon-Derived Organoids to Develop a Model of Traditional Serrated Adenoma. Gastroenterology. 2020;158:638-651.e8. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 62] [Cited by in RCA: 60] [Article Influence: 12.0] [Reference Citation Analysis (0)] |
| 54. | Itatani Y, Kawada K, Sakai Y. Treatment of Elderly Patients with Colorectal Cancer. Biomed Res Int. 2018;2018:2176056. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 49] [Cited by in RCA: 69] [Article Influence: 9.9] [Reference Citation Analysis (0)] |
| 55. | Ladabaum U. You Should Get Screened for Colon Cancer, Really. JAMA Netw Open. 2019;2:e1910452. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1] [Cited by in RCA: 1] [Article Influence: 0.2] [Reference Citation Analysis (0)] |
| 56. | US Preventive Services Task Force, Bibbins-Domingo K, Grossman DC, Curry SJ, Davidson KW, Epling JW Jr, García FAR, Gillman MW, Harper DM, Kemper AR, Krist AH, Kurth AE, Landefeld CS, Mangione CM, Owens DK, Phillips WR, Phipps MG, Pignone MP, Siu AL. Screening for Colorectal Cancer: US Preventive Services Task Force Recommendation Statement. JAMA. 2016;315:2564-2575. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1249] [Cited by in RCA: 1385] [Article Influence: 153.9] [Reference Citation Analysis (1)] |
| 57. | Siegel RL, Miller KD, Fedewa SA, Ahnen DJ, Meester RGS, Barzi A, Jemal A. Colorectal cancer statistics, 2017. CA Cancer J Clin. 2017;67:177-193. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2526] [Cited by in RCA: 2911] [Article Influence: 363.9] [Reference Citation Analysis (3)] |
| 58. | Yamada M, Saito Y, Imaoka H, Saiko M, Yamada S, Kondo H, Takamaru H, Sakamoto T, Sese J, Kuchiba A, Shibata T, Hamamoto R. Development of a real-time endoscopic image diagnosis support system using deep learning technology in colonoscopy. Sci Rep. 2019;9:14465. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 114] [Cited by in RCA: 161] [Article Influence: 26.8] [Reference Citation Analysis (0)] |
| 59. | Yang YJ, Cho BJ, Lee MJ, Kim JH, Lim H, Bang CS, Jeong HM, Hong JT, Baik GH. Automated Classification of Colorectal Neoplasms in White-Light Colonoscopy Images via Deep Learning. J Clin Med. 2020;9:1593. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 33] [Cited by in RCA: 30] [Article Influence: 6.0] [Reference Citation Analysis (0)] |
| 60. | Hashiguchi Y, Muro K, Saito Y, Ito Y, Ajioka Y, Hamaguchi T, Hasegawa K, Hotta K, Ishida H, Ishiguro M, Ishihara S, Kanemitsu Y, Kinugasa Y, Murofushi K, Nakajima TE, Oka S, Tanaka T, Taniguchi H, Tsuji A, Uehara K, Ueno H, Yamanaka T, Yamazaki K, Yoshida M, Yoshino T, Itabashi M, Sakamaki K, Sano K, Shimada Y, Tanaka S, Uetake H, Yamaguchi S, Yamaguchi N, Kobayashi H, Matsuda K, Kotake K, Sugihara K; Japanese Society for Cancer of the Colon and Rectum. Japanese Society for Cancer of the Colon and Rectum (JSCCR) guidelines 2019 for the treatment of colorectal cancer. Int J Clin Oncol. 2020;25:1-42. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1024] [Cited by in RCA: 1308] [Article Influence: 261.6] [Reference Citation Analysis (1)] |
| 61. | Ito N, Kawahira H, Nakashima H, Uesato M, Miyauchi H, Matsubara H. Endoscopic Diagnostic Support System for cT1b Colorectal Cancer Using Deep Learning. Oncology. 2019;96:44-50. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 76] [Cited by in RCA: 59] [Article Influence: 8.4] [Reference Citation Analysis (0)] |
| 62. | Hoerter N, Gross SA, Liang PS. Artificial Intelligence and Polyp Detection. Curr Treat Options Gastroenterol. 2020;epub ahead of print. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 28] [Cited by in RCA: 28] [Article Influence: 5.6] [Reference Citation Analysis (0)] |
| 63. | Vinsard DG, Mori Y, Misawa M, Kudo SE, Rastogi A, Bagci U, Rex DK, Wallace MB. Quality assurance of computer-aided detection and diagnosis in colonoscopy. Gastrointest Endosc. 2019;90:55-63. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 92] [Cited by in RCA: 95] [Article Influence: 15.8] [Reference Citation Analysis (0)] |
| 64. | Zhou G, Xiao X, Tu M, Liu P, Yang D, Liu X, Zhang R, Li L, Lei S, Wang H, Song Y, Wang P. Computer aided detection for laterally spreading tumors and sessile serrated adenomas during colonoscopy. PloS One. 2020;15:e0231880. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 11] [Cited by in RCA: 18] [Article Influence: 3.6] [Reference Citation Analysis (0)] |
| 65. | Repici A, Badalamenti M, Maselli R, Correale L, Radaelli F, Rondonotti E, Ferrara E, Spadaccini M, Alkandari A, Fugazza A, Anderloni A, Galtieri PA, Pellegatta G, Carrara S, Di Leo M, Craviotto V, Lamonaca L, Lorenzetti R, Andrealli A, Antonelli G, Wallace M, Sharma P, Rosch T, Hassan C. Efficacy of Real-Time Computer-Aided Detection of Colorectal Neoplasia in a Randomized Trial. Gastroenterology. 2020;159:512-520.e7. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 237] [Cited by in RCA: 391] [Article Influence: 78.2] [Reference Citation Analysis (0)] |
| 66. | Wang P, Berzin TM, Glissen Brown JR, Bharadwaj S, Becq A, Xiao X, Liu P, Li L, Song Y, Zhang D, Li Y, Xu G, Tu M, Liu X. Real-time automatic detection system increases colonoscopic polyp and adenoma detection rates: a prospective andomized controlled study. Gut. 2019;68:1813-1819. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 398] [Cited by in RCA: 547] [Article Influence: 91.2] [Reference Citation Analysis (0)] |
| 67. | Stidham RW, Liu W, Bishu S, Rice MD, Higgins PDR, Zhu J, Nallamothu BK, Waljee AK. Performance of a Deep Learning Model vs Human Reviewers in Grading Endoscopic Disease Severity of Patients With Ulcerative Colitis. JAMA Netw Open. 2019;2:e193963. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 116] [Cited by in RCA: 173] [Article Influence: 28.8] [Reference Citation Analysis (0)] |
| 68. | Waljee AK, Wallace BI, Cohen-Mekelburg S, Liu Y, Liu B, Sauder K, Stidham RW, Zhu J, Higgins PDR. Development and Validation of Machine Learning Models in Prediction of Remission in Patients With Moderate to Severe Crohn Disease. JAMA Netw Open. 2019;2:e193721. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 56] [Cited by in RCA: 74] [Article Influence: 12.3] [Reference Citation Analysis (0)] |
| 69. | Thakur N, Yoon H, Chong Y. Current Trends of Artificial Intelligence for Colorectal Cancer Pathology Image Analysis: A Systematic Review. Cancers (Basel). 2020;12:1884. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 60] [Cited by in RCA: 53] [Article Influence: 10.6] [Reference Citation Analysis (0)] |
| 70. | Sena P, Fioresi R, Faglioni F, Losi L, Faglioni G, Roncucci L. Deep learning techniques for detecting preneoplastic and neoplastic lesions in human colorectal histological images. Oncol Lett. 2019;18:6101-6107. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 17] [Cited by in RCA: 22] [Article Influence: 3.7] [Reference Citation Analysis (0)] |
| 71. | Takamatsu M, Yamamoto N, Kawachi H, Chino A, Saito S, Ueno M, Ishikawa Y, Takazawa Y, Takeuchi K. Prediction of early colorectal cancer metastasis by machine learning using digital slide images. Comput Methods Programs Biomed. 2019;178:155-161. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 35] [Cited by in RCA: 46] [Article Influence: 7.7] [Reference Citation Analysis (0)] |
| 72. | Guo L, Xiao X, Wu C, Zeng X, Zhang Y, Du J, Bai S, Xie J, Zhang Z, Li Y, Wang X, Cheung O, Sharma M, Liu J, Hu B. Real-time automated diagnosis of precancerous lesions and early esophageal squamous cell carcinoma using a deep learning model (with videos). Gastrointest Endosc. 2020;91:41-51. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 161] [Cited by in RCA: 149] [Article Influence: 29.8] [Reference Citation Analysis (0)] |
| 73. | Cai SL, Li B, Tan WM, Niu XJ, Yu HH, Yao LQ, Zhou PH, Yan B, Zhong YS. Using a deep learning system in endoscopy for screening of early esophageal squamous cell carcinoma (with video). Gastrointest Endosc. 2019;90:745-753.e2. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 74] [Cited by in RCA: 110] [Article Influence: 18.3] [Reference Citation Analysis (0)] |
| 74. | Zhao YY, Xue DX, Wang YL, Zhang R, Sun B, Cai YP, Feng H, Cai Y, Xu JM. Computer-assisted diagnosis of early esophageal squamous cell carcinoma using narrow-band imaging magnifying endoscopy. Endoscopy. 2019;51:333-341. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 66] [Cited by in RCA: 86] [Article Influence: 14.3] [Reference Citation Analysis (0)] |
| 75. | Sakai Y, Takemoto S, Hori K, Nishimura M, Ikematsu H, Yano T, Yokota H. Automatic detection of early gastric cancer in endoscopic images using a transferring convolutional neural network. Annu Int Conf IEEE Eng Med Biol Soc. 2018;2018:4138-4141. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 39] [Cited by in RCA: 48] [Article Influence: 8.0] [Reference Citation Analysis (0)] |
| 76. | Zhang L, Zhang Y, Wang L, Wang J, Liu Y. Diagnosis of gastric lesions through a deep convolutional neural network. Dig Endosc. 2021;33:788-796. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 20] [Cited by in RCA: 20] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 77. | Li L, Chen Y, Shen Z, Zhang X, Sang J, Ding Y, Yang X, Li J, Chen M, Jin C, Chen C, Yu C. Convolutional neural network for the diagnosis of early gastric cancer based on magnifying narrow band imaging. Gastric Cancer. 2020;23:126-132. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 168] [Cited by in RCA: 137] [Article Influence: 27.4] [Reference Citation Analysis (0)] |
| 78. | Wu L, Zhou W, Wan X, Zhang J, Shen L, Hu S, Ding Q, Mu G, Yin A, Huang X, Liu J, Jiang X, Wang Z, Deng Y, Liu M, Lin R, Ling T, Li P, Wu Q, Jin P, Chen J, Yu H. A deep neural network improves endoscopic detection of early gastric cancer without blind spots. Endoscopy. 2019;51:522-531. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 194] [Cited by in RCA: 156] [Article Influence: 26.0] [Reference Citation Analysis (0)] |
| 79. | Ikenoyama Y, Hirasawa T, Ishioka M, Namikawa K, Yoshimizu S, Horiuchi Y, Ishiyama A, Yoshio T, Tsuchida T, Takeuchi Y, Shichijo S, Katayama N, Fujisaki J, Tada T. Detecting early gastric cancer: Comparison between the diagnostic ability of convolutional neural networks and endoscopists. Dig Endosc. 2021;33:141-150. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 129] [Cited by in RCA: 103] [Article Influence: 25.8] [Reference Citation Analysis (0)] |
| 80. | Cho BJ, Bang CS, Park SW, Yang YJ, Seo SI, Lim H, Shin WG, Hong JT, Yoo YT, Hong SH, Choi JH, Lee JJ, Baik GH. Automated classification of gastric neoplasms in endoscopic images using a convolutional neural network. Endoscopy. 2019;51:1121-1129. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 110] [Cited by in RCA: 96] [Article Influence: 16.0] [Reference Citation Analysis (1)] |
| 81. | Urban G, Tripathi P, Alkayali T, Mittal M, Jalali F, Karnes W, Baldi P. Deep Learning Localizes and Identifies Polyps in Real Time With 96% Accuracy in Screening Colonoscopy. Gastroenterology. 2018;155:1069-1078.e8. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 398] [Cited by in RCA: 430] [Article Influence: 61.4] [Reference Citation Analysis (1)] |
| 82. | Wang P, Liu X, Berzin TM, Glissen Brown JR, Liu P, Zhou C, Lei L, Li L, Guo Z, Lei S, Xiong F, Wang H, Song Y, Pan Y, Zhou G. Effect of a deep-learning computer-aided detection system on adenoma detection during colonoscopy (CADe-DB trial): a double-blind andomized study. Lancet Gastroenterol Hepatol. 2020;5:343-351. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 164] [Cited by in RCA: 293] [Article Influence: 58.6] [Reference Citation Analysis (0)] |
| 83. | Park CH, Yang DH, Kim JW, Kim JH, Min YW, Lee SH, Bae JH, Chung H, Choi KD, Park JC, Lee H, Kwak MS, Kim B, Lee HJ, Lee HS, Choi M, Park DA, Lee JY, Byeon JS, Park CG, Cho JY, Lee ST, Chun HJ. Clinical Practice Guideline for Endoscopic Resection of Early Gastrointestinal Cancer. Clin Endosc. 2020;53:142-166. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 41] [Cited by in RCA: 108] [Article Influence: 21.6] [Reference Citation Analysis (0)] |
| 84. | Lee JK, Jensen CD, Levin TR, Zauber AG, Doubeni CA, Zhao WK, Corley DA. Accurate Identification of Colonoscopy Quality and Polyp Findings Using Natural Language Processing. J Clin Gastroenterol. 2019;53:e25-e30. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 18] [Cited by in RCA: 26] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 85. | Rutter MD, Rees CJ. Quality in gastrointestinal endoscopy. Endoscopy. 2014;46:526-528. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 28] [Cited by in RCA: 36] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 86. | Su JR, Li Z, Shao XJ, Ji CR, Ji R, Zhou RC, Li GC, Liu GQ, He YS, Zuo XL, Li YQ. Impact of a real-time automatic quality control system on colorectal polyp and adenoma detection: a prospective randomized controlled study (with videos). Gastrointest Endosc. 2020;91:415-424.e4. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 153] [Cited by in RCA: 207] [Article Influence: 41.4] [Reference Citation Analysis (0)] |
| 87. | Byrne MF, Chapados N, Soudan F, Oertel C, Linares Pérez M, Kelly R, Iqbal N, Chandelier F, Rex DK. Real-time differentiation of adenomatous and hyperplastic diminutive colorectal polyps during analysis of unaltered videos of standard colonoscopy using a deep learning model. Gut. 2019;68:94-100. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 363] [Cited by in RCA: 410] [Article Influence: 68.3] [Reference Citation Analysis (0)] |