Published online Dec 28, 2022. doi: 10.35711/aimi.v3.i4.87
Peer-review started: September 21, 2022
First decision: November 30, 2022
Revised: December 8, 2022
Accepted: December 21, 2022
Article in press: December 21, 2022
Published online: December 28, 2022
Processing time: 99 Days and 5.8 Hours
Noninvasive imaging (computed tomography, magnetic resonance imaging, endoscopic ultrasonography, and positron emission tomography) as an important part of the clinical workflow in the clinic, but it still provides limited information for diagnosis, treatment effect evaluation and prognosis prediction. In addition, judgment and diagnoses made by experts are usually based on multiple years of experience and subjective impression which lead to variable results in the same case. With accumulation of medical imaging data, radiomics emerges as a relatively new approach for analysis. Via artificial intelligence techniques, high-throughput quantitative data which is invisible to the naked eyes extracted from original images can be used in the process of patients’ management. Several studies have evaluated radiomics combined with clinical factors, pathological, or genetic information would assist in the diagnosis, particularly in the prediction of biological characteristics, risk of recurrence, and survival with encouraging results. In various clinical settings, there are limitations and challenges needing to be overcome before transformation. Therefore, we summarize the concepts and method of radiomics including image acquisition, region of interest segmentation, feature extraction and model development. We also set forth the current applications of radiomics in clinical routine. At last, the limitations and related deficiencies of radiomics are pointed out to direct the future opportunities and development.
Core Tip: Radiomics is widespread applied in clinical researches through extracting high-dimensional quantitative imaging features as a relatively emerging and mature technique based on medical imaging. The basic principles and methodologies of radiomics were reviewed to make it easy to understand from the relatively fixed processes. The representative clinical utilizations were declared to show the benefits of radiomics in diagnosis, tumor biological features and prognosis. Radiomics has revealed potential of clinical applications, while there are still many limitations to resolve in the further researches.
- Citation: Jiang ZY, Qi LS, Li JT, Cui N, Li W, Liu W, Wang KZ. Radiomics: Status quo and future challenges. Artif Intell Med Imaging 2022; 3(4): 87-96
- URL: https://www.wjgnet.com/2644-3260/full/v3/i4/87.htm
- DOI: https://dx.doi.org/10.35711/aimi.v3.i4.87
Radiomics was first proposed by Lambin et al[1] in 2012, which converts medical images into high-throughput quantitative features. Radiomic features can capture tissue and lesion properties noninvasively, such as shape and heterogeneity, and radiomics acts as a new approach to extract the information underlying the medical images that fail to be appreciated by naked eyes[2]. In the meantime, radiomics also possesses several advantages over molecular assays, such as being non-tissue-destructive, rapid analysis, easily serialized, fairly inexpensive, and being fully compatible with the existing clinical workflows[3]. In 2014, Aerts et al[4] demonstrated the role of radiomics in disease prognostication, promoting the development of radiomic-based signatures. Subsequently, the Pyradiomics framework based on the image biomarker standardization initiative (IBSI) criteria published in 2017 strongly supported the standardized application of radiomics[5].
Radiomics has evolved tremendously in the last decade, with the objective of precision medicine. However, the interpretability of radiomic-based signatures and the correlation with biology and pathology need to be further discussed. Additional multi-center data and prospective validation are also required for verification, in order to improve the confidence of applications[6]. There are still several substantial barriers to realize the objective of transforming artificial intelligence (AI) into the real clinical practice.
In the present study, the basic principles and methodologies of radiomics were reviewed and an outline of the representative clinical utilization was provided to highlight the benefits of radiomics in diagnosis, staging, tumor biological features, and prognosis. Additionally, it is essential to explore the deficiencies of radiomics to achieve a balanced interpretation between AI and clinical practice.
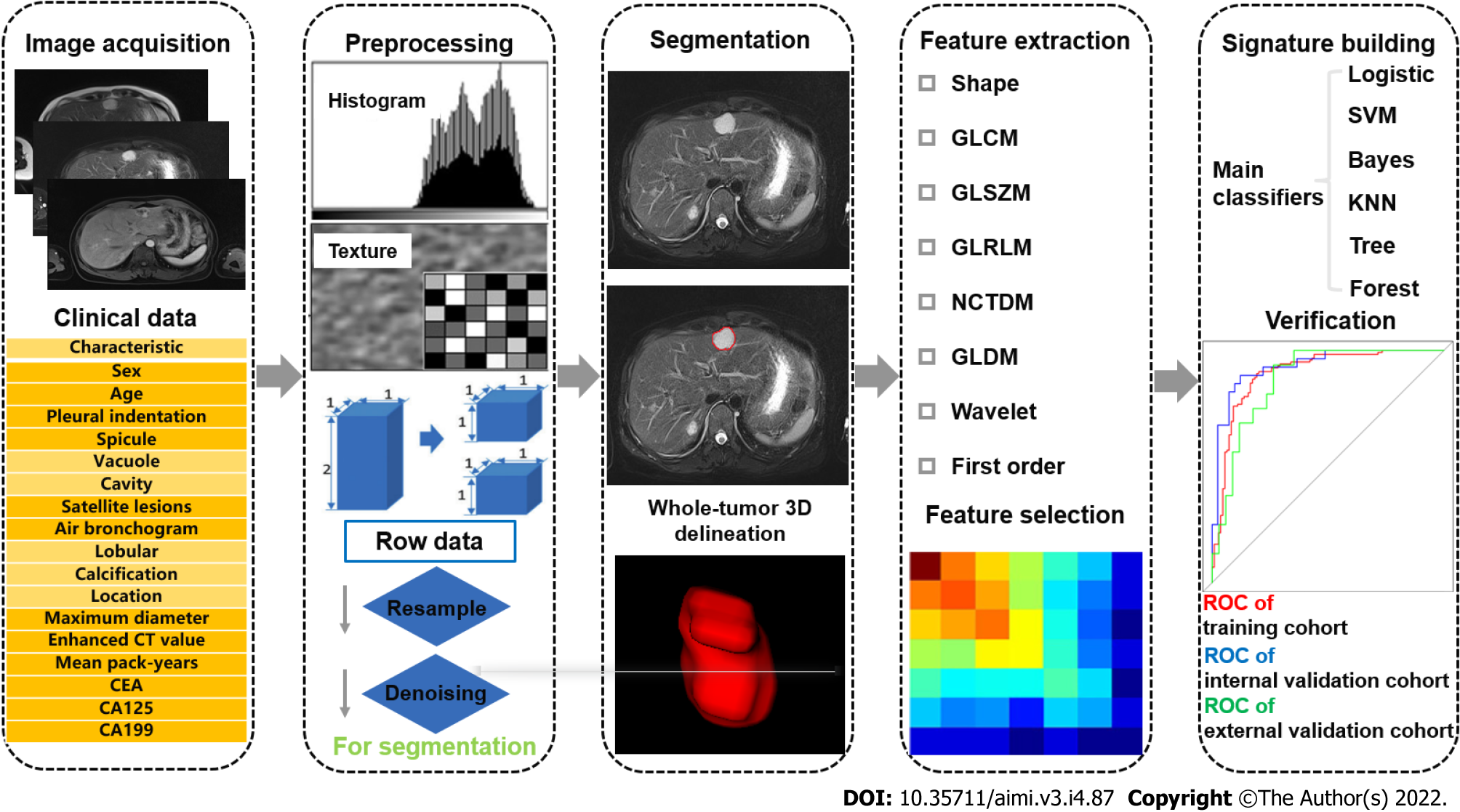
“Radiomics,” a term that describes the “omics” approach for the analysis of imaging data, has emerged as a novel tool for diagnosis and prognosis[2]. Using advanced computational tools, high-throughput quantitative imaging features beyond inspections of naked human eyes are extracted and the desensitized medical images are transformed into multiple textural features for quantitative assessment[7-9]. With semantic features, radiomics enables clinicians to make more objective and accurate clinical decisions in diagnosis and prognosis[10,11]. The workflow of radiomics analysis, consisting of several steps, is illustrated in Figure 1.
Image acquisition is approved by the ethics committee and informed consent form is signed by participants or their close relatives. The right to know patients is protected by relevant regulations. As the research of radiomics concentrated on human participants, it complies with the basic principles of 1964, Helsinki Manifesto and its later revisions. Sensitive information is erased from medical imaging data exported from imaging databases, including but not limited to organization name, organization address, physician’s name, patient’s name, patient’s birthday, etc. Besides, personal data are kept confidential, such as ID number, home address, contact information, medical insurance information, etc. Acquisition, transmission, and use of data should meet relevant legal requirements.
In addition, medical imaging data, which are consistent with standard imaging protocols, are the foundation of radiomics[12,13]. It can be single- or multi-center, and retrospective or prospective. Although there are various types of imaging examinations, including computed tomography (CT), magnetic resonance imaging (MRI), positron emission tomography (PET), ultrasound, etc.[11,14-16] for different research purposes, the dominant examination methods or sequences are more recommended. Hence, more eligible cases are included to find out common features, which may contribute to the stability of models[17]. There is no general standard for the medical imaging data from different examination methods using different imaging methods, acquisition methods, imaging parameters, and imaging quality that may affect the subsequent analysis. Therefore, how to normalize the data and conform to the imaging standard is the focus of radiomics studies at present.
After data collection, the data need to be checked and confirmed, in order to correct or eliminate unqualified data. The specific inspection content includes the validity of the file format, the integrity of the sequence, and the correctness of the image content, in order to exclude unrecognizable images, sequence deletion, and wrong image layers. More detailed image quality specifications can also be formed according to specific research requirements. In the process of image quality control, it is necessary to sort out the imaging problems encountered, so that the data can be traced back when the inclusion and exclusion criteria are defined.
Because of different scanning parameters, reconstruction procedures (slice thickness, voxel size, and reconstruction algorithm), and inconsistent imaging acquisition of multi-brand manufactories, it has a significant influence on distribution of features[18,19]. In order to decrease this discrepancy, preprocessing of the collected imaging data is essential. At present, the most common methods include resampling, gray-level discretization, and intensity normalization. Image resampling involves generation of equal-size voxels by applying the linear interpolation algorithm to improve image quality and to eliminate bias introduced by non-uniform imaging resolution[20]. Gray-level discretization refers to the bundling of pixels based on their density, either by relative discretization (fixed number) or absolute discretization (fixed size)[21]. Image intensity normalization is used to correct inter-subject intensity variation by transforming all images from original greyscale into a standard greyscale. Furthermore, image enhancement approaches, such as image flipping, image rotation, image distortion, image transformation, and image scaling, can enrich data diversity, improve model generalization ability, and reduce the risk of model overfitting.
In addition to the above-mentioned methods, not only for images, we also need to preprocess clinical data. Deidentification of data is beneficial to protect personal information and query data among multiple departments. Hospital number is advised to be the unique identification, realizing the mapping of images. In order to effectively eliminate the deficiency of data inconsistency and bias in multi-center studies, it is necessary to conduct data consistency processing, which is advantageous to realize cross-center data modeling and verification. The methods of data consistency processing include: (1) Standardization of data collection: Data are collected according to the unified data acquisition standard in each center; (2) Consistency processing based on extracted features: The method of Z-score can be used to standardize data; and (3) Consistency processing based on image domain: According to the annotated information, the size of region of interest (ROI) is kept consistent.
Segmentation of ROI can be divided into manual and semiautomatic/automatic segmentation, two-dimensional (2D) and three-dimensional (3D) segmentation, and intratumoral and peritumoral segmentation[22-26]. This process is relatively tedious and requires open-source or dedicated software to support[12]. The process at least needs one labeling physician and one senior physician. The knowledge of relevant anatomy and imaging should be well known by labeling physicians and they must be familiar with the sketching software. In addition, for manual segmentation, intra-class correlation coefficient and concordance correlation coefficient can be advantageous to reduce the discrepancy of subjective judgement and the intra- and inter-reader variability[17,27]. Due to the rapid development of computer science, semiautomatic/automatic segmentation has been frequently applied. Automatic segmentation aims to draw ROIs automatically[28], while semiautomatic segmentation still requires partially manual intervention to mark the center of the lesion before automatic segmentation[29]. They both decrease instability to a certain extent, however, they are less applied because of technical restriction. At present, automatic segmentation can be summarized into three categories[30]: (1) Algorithms based on intensity thresholds and regions; (2) algorithms based on statistical approaches and deformable models; and (3) algorithms incorporating empirical knowledge into the segmentation process.
Features are extracted from ROIs using different software with the similar code, which consist of first-order, second-order, and higher-order features. First-order features describe the geometric attributes and the distribution of voxel intensities of the ROIs, including mean, median, maximum, and minimum values, as well as the skewness, kurtosis, and entropy. Second-order features represent the relationships between adjacent voxels to measure features[31]. Second-order textural features describe the gray-scale alterations and are extracted by different algorithms. Higher-order features are extracted via wavelet, Laplacian, and Gaussian filters from multiple dimensions[32]. With the combination of multiple omics, semantic features, which are based on the experience and knowledge of radiologists, pathological features, genetic features, etc., all promote the transformation of radiomics into clinical practice. In recent years, depiction of deep learning (DL)-based features, which are supplementary high-dimensional features, by observers has been reported as a challenge[33]. Although DL-based features reveal certain advantages in terms of estimating prognosis of malignancies, it is enslaved to be widely used by data size and technological development.
According to the fourth step (feature extraction), the great number of extracted features is achieved, and how to select the most relevant features is the key to establish a robust radiomics model. This process simplifies the mathematical problem by decreasing the number of parameters and also reduces the risk of overfitting. Specific methods include univariate, the least absolute shrinkage and selection operator (LASSO), RELIEF algorithm, redundancy maximum relevance (MRMR), etc[34].
The ultimate objective of radiomics is to establish an effective model for classification and prediction. The data should be clustered into training and validation datasets. Different classifiers, including logistics, support vector machine, Bayes, k-Nearest Neighbor algorithm, Tree and Forest, are used to set up models and to select the most effective model by seed circling for clinical transformation[35]. Meanwhile, the predictive performance of the final model should be verified on a separate cohort, and an external validation cohort is highly appropriate to confirm its generalization. Owing to the lack of data sharing, obtaining the results of external validation of the model is a challenge at this stage.
In previous studies, radiomics has shown a great potential in the diagnosis and staging of different diseases. Although the diagnosis of some lesions is easy according to imaging manifestations, radiomics can improve physicians’ diagnostic confidence and patients’ examination strategies. In a plain CT study, 168 patients with hepatocellular carcinoma (HCC) and 117 patients with hepatic hemangioma were analyzed. Textural features were extracted from plain CT images and 13 features were selected from 1223 candidate features to constitute the radiomics signature, in order to establish a logistic regression model to classify benign and malignant liver tumors. The final model achieved an average area under the curve (AUC) of 0.87. In spite of the lack of innovation, it helps patients who cannot successfully undergo contrast-enhanced CT (CECT) because of iodine contrast agent allergy for a relatively accurate diagnosis[36].
In another study, Ding et al[37] explored the capacity of the combined model for differentiating HCC from focal nodular hyperplasia (FNH) in non-cirrhotic livers using Gd-DTPA contrast-enhanced MRI. For this purpose, 8 radiomics features were selected for the radiomics model, and 4 clinical factors (age, gender, hepatitis B surface antigen (HbsAg), and enhancement pattern) were chosen for the clinical model. The combined model was established using the factors from the previous models. The classification accuracy of the combined model that differentiated HCC from FNH in both the training and validation datasets was 0.956 and 0.941, respectively. The model could support clinicians to make more reliable clinical decisions.
Serous cystadenomas (SCN) are considered as mostly benign cystic neoplasm in the pancreas. Mucinous cystic neoplasm (MCN) is an easily misdiagnosed lesion of SCN, which is associated with the risk of malignant transformation[38]. Therefore, Xie et al[39] confirmed the value of CT-based radiomics analysis in preoperatively discriminating pancreatic MSN and SCN. A total of 103 MCN and 113 SCN patients who underwent surgery were retrospectively enrolled. The Rad-score model was proved to be robust and reliable (average AUC, 0.784; sensitivity, 0.847; specificity, 0.745; positive-predictive value (PPV), 0.767; negative-predictive value, 0.849; accuracy, 0.793), which could serve as a novel tool for guiding clinical decision-making.
In another multi-center study, researchers took advantages of radiomics to develop a nomogram for preoperatively predicting grade 1 and grade 2/3 tumors in patients with pancreatic neuroendocrine tumors (PNETs). Totally, 138 patients from two institutions with pathologically confirmed PNETs were included in that retrospective study. The nomogram integrating an independent risk factor of tumor margin and fusion radiomic signature showed a strong discrimination with an AUC of 0.974 (95% confidence interval (CI): 0.950–0.998) in the training cohort and 0.902 (95% CI: 0.798–1.000) in the validation cohort, with a satisfactory calibration. Decision curve analysis (DCA) verified the clinical applicability of the predictive nomogram[40].
Concurrent advancements in imaging and genomic biomarkers have facilitated identification of noninvasive imaging surrogates of molecular phenotypes. Villanueva et al[41] investigated the genomic features of HCC and peritumoral tissues that were associated with patients’ outcomes, and they explored the relationship between imaging traits and genomic signatures. Patients who underwent pre-operative CT or MRI and transcriptome profiling were assessed using 11 qualitative and 4 quantitative (size, enhancement ratio, wash-out ratio, tumor-to-liver contrast ratio) imaging traits. Several imaging traits, including infiltrative pattern and macrovascular invasion were found to be associated with gene signatures of aggressive HCC phenotype, such as proliferative signatures and CK19 signature.
Microvascular invasion (MVI) is one of the strongest predictors of hepatic transplantation or hepatectomy for HCC, which is one of the independent factors for early recurrence and poor prognosis[42]. MVI could be diagnosed postoperatively and it was defined as the presence of tumor within microscopic vessels of the portal vein, hepatic artery, and lymphatic vessels[43]. Conventional imaging methods cannot reveal MVI because of the poor resolution before operation. Therefore, it is important to develop a non-invasive tool to detect MVI for clinical decision-making. Zhu et al[44] proposed a nomogram for the prediction of MVI that included a radiomic score and alpha fetoprotein, tumor type, peritumoral enhancement, arterial rim, and internal arteries. This nomogram was superior to a clinical and radiologic model with an AUC of 0.858 versus 0.729. In another research, Renzulli et al[45] demonstrated that non-smooth tumor margins and peritumoral enhancement, combined with the radio-genomic features were independent predictors for MVI with a PPV of 0.95. In a large-scale study, Xu et al[46] collected CT scan images from 495 patients and developed a combined model which consisted of semantic features (aspartate aminotransferase, alpha fetoprotein (AFP), non-smooth tumor margin, extrahepatic growth, ill-defined pseudocapsule, and peritumoral arterial enhancement) and radiomic features to predict histological MVI, with an AUC of 0.909 and 0.889 in the training cohort and the test cohort, respectively.
Gao et al[47] assessed the preoperative prediction of TP53 status based on multiparametric MRI (mp-MRI) radiomic features extracted from 3D images. In total, 57 patients with pancreatic cancer who underwent preoperative MRI were included. The 3D ADC-ap-DWI-T2WI model with 11 selected features yielded the best performance for differentiating TP53 status, with an accuracy of 0.91 and an AUC of 0.96. The model revealed a good calibration, and the DCA proved the clinical value of the model. The radiomics model derived from mp-MRI provided a non-invasive, quantitative method to predict mutational status of TP53 in patients with pancreatic cancer that might contribute to the precision treatment.
Current guidelines recommend surgical resection as the first-line therapy for patients with HCC[48]. However, postoperative recurrence rate remains high and there is no reliable prediction tool. In a multi-center study, the potential of radiomics coupled with machine learning algorithms was assessed to improve the predictive accuracy for HCC recurrence. Using the machine learning framework, they identified a three-feature signature that demonstrated a favorable prediction of HCC recurrence across all datasets, with C-index of 0.633-0.699. AFP, albumin-bilirubin, hepatic cirrhosis, tumor margin, and radiomic signature were selected for developing a preoperative model; the postoperative model incorporated satellite nodules into the above-mentioned predictors. The two models showed a superior prognostic performance, with C-index of 0.733-0.801 and integrated Brier score of 0.147-0.165, compared with rival models without radiomics, and are widely used in staging systems. Combined with clinical data, a three-feature fusion signature generated by aggregated ML-based framework could accurately predict individual recurrence risk, enabling appropriate management and surveillance of HCC[49]. In another study, CECT with measurement of Gabor and Wavelet radiomics features in patients with a single HCC tumor treated by hepatectomy revealed that several features were associated with both overall survival (OS) and disease-free survival (P values < 0.05)[50]. Similar results were reported by a separate study that risk scores developed from radiomics nomograms obtained from CECT textural data overmatched traditional clinical staging systems in both the training and validation cohorts for both tumor recurrence and OS[51].
Patients with pancreatic cancer have a poor prognosis, therefore, it is necessary to identify tumor characteristics associated with prognosis. Toyama et al[52] enrolled 161 patients with pancreatic cancer who underwent fluorodeoxyglucose (FDG)-PET/CT before treatment. The area of the primary tumor was semi-automatically contoured with a threshold of 40% of the maximum standardized uptake value, and 42 PET-based features were extracted. Among the PET parameters, 10 features showed statistical significance for predicting OS. Multivariate Cox regression analysis revealed gray-level zone length matrix (GLZLM)-gray-level non-uniformity (GLNU) as the only PET parameter showing statistical significance. In the random forest model, GLZLM-GLNU was the most relevant factor for predicting 1-year survival, followed by total lesion glycolysis. Radiomics with machine learning using FDG-PET in patients with pancreatic cancer provided valuable prognostic information.
There is no doubt that radiomics as a newly emerged quantitative technique is burgeoning in disease management. Nevertheless, the majority of the research of radiomics encountered common problems, and whether the radiomic-based signatures can be used in clinical practice needs to be discussed.
Reproducibility is one of the primary challenges that radiomic techniques must overcome for clinical application. At present, imaging protocols are not standardized worldwide, and hence, variability in image acquisition and reconstruction parameters is inevitable in clinical practice. A recent study demonstrated that the quantitative values of radiomic features varied according to imaging protocols[53]. In addition, although IBSI seeks standardization for radiomic extraction, the differences in techniques or platforms adopted in different centers may lead to differences in feature values[5], propagating to the radiomic signatures. Most radiomic signatures have a sharp drop in performance from training cohort to validation cohort. Researchers have adopted data normalization methods to correct for multicenter effects, such as ComBat harmonization[54]. However, whether the radiomic-based signature developed by normalized radiomic features is appropriate for clinical practice has not yet been studied. It is urgent to develop a reproducible radiomic signature that could overcome inherent multicenter effects, which is the basis for clinical individualized application.
Data sharing for independent validation is a challenge for radiomic signatures. To date, studies have mainly developed and validated the radiomic signatures using imaging data derived from their own center or multiple centers according to the same imaging protocols[55]. However, whether the signatures would be effective in completely independent centers needs further validation. Although images are more readily available than tissue molecular assays, the current open radiomic datasets are not enough for the independent validation. To eliminate this deficiency, data sharing among institutes and hospitals around the country or even around the world is important for radiomics, although it presents complex logistical problems. The Cancer Imaging Archive provides a good example of data sharing with a large portion of clinical data[56], and it is still growing with contribution from different institutes and hospitals. A previous study indicated that signatures should be validated using an open dataset that could become the standard to demonstrate their effectiveness[9].
Biological interpretability of radiomic signatures would accelerate their clinical application. Clinical experts mainly assume the radiomic model as a black box that can provide promising prediction results for clinical outcomes, which may make radiomics as a less accepted approach. The problem is further aggravated in the context of deconvolutional neural or DL networks, which even lack the observable model that solely concentrates on maximizing performance. A great number of these so-called “black-box” approaches may be perfectly viable in the diagnostic setting; however, when it comes to radiomic signatures for optimizing treatment, the question of interpretability becomes more paramount because a biomarker-driven treatment decision needs an explanation rooted in pathophysiology[57]. The emergence of radio-genomics provides a bridge for linking the radiomics to the underlying biological progression. The biological interpretability may provide biological evidence for the predictive ability of the radiomic signatures.
Clinical operability is the key in the clinical adoption of prognostic and predictive radiomic tools. To date, radiomic-based studies have mainly concentrated on developing robust signatures, and their application details in clinical practice are lack. Therefore, translating the computer language into a simple software or system may be an effective method to promote clinical application of radiomics.
In conclusion, the current researches have achieved encouraging results of radiomics and revealed potential of clinical applications, while poor standardization and generalization of radiomics limit the further translation of this method into clinical routine. How to make reproducibility of data, multi-center data sharing, biological interpretability of radiomic signatures and clinical operability come true, will become the crucial issue for development of radiomics. Only then will radiomics be more comparable and increase reliability to get clinician's approval. In foreseeable future, the development of radiomics will occupy a significant position in personalization and precision medicine. At present, it is more important to make clinical participants be conscious of benefits and limitations of radiomics in order to obtain reasonable decision towards clinical practice.
Provenance and peer review: Invited article; Externally peer reviewed.
Peer-review model: Single blind
Specialty type: Radiology, nuclear medicine and medical imaging
Country/Territory of origin: China
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): 0
Grade C (Good): C, C
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Anzola F LK, Colombia; Litvin A, Russia S-Editor: Liu JH L-Editor: A P-Editor: Liu JH
| 1. | Lambin P, Rios-Velazquez E, Leijenaar R, Carvalho S, van Stiphout RG, Granton P, Zegers CM, Gillies R, Boellard R, Dekker A, Aerts HJ. Radiomics: extracting more information from medical images using advanced feature analysis. Eur J Cancer. 2012;48:441-446. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2415] [Cited by in RCA: 3853] [Article Influence: 296.4] [Reference Citation Analysis (2)] |
| 2. | Lambin P, Leijenaar RTH, Deist TM, Peerlings J, de Jong EEC, van Timmeren J, Sanduleanu S, Larue RTHM, Even AJG, Jochems A, van Wijk Y, Woodruff H, van Soest J, Lustberg T, Roelofs E, van Elmpt W, Dekker A, Mottaghy FM, Wildberger JE, Walsh S. Radiomics: the bridge between medical imaging and personalized medicine. Nat Rev Clin Oncol. 2017;14:749-762. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1825] [Cited by in RCA: 3564] [Article Influence: 445.5] [Reference Citation Analysis (0)] |
| 3. | Verma V, Simone CB 2nd, Krishnan S, Lin SH, Yang J, Hahn SM. The Rise of Radiomics and Implications for Oncologic Management. J Natl Cancer Inst. 2017;109. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 70] [Cited by in RCA: 97] [Article Influence: 12.1] [Reference Citation Analysis (0)] |
| 4. | Aerts HJ, Velazquez ER, Leijenaar RT, Parmar C, Grossmann P, Carvalho S, Bussink J, Monshouwer R, Haibe-Kains B, Rietveld D, Hoebers F, Rietbergen MM, Leemans CR, Dekker A, Quackenbush J, Gillies RJ, Lambin P. Decoding tumour phenotype by noninvasive imaging using a quantitative radiomics approach. Nat Commun. 2014;5:4006. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2262] [Cited by in RCA: 3249] [Article Influence: 295.4] [Reference Citation Analysis (0)] |
| 5. | Zwanenburg A, Vallières M, Abdalah MA, Aerts HJWL, Andrearczyk V, Apte A, Ashrafinia S, Bakas S, Beukinga RJ, Boellaard R, Bogowicz M, Boldrini L, Buvat I, Cook GJR, Davatzikos C, Depeursinge A, Desseroit MC, Dinapoli N, Dinh CV, Echegaray S, El Naqa I, Fedorov AY, Gatta R, Gillies RJ, Goh V, Götz M, Guckenberger M, Ha SM, Hatt M, Isensee F, Lambin P, Leger S, Leijenaar RTH, Lenkowicz J, Lippert F, Losnegård A, Maier-Hein KH, Morin O, Müller H, Napel S, Nioche C, Orlhac F, Pati S, Pfaehler EAG, Rahmim A, Rao AUK, Scherer J, Siddique MM, Sijtsema NM, Socarras Fernandez J, Spezi E, Steenbakkers RJHM, Tanadini-Lang S, Thorwarth D, Troost EGC, Upadhaya T, Valentini V, van Dijk LV, van Griethuysen J, van Velden FHP, Whybra P, Richter C, Löck S. The Image Biomarker Standardization Initiative: Standardized Quantitative Radiomics for High-Throughput Image-based Phenotyping. Radiology. 2020;295:328-338. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2090] [Cited by in RCA: 2317] [Article Influence: 463.4] [Reference Citation Analysis (0)] |
| 6. | Hu W, Yang H, Xu H, Mao Y. Radiomics based on artificial intelligence in liver diseases: where we are? Gastroenterol Rep (Oxf). 2020;8:90-97. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 29] [Cited by in RCA: 30] [Article Influence: 6.0] [Reference Citation Analysis (0)] |
| 7. | Gillies RJ, Kinahan PE, Hricak H. Radiomics: Images Are More than Pictures, They Are Data. Radiology. 2016;278:563-577. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 4541] [Cited by in RCA: 5550] [Article Influence: 616.7] [Reference Citation Analysis (3)] |
| 8. | Limkin EJ, Sun R, Dercle L, Zacharaki EI, Robert C, Reuzé S, Schernberg A, Paragios N, Deutsch E, Ferté C. Promises and challenges for the implementation of computational medical imaging (radiomics) in oncology. Ann Oncol. 2017;28:1191-1206. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 345] [Cited by in RCA: 553] [Article Influence: 79.0] [Reference Citation Analysis (0)] |
| 9. | Liu Z, Wang S, Dong D, Wei J, Fang C, Zhou X, Sun K, Li L, Li B, Wang M, Tian J. The Applications of Radiomics in Precision Diagnosis and Treatment of Oncology: Opportunities and Challenges. Theranostics. 2019;9:1303-1322. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 558] [Cited by in RCA: 608] [Article Influence: 101.3] [Reference Citation Analysis (0)] |
| 10. | Paul R, Schabath M, Balagurunathan Y, Liu Y, Li Q, Gillies R, Hall LO, Goldgof DB. Explaining Deep Features Using Radiologist-Defined Semantic Features and Traditional Quantitative Features. Tomography. 2019;5:192-200. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 21] [Cited by in RCA: 19] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 11. | Fu S, Wei J, Zhang J, Dong D, Song J, Li Y, Duan C, Zhang S, Li X, Gu D, Chen X, Hao X, He X, Yan J, Liu Z, Tian J, Lu L. Selection Between Liver Resection Versus Transarterial Chemoembolization in Hepatocellular Carcinoma: A Multicenter Study. Clin Transl Gastroenterol. 2019;10:e00070. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 15] [Cited by in RCA: 14] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 12. | Park S, Chu LC, Hruban RH, Vogelstein B, Kinzler KW, Yuille AL, Fouladi DF, Shayesteh S, Ghandili S, Wolfgang CL, Burkhart R, He J, Fishman EK, Kawamoto S. Differentiating autoimmune pancreatitis from pancreatic ductal adenocarcinoma with CT radiomics features. Diagn Interv Imaging. 2020;101:555-564. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 40] [Cited by in RCA: 75] [Article Influence: 15.0] [Reference Citation Analysis (0)] |
| 13. | Dalal V, Carmicheal J, Dhaliwal A, Jain M, Kaur S, Batra SK. Radiomics in stratification of pancreatic cystic lesions: Machine learning in action. Cancer Lett. 2020;469:228-237. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 37] [Cited by in RCA: 68] [Article Influence: 11.3] [Reference Citation Analysis (0)] |
| 14. | Zhou T, Xie CL, Chen Y, Deng Y, Wu JL, Liang R, Yang GD, Zhang XM. Magnetic Resonance Imaging-Based Radiomics Models to Predict Early Extrapancreatic Necrosis in Acute Pancreatitis. Pancreas. 2021;50:1368-1375. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3] [Cited by in RCA: 9] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 15. | Gao X, Wang X. Performance of deep learning for differentiating pancreatic diseases on contrast-enhanced magnetic resonance imaging: A preliminary study. Diagn Interv Imaging. 2020;101:91-100. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 15] [Cited by in RCA: 29] [Article Influence: 4.8] [Reference Citation Analysis (0)] |
| 16. | Wei M, Gu B, Song S, Zhang B, Wang W, Xu J, Yu X, Shi S. A Novel Validated Recurrence Stratification System Based on (18)F-FDG PET/CT Radiomics to Guide Surveillance After Resection of Pancreatic Cancer. Front Oncol. 2021;11:650266. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 7] [Cited by in RCA: 3] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 17. | Li Y, Reyhan M, Zhang Y, Wang X, Zhou J, Yue NJ, Nie K. The impact of phantom design and material-dependence on repeatability and reproducibility of CT-based radiomics features. Med Phys. 2022;49:1648-1659. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 23] [Reference Citation Analysis (0)] |
| 18. | Duron L, Balvay D, Vande Perre S, Bouchouicha A, Savatovsky J, Sadik JC, Thomassin-Naggara I, Fournier L, Lecler A. Gray-level discretization impacts reproducible MRI radiomics texture features. PLoS One. 2019;14:e0213459. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 109] [Cited by in RCA: 136] [Article Influence: 22.7] [Reference Citation Analysis (0)] |
| 19. | Moradmand H, Aghamiri SMR, Ghaderi R. Impact of image preprocessing methods on reproducibility of radiomic features in multimodal magnetic resonance imaging in glioblastoma. J Appl Clin Med Phys. 2020;21:179-190. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 58] [Cited by in RCA: 106] [Article Influence: 17.7] [Reference Citation Analysis (0)] |
| 20. | Larue RTHM, van Timmeren JE, de Jong EEC, Feliciani G, Leijenaar RTH, Schreurs WMJ, Sosef MN, Raat FHPJ, van der Zande FHR, Das M, van Elmpt W, Lambin P. Influence of gray level discretization on radiomic feature stability for different CT scanners, tube currents and slice thicknesses: a comprehensive phantom study. Acta Oncol. 2017;56:1544-1553. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 119] [Cited by in RCA: 176] [Article Influence: 22.0] [Reference Citation Analysis (0)] |
| 21. | Zhuge Y, Udupa JK, Liu J, Saha PK. Image background inhomogeneity correction in MRI via intensity standardization. Comput Med Imaging Graph. 2009;33:7-16. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 31] [Cited by in RCA: 20] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 22. | Peng J, Zhang J, Zhang Q, Xu Y, Zhou J, Liu L. A radiomics nomogram for preoperative prediction of microvascular invasion risk in hepatitis B virus-related hepatocellular carcinoma. Diagn Interv Radiol. 2018;24:121-127. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 98] [Cited by in RCA: 145] [Article Influence: 20.7] [Reference Citation Analysis (0)] |
| 23. | Huang X, Long L, Wei J, Li Y, Xia Y, Zuo P, Chai X. Radiomics for diagnosis of dual-phenotype hepatocellular carcinoma using Gd-EOB-DTPA-enhanced MRI and patient prognosis. J Cancer Res Clin Oncol. 2019;145:2995-3003. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 20] [Cited by in RCA: 43] [Article Influence: 7.2] [Reference Citation Analysis (0)] |
| 24. | Ciaravino V, Cardobi N, DE Robertis R, Capelli P, Melisi D, Simionato F, Marchegiani G, Salvia R, D'Onofrio M. CT Texture Analysis of Ductal Adenocarcinoma Downstaged After Chemotherapy. Anticancer Res. 2018;38:4889-4895. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 24] [Cited by in RCA: 24] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 25. | D'Onofrio M, Ciaravino V, Cardobi N, De Robertis R, Cingarlini S, Landoni L, Capelli P, Bassi C, Scarpa A. CT Enhancement and 3D Texture Analysis of Pancreatic Neuroendocrine Neoplasms. Sci Rep. 2019;9:2176. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 41] [Cited by in RCA: 58] [Article Influence: 9.7] [Reference Citation Analysis (1)] |
| 26. | Braman NM, Etesami M, Prasanna P, Dubchuk C, Gilmore H, Tiwari P, Plecha D, Madabhushi A. Erratum to: Intratumoral and peritumoral radiomics for the pretreatment prediction of pathological complete response to neoadjuvant chemotherapy based on breast DCE-MRI. Breast Cancer Res. 2017;19:80. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 17] [Cited by in RCA: 21] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 27. | Rios Velazquez E, Aerts HJ, Gu Y, Goldgof DB, De Ruysscher D, Dekker A, Korn R, Gillies RJ, Lambin P. A semiautomatic CT-based ensemble segmentation of lung tumors: comparison with oncologists' delineations and with the surgical specimen. Radiother Oncol. 2012;105:167-173. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 91] [Cited by in RCA: 83] [Article Influence: 6.4] [Reference Citation Analysis (0)] |
| 28. | Häme Y, Pollari M. Semi-automatic liver tumor segmentation with hidden Markov measure field model and non-parametric distribution estimation. Med Image Anal. 2012;16:140-149. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 59] [Cited by in RCA: 34] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 29. | Permuth JB, Choi J, Balarunathan Y, Kim J, Chen DT, Chen L, Orcutt S, Doepker MP, Gage K, Zhang G, Latifi K, Hoffe S, Jiang K, Coppola D, Centeno BA, Magliocco A, Li Q, Trevino J, Merchant N, Gillies R, Malafa M; Florida Pancreas Collaborative. Combining radiomic features with a miRNA classifier may improve prediction of malignant pathology for pancreatic intraductal papillary mucinous neoplasms. Oncotarget. 2016;7:85785-85797. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 81] [Cited by in RCA: 102] [Article Influence: 14.6] [Reference Citation Analysis (0)] |
| 30. | Chen W, Liu B, Peng S, Sun J, Qiao X. Computer-Aided Grading of Gliomas Combining Automatic Segmentation and Radiomics. Int J Biomed Imaging. 2018;2018:2512037. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 47] [Cited by in RCA: 46] [Article Influence: 6.6] [Reference Citation Analysis (0)] |
| 31. | Li J, Lu J, Liang P, Li A, Hu Y, Shen Y, Hu D, Li Z. Differentiation of atypical pancreatic neuroendocrine tumors from pancreatic ductal adenocarcinomas: Using whole-tumor CT texture analysis as quantitative biomarkers. Cancer Med. 2018;7:4924-4931. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 35] [Cited by in RCA: 51] [Article Influence: 7.3] [Reference Citation Analysis (0)] |
| 32. | Nougaret S, Tardieu M, Vargas HA, Reinhold C, Vande Perre S, Bonanno N, Sala E, Thomassin-Naggara I. Ovarian cancer: An update on imaging in the era of radiomics. Diagn Interv Imaging. 2019;100:647-655. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 43] [Cited by in RCA: 67] [Article Influence: 11.2] [Reference Citation Analysis (0)] |
| 33. | Thawani R, McLane M, Beig N, Ghose S, Prasanna P, Velcheti V, Madabhushi A. Radiomics and radiogenomics in lung cancer: A review for the clinician. Lung Cancer. 2018;115:34-41. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 235] [Cited by in RCA: 319] [Article Influence: 45.6] [Reference Citation Analysis (0)] |
| 34. | Matzner-Lober E, Suehs CM, Dohan A, Molinari N. Thoughts on entering correlated imaging variables into a multivariable model: Application to radiomics and texture analysis. Diagn Interv Imaging. 2018;99:269-270. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 15] [Cited by in RCA: 18] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 35. | Rizzo S, Botta F, Raimondi S, Origgi D, Fanciullo C, Morganti AG, Bellomi M. Radiomics: the facts and the challenges of image analysis. Eur Radiol Exp. 2018;2:36. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 413] [Cited by in RCA: 683] [Article Influence: 97.6] [Reference Citation Analysis (0)] |
| 36. | Yin J, Qiu JJ, Qian W, Ji L, Yang D, Jiang JW, Wang JR, Lan L. A radiomics signature to identify malignant and benign liver tumors on plain CT images. J Xray Sci Technol. 2020;28:683-694. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 5] [Cited by in RCA: 5] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 37. | Ding Z, Lin K, Fu J, Huang Q, Fang G, Tang Y, You W, Lin Z, Pan X, Zeng Y. An MR-based radiomics model for differentiation between hepatocellular carcinoma and focal nodular hyperplasia in non-cirrhotic liver. World J Surg Oncol. 2021;19:181. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 22] [Cited by in RCA: 19] [Article Influence: 4.8] [Reference Citation Analysis (0)] |
| 38. | Fernández-del Castillo C. Mucinous cystic neoplasms. J Gastrointest Surg. 2008;12:411-413. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 41] [Cited by in RCA: 32] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 39. | Xie T, Wang X, Zhang Z, Zhou Z. CT-Based Radiomics Analysis for Preoperative Diagnosis of Pancreatic Mucinous Cystic Neoplasm and Atypical Serous Cystadenomas. Front Oncol. 2021;11:621520. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 4] [Cited by in RCA: 15] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 40. | Gu D, Hu Y, Ding H, Wei J, Chen K, Liu H, Zeng M, Tian J. CT radiomics may predict the grade of pancreatic neuroendocrine tumors: a multicenter study. Eur Radiol. 2019;29:6880-6890. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 60] [Cited by in RCA: 115] [Article Influence: 19.2] [Reference Citation Analysis (0)] |
| 41. | Villanueva A, Hoshida Y, Battiston C, Tovar V, Sia D, Alsinet C, Cornella H, Liberzon A, Kobayashi M, Kumada H, Thung SN, Bruix J, Newell P, April C, Fan JB, Roayaie S, Mazzaferro V, Schwartz ME, Llovet JM. Combining clinical, pathology, and gene expression data to predict recurrence of hepatocellular carcinoma. Gastroenterology. 2011;140:1501-12.e2. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 334] [Cited by in RCA: 334] [Article Influence: 23.9] [Reference Citation Analysis (0)] |
| 42. | Mazzaferro V, Llovet JM, Miceli R, Bhoori S, Schiavo M, Mariani L, Camerini T, Roayaie S, Schwartz ME, Grazi GL, Adam R, Neuhaus P, Salizzoni M, Bruix J, Forner A, De Carlis L, Cillo U, Burroughs AK, Troisi R, Rossi M, Gerunda GE, Lerut J, Belghiti J, Boin I, Gugenheim J, Rochling F, Van Hoek B, Majno P; Metroticket Investigator Study Group. Predicting survival after liver transplantation in patients with hepatocellular carcinoma beyond the Milan criteria: a retrospective, exploratory analysis. Lancet Oncol. 2009;10:35-43. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1267] [Cited by in RCA: 1572] [Article Influence: 92.5] [Reference Citation Analysis (1)] |
| 43. | Lim KC, Chow PK, Allen JC, Chia GS, Lim M, Cheow PC, Chung AY, Ooi LL, Tan SB. Microvascular invasion is a better predictor of tumor recurrence and overall survival following surgical resection for hepatocellular carcinoma compared to the Milan criteria. Ann Surg. 2011;254:108-113. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 284] [Cited by in RCA: 414] [Article Influence: 29.6] [Reference Citation Analysis (0)] |
| 44. | Zhu YJ, Feng B, Wang S, Wang LM, Wu JF, Ma XH, Zhao XM. Model-based three-dimensional texture analysis of contrast-enhanced magnetic resonance imaging as a potential tool for preoperative prediction of microvascular invasion in hepatocellular carcinoma. Oncol Lett. 2019;18:720-732. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 10] [Cited by in RCA: 16] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 45. | Renzulli M, Brocchi S, Cucchetti A, Mazzotti F, Mosconi C, Sportoletti C, Brandi G, Pinna AD, Golfieri R. Can Current Preoperative Imaging Be Used to Detect Microvascular Invasion of Hepatocellular Carcinoma? Radiology. 2016;279:432-442. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 180] [Cited by in RCA: 293] [Article Influence: 29.3] [Reference Citation Analysis (0)] |
| 46. | Xu X, Zhang HL, Liu QP, Sun SW, Zhang J, Zhu FP, Yang G, Yan X, Zhang YD, Liu XS. Radiomic analysis of contrast-enhanced CT predicts microvascular invasion and outcome in hepatocellular carcinoma. J Hepatol. 2019;70:1133-1144. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 278] [Cited by in RCA: 495] [Article Influence: 82.5] [Reference Citation Analysis (0)] |
| 47. | Gao J, Chen X, Li X, Miao F, Fang W, Li B, Qian X, Lin X. Differentiating TP53 Mutation Status in Pancreatic Ductal Adenocarcinoma Using Multiparametric MRI-Derived Radiomics. Front Oncol. 2021;11:632130. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2] [Cited by in RCA: 7] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 48. | Vogel A, Cervantes A, Chau I, Daniele B, Llovet JM, Meyer T, Nault JC, Neumann U, Ricke J, Sangro B, Schirmacher P, Verslype C, Zech CJ, Arnold D, Martinelli E. Hepatocellular carcinoma: ESMO Clinical Practice Guidelines for diagnosis, treatment and follow-up. Ann Oncol. 2019;30:871-873. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 152] [Cited by in RCA: 188] [Article Influence: 31.3] [Reference Citation Analysis (0)] |
| 49. | Ji GW, Zhu FP, Xu Q, Wang K, Wu MY, Tang WW, Li XC, Wang XH. Machine-learning analysis of contrast-enhanced CT radiomics predicts recurrence of hepatocellular carcinoma after resection: A multi-institutional study. EBioMedicine. 2019;50:156-165. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 154] [Cited by in RCA: 145] [Article Influence: 24.2] [Reference Citation Analysis (0)] |
| 50. | Chen S, Zhu Y, Liu Z, Liang C. Texture analysis of baseline multiphasic hepatic computed tomography images for the prognosis of single hepatocellular carcinoma after hepatectomy: A retrospective pilot study. Eur J Radiol. 2017;90:198-204. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 30] [Cited by in RCA: 41] [Article Influence: 5.1] [Reference Citation Analysis (1)] |
| 51. | Zheng BH, Liu LZ, Zhang ZZ, Shi JY, Dong LQ, Tian LY, Ding ZB, Ji Y, Rao SX, Zhou J, Fan J, Wang XY, Gao Q. Radiomics score: a potential prognostic imaging feature for postoperative survival of solitary HCC patients. BMC Cancer. 2018;18:1148. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 112] [Cited by in RCA: 107] [Article Influence: 15.3] [Reference Citation Analysis (0)] |
| 52. | Toyama Y, Hotta M, Motoi F, Takanami K, Minamimoto R, Takase K. Prognostic value of FDG-PET radiomics with machine learning in pancreatic cancer. Sci Rep. 2020;10:17024. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 36] [Cited by in RCA: 49] [Article Influence: 9.8] [Reference Citation Analysis (0)] |
| 53. | Midya A, Chakraborty J, Gönen M, Do RKG, Simpson AL. Influence of CT acquisition and reconstruction parameters on radiomic feature reproducibility. J Med Imaging (Bellingham). 2018;5:011020. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 30] [Cited by in RCA: 56] [Article Influence: 8.0] [Reference Citation Analysis (0)] |
| 54. | Orlhac F, Frouin F, Nioche C, Ayache N, Buvat I. Validation of A Method to Compensate Multicenter Effects Affecting CT Radiomics. Radiology. 2019;291:53-59. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 157] [Cited by in RCA: 171] [Article Influence: 28.5] [Reference Citation Analysis (0)] |
| 55. | Wu G, Woodruff HC, Shen J, Refaee T, Sanduleanu S, Ibrahim A, Leijenaar RTH, Wang R, Xiong J, Bian J, Wu J, Lambin P. Diagnosis of Invasive Lung Adenocarcinoma Based on Chest CT Radiomic Features of Part-Solid Pulmonary Nodules: A Multicenter Study. Radiology. 2020;297:451-458. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 31] [Cited by in RCA: 74] [Article Influence: 14.8] [Reference Citation Analysis (0)] |
| 56. | Lu L, Sun SH, Yang H, E L, Guo P, Schwartz LH, Zhao B. Radiomics Prediction of EGFR Status in Lung Cancer-Our Experience in Using Multiple Feature Extractors and The Cancer Imaging Archive Data. Tomography. 2020;6:223-230. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 17] [Cited by in RCA: 25] [Article Influence: 6.3] [Reference Citation Analysis (0)] |
| 57. | Bera K, Braman N, Gupta A, Velcheti V, Madabhushi A. Predicting cancer outcomes with radiomics and artificial intelligence in radiology. Nat Rev Clin Oncol. 2022;19:132-146. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 501] [Cited by in RCA: 421] [Article Influence: 140.3] [Reference Citation Analysis (1)] |