Published online Sep 8, 2023. doi: 10.35712/aig.v4.i2.28
Peer-review started: March 6, 2023
First decision: May 9, 2023
Revised: May 17, 2023
Accepted: June 12, 2023
Article in press: June 12, 2023
Published online: September 8, 2023
Processing time: 184 Days and 22.5 Hours
The application of artificial intelligence (AI) in gastrointestinal endoscopy has gained significant traction over the last decade. One of the more recent applications of AI in this field includes the detection of dysplasia and cancer in Barrett’s esophagus (BE). AI using deep learning methods has shown promise as an adjunct to the endoscopist in detecting dysplasia and cancer. Apart from visual detection and diagnosis, AI may also aid in reducing the considerable interobserver variability in identifying and distinguishing dysplasia on whole slide images from digitized BE histology slides. This review aims to provide a comprehensive summary of the key studies thus far as well as providing an insight into the future role of AI in Barrett’s esophagus.
Core Tip: Barrett’s esophagus is a significant precursor to esophageal adenocarcinoma. Detection of dysplasia or neoplastic changes in Barrett’s esophagus can often be difficult as endoscopic changes can be subtle. Artificial intelligence has the potential to aid endoscopist in detecting such lesions endoscopically and also reduce the inter-observer variability in detecting dysplasia in Barrett’s esophagus histologically.
- Citation: Tee CHN, Ravi R, Ang TL, Li JW. Role of artificial intelligence in Barrett’s esophagus. Artif Intell Gastroenterol 2023; 4(2): 28-35
- URL: https://www.wjgnet.com/2644-3236/full/v4/i2/28.htm
- DOI: https://dx.doi.org/10.35712/aig.v4.i2.28
“Artificial Intelligence” (AI) is a generic term used to denote the ability of a computer program to learn and solve problems autonomously[1]. AI uses input data to learn with the intention of refining the ability to process new data samples that are not part of the original set of training data. This process of “machine learning” (ML) uses mathematical algorithms to capture structure and patterns in large data sets, often in a way that allows the learned function to be applied to new data. Machine learning can be supervised or unsupervised depending on whether the algorithms were trained with known patterns or unknown patterns respectively[2]. Deep learning is a subtype of machine learning in which a convolutional neural network (CNN) receives input (e.g., endoscopic images), learns specific patterns (e.g., mucosal surface/vascular pattern) and processes this information through the multi-layered network to produce an output (e.g., presence or absence of neoplasia). This form of deep learning algorithm is the main driver for the rapidly advancing role of computer-aided diagnosis (CAD) in detection and characterization of lesions during endoscopy[3].
The greatest impact of AI in gastrointestinal (GI) endoscopy has been made in the area of colonic polyp and adenoma detection[4]. Several clinical studies and meta-analyses have shown the potential and at times, the superiority of AI in colonic adenoma detection rate compared to the endoscopist[5-8]. The crux of AI research in GI endoscopy has focused primarily on three domains which include detection, classification and delineation of lesions or disease entities[9]. There is an increasing amount of research in all three domains with regards to the application of AI in BE.
Barrett’s esophagus (BE) is defined as a change in the squamous lining of the distal esophagus to metaplastic columnar epithelium with goblet cells[10]. This is typically associated with chronic gastroesophageal reflux disease (GERD) with as much as 12% of patients with GERD symptoms harboring BE[11]. While there are variances in how BE is defined between different guidelines[12-14], there is definitive data that it is a precursor that increases the risk of esophageal adenocarcinoma (EAC)[15]. Hence early detection of dysplasia within BE is crucial to institute definitive treatment where possible and prevent further progression into neoplasia. However, this remains a challenge as endoscopic changes indicating dysplasia or early neoplasia can be subtle and be easily missed[16]. Even when there is no visible dysplasia and biopsies are done as per the Seattle protocol, sampling error can lead to areas of concern being missed[17]. The endoscopic diagnosis of BE dysplasia is generally a two-step process of primary detection in overview, followed by detailed inspection of these visible abnormalities for characterization[18]. This process relies on the individual experience of the endoscopist, which might further introduce variations and bias, leading to misjudgment and potentially delay in diagnosis and treatment.
Initially there was great interest in image enhancement technologies to overcome these challenges but to date only virtual and dye based chromoendoscopy have met the parameters outlined in American Society for Gastrointestinal Endoscopy’s (ASGE) Preservation and Incorporation of Valuable Endoscopic Innovations (PIVI). Specific to BE imaging[19], PIVI recommends that imaging technology with targeted biopsies should have a per-patient sensitivity of 90% or greater and a specificity of 80% or higher to allow reduction in the number of biopsies.
AI has since emerged as a promising adjunct on this front. AI uses various ML algorithms including CNN to identify and process real time endoscopic data to overcome the inherent limitations of an endoscopist.
This review will provide a comprehensive summary of the present evidence, recent research advances and future perspectives regarding the utility of AI in BE endoscopy. AI may overcome the human limitations related to poor intra- and inter-observer agreement, a burden that affects many aspects of medical imaging and endoscopy. If a CAD system was to be trained to distinguish between neoplastic and non-neoplastic BE macroscopically on endoscopy and microscopically on histology with almost-perfection, it seems logical these limitations can be overcomed and better tailored medical management can be rendered.
A comprehensive electronic literature search was performed in the PubMed, MEDLINE and EMBASE databases from inception to the 1st September 2022 using the following key search terms “artificial intelligence” OR “AI” OR “convolutional neural network” OR “deep learning” OR “computer-aided detection” OR “computer-aided diagnosis” AND “Barrett’s esophagus.” The search was limited to human studies.
Titles and abstracts were screened to exclude studies that did not address the purpose of this review article. The titles of all the identified studies were screened by two reviewers (TNCH and RR) to exclude studies not related to the study topic. The full texts of the screened studies were then assessed for inclusion. Review articles and letters to the editor were excluded. Studies that used other endoscopic techniques such as volumetric laser endomicroscopy were also excluded. Any disagreements were resolved through discussion with senior author LJW until consensus was achieved.
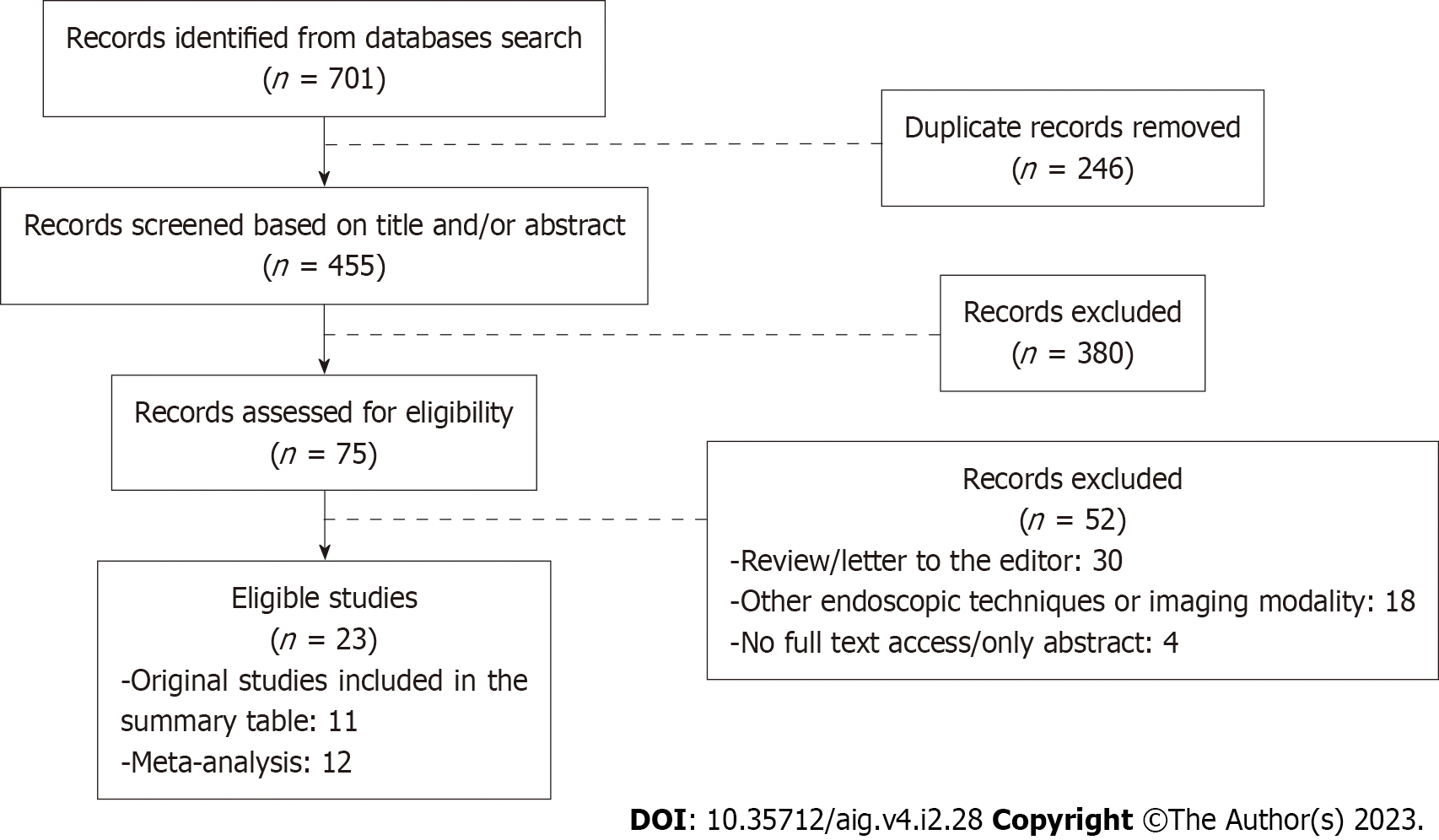
Eligible studies including 12 meta-analyses which reported the use of AI in Barrett’s endoscopy and histopathology were included in the review. The analysis flow chart of the included studies is shown in Figure 1.
Pan et al[20] developed a DL algorithm using 443 endoscopic images from 187 patients to automatically identify and segment the gastroesophageal junction and squamous-columnar junction of BE. The performance of this automated segmentation algorithm demonstrated satisfactory agreement with expert annotations as measured by intersection over union. This study demonstrates the potential of DL in automating the identification and the classification of BE according to the Prague C&M classification[21] while reducing the inter-observer variability.
Following successful identification and classification of BE, the subsequent detection of dysplasia or EAC can be clinically challenging, particularly for non-experts[22]. Further differentiation between non-dysplasia, low-grade dysplasia, high-grade dysplasia and EAC can be subjective and difficult when a focal lesion has been detected. Ebigbo et al[23] managed to demonstrate that a CAD system using deep learning of still images [248 high-definition white light images and 74 narrow band images (NBI)] from two databases, was able to diagnose EAC with sensitivity of 97% and 92% as well as specificity of 88% and 100% for white light images (WLI) in both databases respectively. Additionally, the CAD system was able to achieve sensitivity of 94% and specificity of 80% for NBI images. This study demonstrated that a CNN algorithm was able to accurately identify EAC in still endoscopic images, all validated by expert pathologists, at a sufficiently high sensitivity and specificity to meet the PIVI standards mentioned previously.
Due to the limitations of using still endoscopic images, further steps were taken to validate the AI system in real-time by assessing captured endoscopic images taken by an expert BE endoscopist to differentiate between EAC and normal BE with a sensitivity and specificity of 83.7% and 100% respectively with an overall accuracy of 89.9%[24].
Concurrently, de Groof et al[25] conducted a pilot study to develop a CAD system using white light endoscopic images from 60 patients to delineate between BE neoplasm and non-dysplastic BE. One endoscopic image from each patient was included in the CAD system with per-image analysis demonstrating sensitivity of 95%, specificity of 85% and diagnostic accuracy of 91.7%. The CAD system was not only able to delineate a BE neoplastic lesion but also able to indicate the most abnormal area within that delineation to obtain a targeted biopsy. Additionally, it took an average of 1.051 s for the algorithm to analyze an endoscopic image and subsequently produce its lesion delineation, signaling the potential to be used in a real-time, automated setting.
The same group of investigators went on further to develop a deep-learning CAD system for primary detection of neoplasia in patients with BE using a CNN model. The system was initially pretrained with a large dataset of 494364 Labeled endoscopic images from multiple locations of the GI tract using a supervised learning approach. The system was then subsequently trained with BE-specific endoscopic images containing a total of 1544 images of BE neoplasia and non-dysplastic BE before being validated using two separate external datasets. The CAD system managed to classify images as containing neoplasms or non-dysplastic BE with 89% accuracy, 90% sensitivity and 88% specificity. Performance was also benchmarked against 53 general endoscopists with a wide range of experience. When compared to the endoscopists, the CAD system managed to achieve higher diagnostic accuracy of 88% vs 73%, sensitivity of 93% vs 72% and specificity of 83% vs 74%[26]. Apart from the large databases used to develop and validate the CNN based algorithm, the computational speed per image analysis was 0.24 s which was a significant improvement from their previous CAD system, paving the way for the incorporation of the CAD system during live endoscopic procedures to help delineate BE neoplastic lesions.
For further validation, the CAD system was tested during live endoscopic procedures in 10 patients with non-dysplastic BE and 10 patients with confirmed BE neoplasia. White light endoscopic images were obtained at every 2 cm level of the Barrett’s segment and analyzed by the CAD system. The per-level analysis was 90% accurate with a 91% sensitivity and 89% specificity[27], highlighting the comparable diagnostic performance of the CAD system in both real-time and “offline” settings.
Another study also utilized image databases to develop an AI algorithm using 132 high-definition white light endoscopic images from 46 lesions of histologically confirmed Barrett’s neoplasia and 119 images on non-dysplastic Barrett’s from 20 patients. The images were used for training, validation and testing of a CNN algorithm to detect Barrett’s neoplasia with a sensitivity of 93%, specificity of 78% and accuracy of 83%[28].
The utility of AI in CAD of BE neoplasia was further highlighted in another pilot study, in which Hashimoto et al[29] developed a CNN algorithm using 916 images of histology-proven early BE neoplasia containing high-grade dysplasia or T1 stage adenocarcinoma and 919 control images of BE without high-grade dysplasia. The trained CNN algorithm managed to correctly detect early neoplasia in a total of 458 test images with sensitivity of 96.4%, specificity of 94.2% and accuracy of 95.4%.
With the widespread use of image enhanced endoscopy like NBI in routine endoscopic practice for further lesion characterization, it is only natural that a deep learning algorithm would be developed to interpret NBI images and to aid in the diagnosis of BE neoplasia. A study was conducted using a trained CAD system to interpret 183 NBI zoom images and 157 NBI zoom videos with similar diagnostic accuracy of 84%-85%[30].
As most AI studies were largely image database studies and relatively small in number, a meta-analysis of the studies on the performance of AI in detection and characterization of upper GI neoplasia was performed by Arribas et al[34], which included nine studies[22-26,28,31-33] on BE neoplasia detection (total of 12909 images from 1506 patients used for training and a total of 2340 images from 445 patients used for testing). The pooled sensitivity and specificity of BE neoplasia detection was 89% and 88% respectively.
A more recent meta-analysis of twelve studies[22,24-28,30,35-38] was conducted to evaluate the diagnostic performance of AI in detecting BE neoplasia comprising 1361 patients and utilizing 532328 images for training. Pooled sensitivity was 90.3% while pooled specificity was 84.4%. Further subgroup analysis demonstrated that pooled sensitivity and specificity were also similar in six studies that used WLI as the main mode of modality[39]. An interesting observation from the meta-analysis was that there was significant heterogeneity amongst the included studies with I2 of > 50% but the area under the summary of receiver operating characteristics curve was 0.94 (95%CI: 0.92-0.96). Upon further assessment of these studies, multiple factors such as the definitions of dysplastic Barrett’s, the different types of AI algorithm and imaging modality used are very likely to contribute to the heterogeneity of the study outcome. This highlights the importance for further standardization of future study protocols with regards to the definition of BE neoplastic lesions and imaging modality used.
Apart from detection and classification of neoplasia in BE, the application of AI has shown promise in predicting submucosal invasion in Barrett’s cancer. The identification of submucosal invasion (T1b) in Barrett’s cancer is important as it has implications for the choice of treatment. Lesions with suspected submucosal invasion should be treated with endoscopic submucosal dissection (ESD) instead of the conventional endoscopic mucosal resection (EMR). ESD is a viable alternative to surgery and considered curative if the resected specimen fulfills the necessary criteria including submucosal invasion depth < 500 µm, good to moderate differentiation and no lympho-vascular invasion[13,40]. A retrospective, multicenter study was conducted to evaluate the diagnostic performance of a CNN based algorithm using a total of 230 white-light endoscopic still images to discriminate between mucosal (T1a) and submucosal (T1b) Barrett’s cancer. The trained AI algorithm was able to predict submucosal invasion and differentiate between T1a and T1b carcinoma with a sensitivity of 77%, specificity of 64% and an accuracy of 71%. The AI algorithm demonstrated comparable performance to five international Barrett’s expert endoscopists who evaluated the same set of images[41]. This study brings to light the potential for AI to support the clinical decision-making process with regards to the endoscopic vs surgical resection of precancerous lesions by predicting the submucosal invasion in Barrett’s cancer.
Interobserver agreement between pathologists can be variable with regards to interpretation of BE histology, a recognized issue particularly for low grade dysplasia (LGD) and indefinite dysplasia (IND)[42]. A study showed that concordance between pathologists progressively decreased from non-dysplastic BE (79%), high grade dysplasia (71%), LGD (42%) to IND (23%)[43]. Given that a diagnosis of dysplasia has significant implications on surveillance schedule and BE therapy, American College of Gastroenterology recommends confirmation by a second GI pathologist for dysplasia of any grade detected on biopsy[12].
Attempts have been made to use AI to complement the pathologist and improve interpretation of BE histology. It has been made possible with the rapid advancement in the field of digital pathology and the subsequent incorporation of image analysis using AI. Since the introduction of commercial digital slide scanners, it was possible to digitize glass histology slides into whole-slide images (WSI), to facilitate slide-sharing and clinical discussion, archiving of digitized slides and extraction of histopathological features using deep learning methods for image analysis[44].
A study utilized an attention-based CNN algorithm to analyze BE and esophageal adenocarcinoma using high-resolution WSI and achieved a mean classification accuracy of 83%[45]. Similarly, another study trained and validated a deep learning model using WSI from 542 patients that managed to demonstrate sensitivity and specificity > 90% at the various grades of dysplasia (non-dysplastic BE, LGD and HGD)[46]. In time, we expect more studies and advances in this field that can improve interpretation of BE histology with reproducible reliability.
GI endoscopy has seen remarkable progress throughout the last few decades with incremental step-wise progress through incorporation of breakthrough technology and medical device innovation. AI has the potential to push the innovation boundary of GI endoscopy by leveraging on existing and new information as well as vast databases to formulate algorithms, and to support the clinician in identifying and characterizing suspicious lesions. In practice, live upper endoscopic images can be sent locally or remotely to the AI system and be analyzed in real time. Based on the available data and capability, the system will be able to detect suspicious lesions for neoplasia and alert the endoscopists to those lesions either with a screen alert or location box. The endoscopist can then decide on the management of the highlighted lesion based on the characterization provided by the system.
AI can lead to earlier detection of neoplasia in BE, improvement in prognosis and reduction of mortality due to EAC. AI in BE is still in its infancy and there is no long-term data to determine the impact of AI on reduction of EAC incidence and EAC-related mortality[47]. But it is not difficult to envision that early and correct staging of neoplasia will spare the patient from the grueling experience of esophageal surgery and will enable the possibility of minimally invasive endoscopic treatments. Additionally, as described above, the invasion of depth of detected lesions could be characterized with a higher level of confidence, in particular among less experienced endoscopists, in the differentiation between mucosal and submucosal invasion. This has therapeutic consequences in the endoscopic resection approach; EMR vs ESD. The studies described were summarized in Table 1.
| Ref. | Study objective | Diagnostic modality | Study type | Real time | Dysplasia inclusion | Test images, n | Diagnostic performance |
| Pan et al[20], 2021 | BE segment identification | WLI, NBI | Retrospective | No | NA | 443 | IOU: GEJ 0.56, SCJ 0.82, GEJ+SCJ 0.66 |
| Ebigbo et al[23], 2019 | BE neoplasia detection | WLI, NBI | Retrospective | No | EAC | MICCAI: 100; Ausburg: 148 | Sen 92%, Spec 100%; WLI: Sen 97%, Spec 88%; NBI: Sen 94%, Spec 80% |
| Ebigbo et al[24], 2020 | BE neoplasia detection | WLI | Prospective | Yes | EAC | 191 | Sen 83.7%, Spec 100% |
| de Groof et al[25], 2019 | BE neoplasia detection | WLI | Retrospective | No | HGD, EAC | 60 | Sen 95%, Spec 85% |
| de Groof et al[26], 2020 | BE neoplasia detection | WLI | Retrospective | No | HGD, EAC | 297; 80; 80 | Sen 87.6%, Spec 88.6%; Sen 90%, Spec 87.5%; Sen 92.5%, Spec 82.5% |
| de Groof et al[27], 2020 | BE neoplasia detection | WLI | Prospective | Yes | HGD, EAC | 144 | Sen 91%, Spec 89% |
| Abdelrahim et al[28], 2020 | BE neoplasia detection | WLI | Retrospective | No | NA | 251 | Sen 93%, Spec 78% |
| Hashimoto et al[29], 2020 | BE neoplasia detection | WLI, NBI | Retrospective | No | HGD, EAC | 458 | Sen 96.4%, Spec 94.2% |
| Struyvenberg et al[30], 2021 | BE neoplasia detection | NBI | Retrospective | No | HGD, EAC | 183 zoom images; 157 zoom videos | Sen 88%, Spec 78%; Sen 85%, Spec 83% |
| Ebigbo et al[41], 2021 | BE cancer invasion | WLI | Retrospective | No | EAC (T1a, T2a) | 230 | Sen 77%, Spec 64% |
| Tomita et al[45], 2019 | BE neoplasia histology detection | NA | Retrospective | No | LGD, HGD, EAC | 123 WSI | Mean accuracy 83% |
AI has also the potential to reduce inter-observer variability in interpretation of not only endoscopic images but also of high-resolution, digitized histology slides to ascertain presence of dysplasia or EAC, thereby alleviating the burden of having a second pathologist for confirmation. As AI systems develop and assimilate into clinical practice, it becomes imperative that they are tested and validated in real-world settings, in diverse patient populations, with physicians of varying expertise, with different endoscope types and in different practice settings. There has been a proposal by ASGE AI task force to develop a large open-source image library as a resource to validate AI systems and to moderate data variability[48].
It is also conceivable that a trained AI system will also be able to generate an endoscopy report at the end of a session, including automated Prague C&M measurements, measurements of hiatal hernia and so on to be reviewed by the endoscopist for verification. Extending beyond that, AI has the potential, via a subtype of deep learning called natural language processing[49,50], to automatically extract and analyze keywords from free-text endoscopic and pathology reports, potentially aiding the physician to diagnose, plan and to recommend the appropriate endoscopic surveillance intervals for patients with BE.
AI has made significant progress in diagnostic endoscopy and in the identification of BE pathology using a digital workflow. AI driven systems are likely to become an important tool to detect and to characterize Barrett’s esophagus related dysplasia and early adenocarcinoma as they can present as very subtle lesions on endoscopy. Further development and validation are required before AI can be adopted mainstream in the clinical management of BE.
Provenance and peer review: Invited article; Externally peer reviewed.
Peer-review model: Single blind
Corresponding Author's Membership in Professional Societies: Gastroenterological Society of Singapore.
Specialty type: Gastroenterology and hepatology
Country/Territory of origin: Singapore
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): B
Grade C (Good): C
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Hunasanahalli Giriyappa V, India; Qin Y, China S-Editor: Liu JH L-Editor: A P-Editor: Ju JL
| 1. | Topol E. Hachette Book Group USA; 2019. Deep Medicine. |
| 2. | van der Sommen F, de Groof J, Struyvenberg M, van der Putten J, Boers T, Fockens K, Schoon EJ, Curvers W, de With P, Mori Y, Byrne M, Bergman JJGHM. Machine learning in GI endoscopy: practical guidance in how to interpret a novel field. Gut. 2020;69:2035-2045. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 60] [Cited by in RCA: 86] [Article Influence: 17.2] [Reference Citation Analysis (0)] |
| 3. | Ebigbo A, Palm C, Probst A, Mendel R, Manzeneder J, Prinz F, de Souza LA, Papa JP, Siersema P, Messmann H. A technical review of artificial intelligence as applied to gastrointestinal endoscopy: clarifying the terminology. Endosc Int Open. 2019;7:E1616-E1623. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 27] [Cited by in RCA: 28] [Article Influence: 4.7] [Reference Citation Analysis (0)] |
| 4. | Kudo SE, Ichimasa K, Villard B, Mori Y, Misawa M, Saito S, Hotta K, Saito Y, Matsuda T, Yamada K, Mitani T, Ohtsuka K, Chino A, Ide D, Imai K, Kishida Y, Nakamura K, Saiki Y, Tanaka M, Hoteya S, Yamashita S, Kinugasa Y, Fukuda M, Kudo T, Miyachi H, Ishida F, Itoh H, Oda M, Mori K. Artificial Intelligence System to Determine Risk of T1 Colorectal Cancer Metastasis to Lymph Node. Gastroenterology. 2021;160:1075-1084.e2. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 143] [Cited by in RCA: 129] [Article Influence: 32.3] [Reference Citation Analysis (0)] |
| 5. | Kominami Y, Yoshida S, Tanaka S, Sanomura Y, Hirakawa T, Raytchev B, Tamaki T, Koide T, Kaneda K, Chayama K. Computer-aided diagnosis of colorectal polyp histology by using a real-time image recognition system and narrow-band imaging magnifying colonoscopy. Gastrointest Endosc. 2016;83:643-649. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 157] [Cited by in RCA: 160] [Article Influence: 17.8] [Reference Citation Analysis (0)] |
| 6. | Mori Y, Kudo SE, Misawa M, Saito Y, Ikematsu H, Hotta K, Ohtsuka K, Urushibara F, Kataoka S, Ogawa Y, Maeda Y, Takeda K, Nakamura H, Ichimasa K, Kudo T, Hayashi T, Wakamura K, Ishida F, Inoue H, Itoh H, Oda M, Mori K. Real-Time Use of Artificial Intelligence in Identification of Diminutive Polyps During Colonoscopy: A Prospective Study. Ann Intern Med. 2018;169:357-366. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 412] [Cited by in RCA: 353] [Article Influence: 50.4] [Reference Citation Analysis (1)] |
| 7. | Hassan C, Spadaccini M, Iannone A, Maselli R, Jovani M, Chandrasekar VT, Antonelli G, Yu H, Areia M, Dinis-Ribeiro M, Bhandari P, Sharma P, Rex DK, Rösch T, Wallace M, Repici A. Performance of artificial intelligence in colonoscopy for adenoma and polyp detection: a systematic review and meta-analysis. Gastrointest Endosc. 2021;93:77-85.e6. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 361] [Cited by in RCA: 307] [Article Influence: 76.8] [Reference Citation Analysis (1)] |
| 8. | Urban G, Tripathi P, Alkayali T, Mittal M, Jalali F, Karnes W, Baldi P. Deep Learning Localizes and Identifies Polyps in Real Time With 96% Accuracy in Screening Colonoscopy. Gastroenterology. 2018;155:1069-1078.e8. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 398] [Cited by in RCA: 430] [Article Influence: 61.4] [Reference Citation Analysis (1)] |
| 9. | Kröner PT, Engels MM, Glicksberg BS, Johnson KW, Mzaik O, van Hooft JE, Wallace MB, El-Serag HB, Krittanawong C. Artificial intelligence in gastroenterology: A state-of-the-art review. World J Gastroenterol. 2021;27:6794-6824. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 28] [Cited by in RCA: 78] [Article Influence: 19.5] [Reference Citation Analysis (7)] |
| 10. | BARRETT NR. Chronic peptic ulcer of the oesophagus and 'oesophagitis'. Br J Surg. 1950;38:175-182. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 655] [Cited by in RCA: 545] [Article Influence: 26.0] [Reference Citation Analysis (0)] |
| 11. | Winters C Jr, Spurling TJ, Chobanian SJ, Curtis DJ, Esposito RL, Hacker JF 3rd, Johnson DA, Cruess DF, Cotelingam JD, Gurney MS. Barrett's esophagus. A prevalent, occult complication of gastroesophageal reflux disease. Gastroenterology. 1987;92:118-124. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 490] [Cited by in RCA: 419] [Article Influence: 11.0] [Reference Citation Analysis (0)] |
| 12. | Shaheen NJ, Falk GW, Iyer PG, Souza RF, Yadlapati RH, Sauer BG, Wani S. Diagnosis and Management of Barrett's Esophagus: An Updated ACG Guideline. Am J Gastroenterol. 2022;117:559-587. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 30] [Cited by in RCA: 242] [Article Influence: 80.7] [Reference Citation Analysis (0)] |
| 13. | Weusten B, Bisschops R, Coron E, Dinis-Ribeiro M, Dumonceau JM, Esteban JM, Hassan C, Pech O, Repici A, Bergman J, di Pietro M. Endoscopic management of Barrett's esophagus: European Society of Gastrointestinal Endoscopy (ESGE) Position Statement. Endoscopy. 2017;49:191-198. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 321] [Cited by in RCA: 371] [Article Influence: 46.4] [Reference Citation Analysis (0)] |
| 14. | Fock KM, Talley N, Goh KL, Sugano K, Katelaris P, Holtmann G, Pandolfino JE, Sharma P, Ang TL, Hongo M, Wu J, Chen M, Choi MG, Law NM, Sheu BS, Zhang J, Ho KY, Sollano J, Rani AA, Kositchaiwat C, Bhatia S. Asia-Pacific consensus on the management of gastro-oesophageal reflux disease: an update focusing on refractory reflux disease and Barrett's oesophagus. Gut. 2016;65:1402-1415. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 105] [Cited by in RCA: 150] [Article Influence: 16.7] [Reference Citation Analysis (0)] |
| 15. | Bhat S, Coleman HG, Yousef F, Johnston BT, McManus DT, Gavin AT, Murray LJ. Risk of malignant progression in Barrett's esophagus patients: results from a large population-based study. J Natl Cancer Inst. 2011;103:1049-1057. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 573] [Cited by in RCA: 516] [Article Influence: 36.9] [Reference Citation Analysis (0)] |
| 16. | Visrodia K, Singh S, Krishnamoorthi R, Ahlquist DA, Wang KK, Iyer PG, Katzka DA. Magnitude of Missed Esophageal Adenocarcinoma After Barrett's Esophagus Diagnosis: A Systematic Review and Meta-analysis. Gastroenterology. 2016;150:599-607.e7; quiz e14. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 153] [Cited by in RCA: 133] [Article Influence: 14.8] [Reference Citation Analysis (0)] |
| 17. | Jankowski M, Wani S. Diagnostic and Management Implications of Basic Science Advances in Barrett's Esophagus. Curr Treat Options Gastroenterol. 2015;13:16-29. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 8] [Cited by in RCA: 9] [Article Influence: 0.9] [Reference Citation Analysis (0)] |
| 18. | Peters Y, Al-Kaabi A, Shaheen NJ, Chak A, Blum A, Souza RF, Di Pietro M, Iyer PG, Pech O, Fitzgerald RC, Siersema PD. Barrett oesophagus. Nat Rev Dis Primers. 2019;5:35. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 64] [Cited by in RCA: 90] [Article Influence: 15.0] [Reference Citation Analysis (0)] |
| 19. | Sharma P, Savides TJ, Canto MI, Corley DA, Falk GW, Goldblum JR, Wang KK, Wallace MB, Wolfsen HC; ASGE Technology and Standards of Practice Committee. The American Society for Gastrointestinal Endoscopy PIVI (Preservation and Incorporation of Valuable Endoscopic Innovations) on imaging in Barrett's Esophagus. Gastrointest Endosc. 2012;76:252-254. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 113] [Cited by in RCA: 113] [Article Influence: 8.7] [Reference Citation Analysis (0)] |
| 20. | Pan W, Li X, Wang W, Zhou L, Wu J, Ren T, Liu C, Lv M, Su S, Tang Y. Identification of Barrett's esophagus in endoscopic images using deep learning. BMC Gastroenterol. 2021;21:479. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 13] [Cited by in RCA: 14] [Article Influence: 3.5] [Reference Citation Analysis (3)] |
| 21. | Sharma P, Dent J, Armstrong D, Bergman JJ, Gossner L, Hoshihara Y, Jankowski JA, Junghard O, Lundell L, Tytgat GN, Vieth M. The development and validation of an endoscopic grading system for Barrett's esophagus: the Prague C & M criteria. Gastroenterology. 2006;131:1392-1399. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 731] [Cited by in RCA: 715] [Article Influence: 37.6] [Reference Citation Analysis (0)] |
| 22. | Schölvinck DW, van der Meulen K, Bergman JJGHM, Weusten BLAM. Detection of lesions in dysplastic Barrett's esophagus by community and expert endoscopists. Endoscopy. 2017;49:113-120. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 13] [Cited by in RCA: 23] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 23. | Ebigbo A, Mendel R, Probst A, Manzeneder J, Souza LA Jr, Papa JP, Palm C, Messmann H. Computer-aided diagnosis using deep learning in the evaluation of early oesophageal adenocarcinoma. Gut. 2019;68:1143-1145. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 94] [Cited by in RCA: 102] [Article Influence: 17.0] [Reference Citation Analysis (0)] |
| 24. | Ebigbo A, Mendel R, Probst A, Manzeneder J, Prinz F, de Souza LA Jr, Papa J, Palm C, Messmann H. Real-time use of artificial intelligence in the evaluation of cancer in Barrett's oesophagus. Gut. 2020;69:615-616. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 84] [Cited by in RCA: 123] [Article Influence: 24.6] [Reference Citation Analysis (0)] |
| 25. | de Groof J, van der Sommen F, van der Putten J, Struyvenberg MR, Zinger S, Curvers WL, Pech O, Meining A, Neuhaus H, Bisschops R, Schoon EJ, de With PH, Bergman JJ. The Argos project: The development of a computer-aided detection system to improve detection of Barrett's neoplasia on white light endoscopy. United European Gastroenterol J. 2019;7:538-547. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 83] [Cited by in RCA: 76] [Article Influence: 12.7] [Reference Citation Analysis (0)] |
| 26. | de Groof AJ, Struyvenberg MR, van der Putten J, van der Sommen F, Fockens KN, Curvers WL, Zinger S, Pouw RE, Coron E, Baldaque-Silva F, Pech O, Weusten B, Meining A, Neuhaus H, Bisschops R, Dent J, Schoon EJ, de With PH, Bergman JJ. Deep-Learning System Detects Neoplasia in Patients With Barrett's Esophagus With Higher Accuracy Than Endoscopists in a Multistep Training and Validation Study With Benchmarking. Gastroenterology. 2020;158:915-929.e4. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 172] [Cited by in RCA: 221] [Article Influence: 44.2] [Reference Citation Analysis (0)] |
| 27. | de Groof AJ, Struyvenberg MR, Fockens KN, van der Putten J, van der Sommen F, Boers TG, Zinger S, Bisschops R, de With PH, Pouw RE, Curvers WL, Schoon EJ, Bergman JJGHM. Deep learning algorithm detection of Barrett's neoplasia with high accuracy during live endoscopic procedures: a pilot study (with video). Gastrointest Endosc. 2020;91:1242-1250. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 66] [Cited by in RCA: 89] [Article Influence: 17.8] [Reference Citation Analysis (0)] |
| 28. | Abdelrahim M, Saikou M, Masaike Y, Ohtsuka S, Maeda N, Hossain E, Arndtz S, Bhandari P. Artificial intelligence using convolutional neural networks for detection of early Barrett's neoplasia. United European Gastroenterol J. 2020;8 (Suppl):114. [DOI] [Full Text] |
| 29. | Hashimoto R, Requa J, Dao T, Ninh A, Tran E, Mai D, Lugo M, El-Hage Chehade N, Chang KJ, Karnes WE, Samarasena JB. Artificial intelligence using convolutional neural networks for real-time detection of early esophageal neoplasia in Barrett's esophagus (with video). Gastrointest Endosc. 2020;91:1264-1271.e1. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 172] [Cited by in RCA: 143] [Article Influence: 28.6] [Reference Citation Analysis (0)] |
| 30. | Struyvenberg MR, de Groof AJ, van der Putten J, van der Sommen F, Baldaque-Silva F, Omae M, Pouw R, Bisschops R, Vieth M, Schoon EJ, Curvers WL, de With PH, Bergman JJ. A computer-assisted algorithm for narrow-band imaging-based tissue characterization in Barrett's esophagus. Gastrointest Endosc. 2021;93:89-98. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 27] [Cited by in RCA: 53] [Article Influence: 13.3] [Reference Citation Analysis (0)] |
| 31. | Ghatwary N, Zolgharni M, Ye X. Early esophageal adenocarcinoma detection using deep learning methods. Int J Comput Assist Radiol Surg. 2019;14:611-621. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 38] [Cited by in RCA: 65] [Article Influence: 10.8] [Reference Citation Analysis (0)] |
| 32. | van der Sommen F, Zinger S, Curvers WL, Bisschops R, Pech O, Weusten BL, Bergman JJ, de With PH, Schoon EJ. Computer-aided detection of early neoplastic lesions in Barrett's esophagus. Endoscopy. 2016;48:617-624. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 111] [Cited by in RCA: 126] [Article Influence: 14.0] [Reference Citation Analysis (2)] |
| 33. | van der Sommen F, Zinger S, Schoon EJ, de With PHN. Supportive automatic annotation of early esophageal cancer using local gabor and color features. Neurocomputing. 2014;144:92-106. [RCA] [DOI] [Full Text] [Cited by in Crossref: 40] [Cited by in RCA: 33] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 34. | Arribas J, Antonelli G, Frazzoni L, Fuccio L, Ebigbo A, van der Sommen F, Ghatwary N, Palm C, Coimbra M, Renna F, Bergman JJGHM, Sharma P, Messmann H, Hassan C, Dinis-Ribeiro MJ. Standalone performance of artificial intelligence for upper GI neoplasia: a meta-analysis. Gut. 2020;. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 35] [Cited by in RCA: 45] [Article Influence: 9.0] [Reference Citation Analysis (0)] |
| 35. | Samarasena JB, Konda VJ, Trindade AJ, Cavaliere KR, Chang K, Hashimoto R, et al. Detection of early esophageal neoplasia in barrett’s esophagus using real time artificial intelligence: a multicenter external video validation study. Gastrointest Endosc. 2021;93 (6 Supplement):AB195. [DOI] [Full Text] |
| 36. | Hussein M, Lines D, Puyal JGB, Bowman N, Sehgal V, Toth D, Everson M, Ahmad O, Kader R, Esteban JM, Bisschops R, Banks M, Haefner M, Mountney P, Stoyanov D, Lovat L, Haidry R. Computer aided diagnosis for the characterisation of dysplasia in Barrett’s oesophagus with magnification endoscopy. Endoscopy. 2021;53 (Suppl 1):S10. [DOI] [Full Text] |
| 37. | Hussein M, Puyal JGB, Brandao P, Toth D, Sehgal V, Everson M, Lipman G, Ahmad O, Kader R, Esteban JM, Bisschops R, Banks M, Mountney P, Stoyanov D, Lovat L, Haidry R. Deep neural network for the detection of early neoplasia in Barrett’s oesophagus. Gut. 2021;70 (Suppl 1):A17. [DOI] [Full Text] |
| 38. | Hussein M, Puyal JGB, Lines D, Sehgal V, Toth D, Everson M, Lipman G, Ahmad O, Kader R, Esteban JM, Bisschops R, Banks M, Mountney P, Stoyanov D, Lovat L, Haidry R. Deep neural network for the localisation of early neoplasia in Barrett’s oesophagus with targeted biopsies. Endoscopy. 2021;53 (Suppl 1):S8-S9. [DOI] [Full Text] |
| 39. | Tan JL, Chinnaratha MA, Woodman R, Martin R, Chen HT, Carneiro G, Singh R. Diagnostic Accuracy of Artificial Intelligence (AI) to Detect Early Neoplasia in Barrett's Esophagus: A Non-comparative Systematic Review and Meta-Analysis. Front Med (Lausanne). 2022;9:890720. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 14] [Cited by in RCA: 16] [Article Influence: 5.3] [Reference Citation Analysis (0)] |
| 40. | Sharma P, Shaheen NJ, Katzka D, Bergman JJGHM. AGA Clinical Practice Update on Endoscopic Treatment of Barrett's Esophagus With Dysplasia and/or Early Cancer: Expert Review. Gastroenterology. 2020;158:760-769. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 106] [Cited by in RCA: 132] [Article Influence: 26.4] [Reference Citation Analysis (0)] |
| 41. | Ebigbo A, Mendel R, Rückert T, Schuster L, Probst A, Manzeneder J, Prinz F, Mende M, Steinbrück I, Faiss S, Rauber D, de Souza LA Jr, Papa JP, Deprez PH, Oyama T, Takahashi A, Seewald S, Sharma P, Byrne MF, Palm C, Messmann H. Endoscopic prediction of submucosal invasion in Barrett's cancer with the use of artificial intelligence: a pilot study. Endoscopy. 2021;53:878-883. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 20] [Cited by in RCA: 43] [Article Influence: 10.8] [Reference Citation Analysis (0)] |
| 42. | Montgomery E, Bronner MP, Goldblum JR, Greenson JK, Haber MM, Hart J, Lamps LW, Lauwers GY, Lazenby AJ, Lewin DN, Robert ME, Toledano AY, Shyr Y, Washington K. Reproducibility of the diagnosis of dysplasia in Barrett esophagus: a reaffirmation. Hum Pathol. 2001;32:368-378. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 606] [Cited by in RCA: 566] [Article Influence: 23.6] [Reference Citation Analysis (0)] |
| 43. | van der Wel MJ, Coleman HG, Bergman JJGHM, Jansen M, Meijer SL; BOLERO working group. Histopathologist features predictive of diagnostic concordance at expert level among a large international sample of pathologists diagnosing Barrett's dysplasia using digital pathology. Gut. 2020;69:811-822. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 42] [Cited by in RCA: 38] [Article Influence: 7.6] [Reference Citation Analysis (0)] |
| 44. | Aeffner F, Zarella MD, Buchbinder N, Bui MM, Goodman MR, Hartman DJ, Lujan GM, Molani MA, Parwani AV, Lillard K, Turner OC, Vemuri VNP, Yuil-Valdes AG, Bowman D. Introduction to Digital Image Analysis in Whole-slide Imaging: A White Paper from the Digital Pathology Association. J Pathol Inform. 2019;10:9. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 153] [Cited by in RCA: 209] [Article Influence: 34.8] [Reference Citation Analysis (0)] |
| 45. | Tomita N, Abdollahi B, Wei J, Ren B, Suriawinata A, Hassanpour S. Attention-Based Deep Neural Networks for Detection of Cancerous and Precancerous Esophagus Tissue on Histopathological Slides. JAMA Netw Open. 2019;2:e1914645. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 133] [Cited by in RCA: 100] [Article Influence: 16.7] [Reference Citation Analysis (0)] |
| 46. | Faghani S, Codipilly DC, David Vogelsang, Moassefi M, Rouzrokh P, Khosravi B, Agarwal S, Dhaliwal L, Katzka DA, Hagen C, Lewis J, Leggett CL, Erickson BJ, Iyer PG. Development of a deep learning model for the histologic diagnosis of dysplasia in Barrett's esophagus. Gastrointest Endosc. 2022;96:918-925.e3. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2] [Cited by in RCA: 29] [Article Influence: 9.7] [Reference Citation Analysis (0)] |
| 47. | Then EO, Lopez M, Saleem S, Gayam V, Sunkara T, Culliford A, Gaduputi V. Esophageal Cancer: An Updated Surveillance Epidemiology and End Results Database Analysis. World J Oncol. 2020;11:55-64. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 76] [Cited by in RCA: 195] [Article Influence: 39.0] [Reference Citation Analysis (0)] |
| 48. | Berzin TM, Parasa S, Wallace MB, Gross SA, Repici A, Sharma P. Position statement on priorities for artificial intelligence in GI endoscopy: a report by the ASGE Task Force. Gastrointest Endosc. 2020;92:951-959. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 40] [Cited by in RCA: 63] [Article Influence: 12.6] [Reference Citation Analysis (0)] |
| 49. | Imler TD, Morea J, Kahi C, Sherer EA, Cardwell J, Johnson CS, Xu H, Ahnen D, Antaki F, Ashley C, Baffy G, Cho I, Dominitz J, Hou J, Korsten M, Nagar A, Promrat K, Robertson D, Saini S, Shergill A, Smalley W, Imperiale TF. Multi-center colonoscopy quality measurement utilizing natural language processing. Am J Gastroenterol. 2015;110:543-552. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 53] [Cited by in RCA: 62] [Article Influence: 6.2] [Reference Citation Analysis (0)] |
| 50. | Raju GS, Lum PJ, Slack RS, Thirumurthi S, Lynch PM, Miller E, Weston BR, Davila ML, Bhutani MS, Shafi MA, Bresalier RS, Dekovich AA, Lee JH, Guha S, Pande M, Blechacz B, Rashid A, Routbort M, Shuttlesworth G, Mishra L, Stroehlein JR, Ross WA. Natural language processing as an alternative to manual reporting of colonoscopy quality metrics. Gastrointest Endosc. 2015;82:512-519. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 69] [Cited by in RCA: 69] [Article Influence: 6.9] [Reference Citation Analysis (0)] |