Published online Apr 28, 2022. doi: 10.35712/aig.v3.i2.54
Peer-review started: February 1, 2022
First decision: February 18, 2022
Revised: March 28, 2022
Accepted: April 19, 2022
Article in press: April 19, 2022
Published online: April 28, 2022
Processing time: 87 Days and 8.9 Hours
Pancreatic diseases have a substantial burden on society which is predicted to increase further over the next decades. Endoscopic ultrasonography (EUS) remains the best available diagnostic method to assess the pancreas, however, there remains room for improvement. Artificial intelligence (AI) approaches have been adopted to assess pancreatic diseases for over a decade, but this metho
Core Tip: Several reviews in the literature have discussed the use of artificial intelligence in pancreatic disease. However, this is the first review that focuses on the application of artificial intelligence (AI) specifically to endoscopic ultrasonography (EUS) of the pancreas, including pancreatic cystic lesions, pancreatic cancer, chronic pancreatitis, and autoimmune pancreatitis, where it appears to enhance EUS diagnosis. AI may also offer real-time assistance during procedures to direct biopsy towards the highest yield areas as well augment EUS training.
- Citation: Simsek C, Lee LS. Machine learning in endoscopic ultrasonography and the pancreas: The new frontier? Artif Intell Gastroenterol 2022; 3(2): 54-65
- URL: https://www.wjgnet.com/2644-3236/full/v3/i2/54.htm
- DOI: https://dx.doi.org/10.35712/aig.v3.i2.54
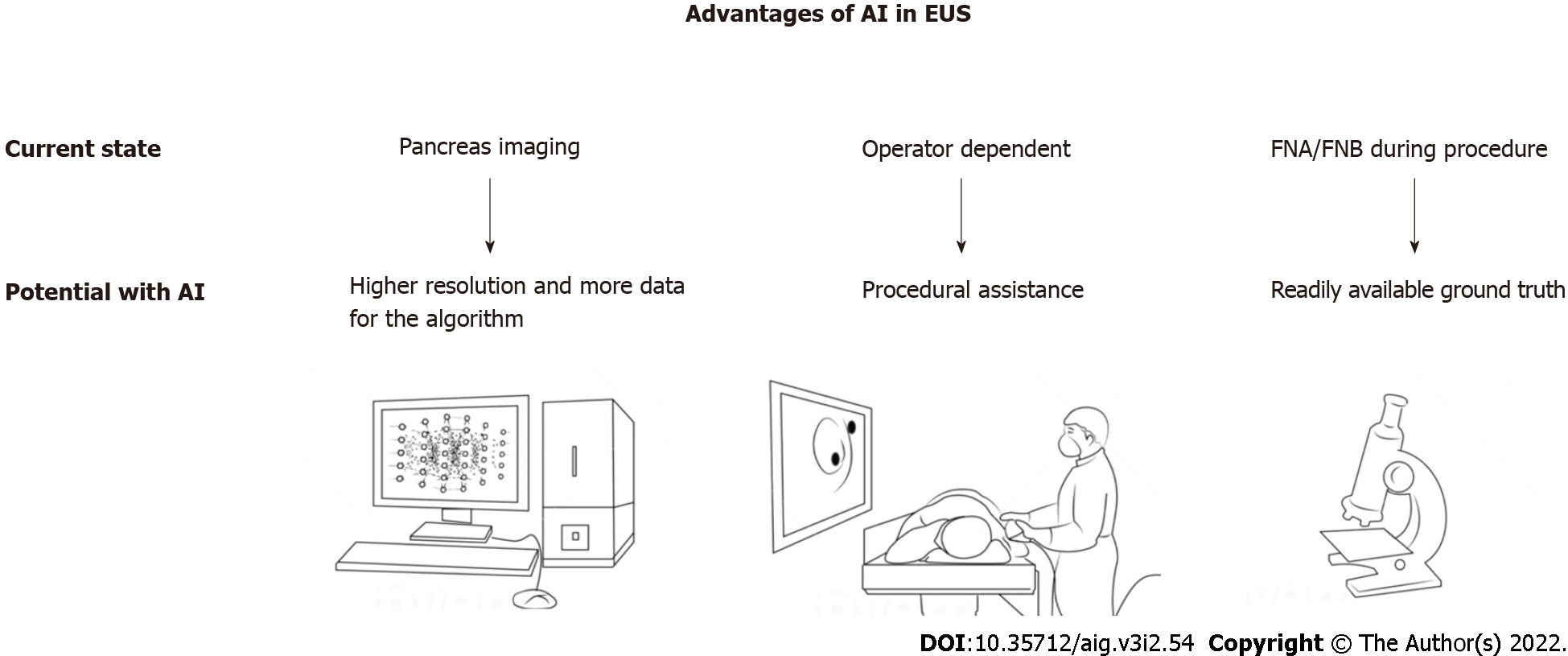
Pancreatic diseases create a substantial burden on society. Pancreatic cancer is the third leading cause of cancer-related death in the United States, and its death count is expected to rise to 460000 by 2040, becoming the second leading cause of cancer related death in 2040[1-3]. Chronic pancreatitis is another cause of the burden with significant morbidity from chronic pain, diabetes mellitus, and even pancreatic cancer[4,5]. Additionally, pancreatic cystic lesions are reported to be detected up to 20% of abdominal imaging studies[6]. Endoscopic ultrasonography (EUS) has surpassed magnetic resonance imaging (MRI), computed tomography (CT) and transabdominal ultrasonography in the diagnosis of pancreatic diseases; however, there remains room for improvement in the diagnostic sensitivity of EUS[7]. In this regard, utilization of artificial intelligence (AI) with EUS has emerged as a promising strategy (Figure 1). Although EUS has better performance than the alternative radiology imaging methods, it is also more operator dependent. The endosonographer’s experience and skills can significantly alter the diagnostic or therapeutic outcomes of an EUS procedure. AI may decrease this operator dependency as it can assist the endosonographer in several tasks that include, but are not limited, to identifying anatomical landmarks, detecting lesions, interpreting sonographic findings, and guiding obtaining optimal tissue biopsy with higher diagnostic yield. Because AI algorithms use higher resolution EUS imaging data, they might distinguish patterns and identify details from the images which may not be recognizable with human detection alone currently. Finally, AI research with EUS is more convenient because imaging data used to train the AI models often have readily available definitive histologic diagnoses.
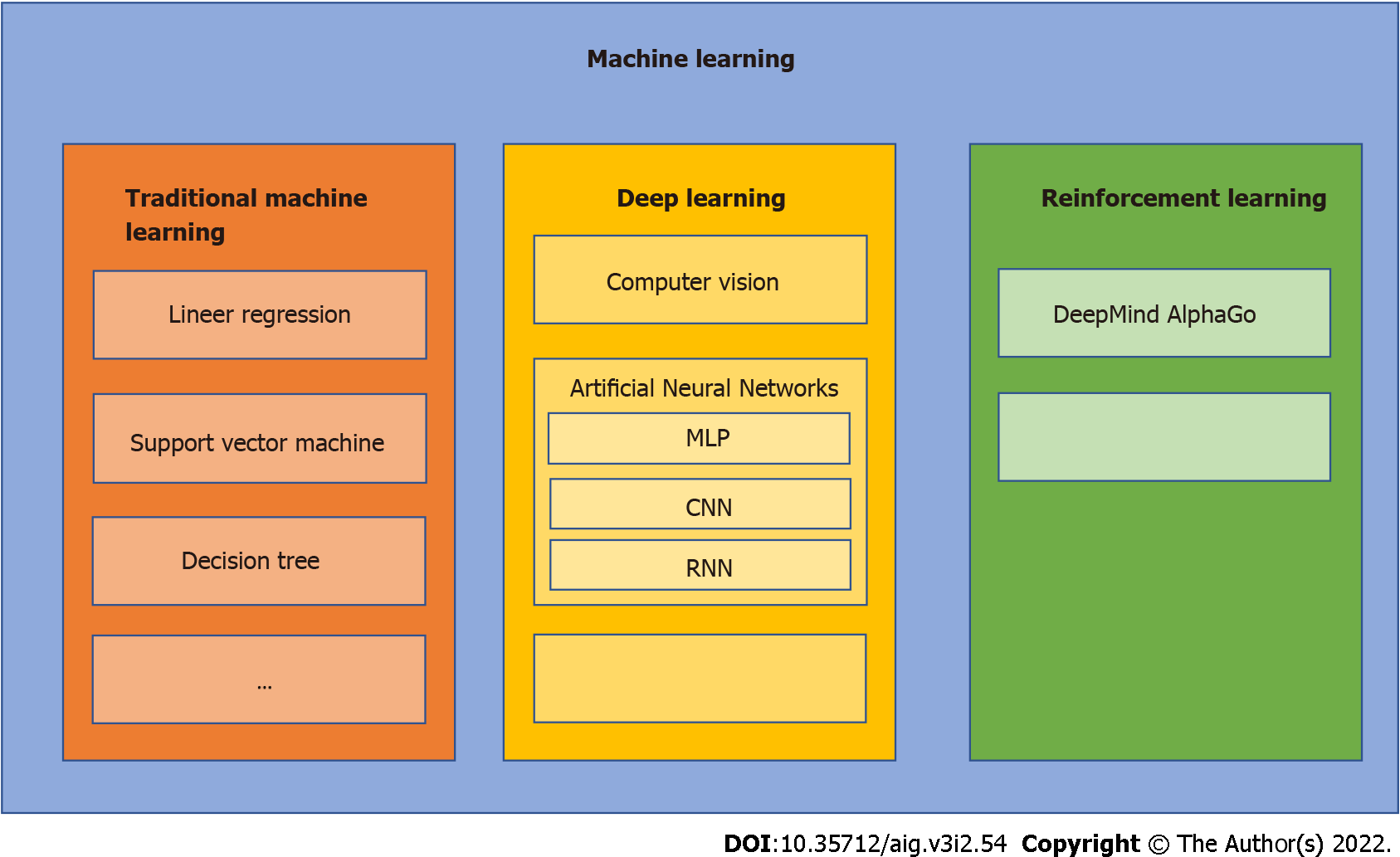
AI is an umbrella term for the computerized performance of complex tasks that normally require human intelligence, such as visual perception, learning, pattern recognition and decision-making[8] (Figure 2). Current medical applications using AI have made significant progress due to advancements in computer technology, data science, and the digitalization of health care. From the development of more complex machine learning algorithms, AI has progressed rapidly to its current front-line role in image-based diagnosis, speech recognition, robotic surgery, drug discovery and patient monitoring[9]. However, the progress of AI in medicine has just begun and has yet to realize its full potential.
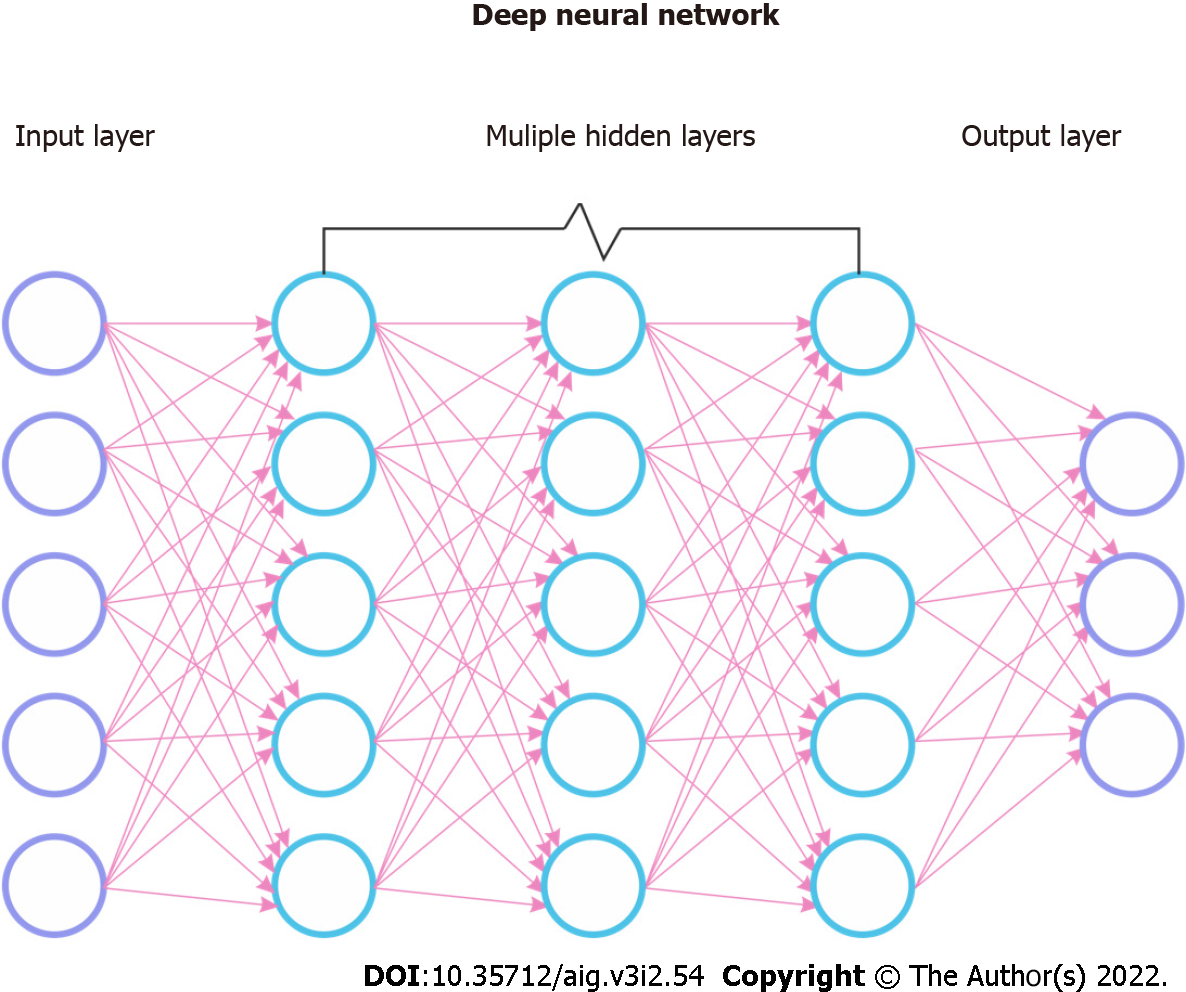
Machine learning (ML) is a field of artificial intelligence in which algorithms learn and improve from interactions with the data, obviating the need for explicit programming. Deep learning (DL) is a subfield of ML inspired by the organization and working principle of the human brain and is made up of individual neurons which form multilayered artificial neural networks (ANN). These networks are comprised of input and output layers each of which can execute simple tasks and sequentially interact with one another to produce a conclusion. Among ANNs, Multi-Layered Perceptron are earlier models that are simpler with fewer layers and can only use linear functions[10]. Convolutional neural networks (CNN) include more layers that can also operate in a non-linear fashion allowing more complex tasks such as image classification and have been the most popular DL algorithm. CNNs were inspired by the human visual cortex and designed to process grid pattern data such as images. They have serial neural network layers to recognize and extract features from the input data, learn the patterns of features, and perform hierarchical organization through the layers to search for the intended output (Figure 3)[11]. Most commonly used CNN algorithms are AlexNet, ResNet, U-Net, which all work using the same principle, and the technical details are beyond the scope of this review[12]. Another type of ANN is recurrent neural network (RNN), which also contains a multi-layered structure. In addition, each neuron in this network has its own internal memory, which taken altogether constitutes a collective memory of the network. This neural network can remember previous input data and use it to process subsequent inputs. Therefore, these algorithms are beneficial in processing sequential data such as before and after an intervention or time series data. An example of RNN is the long short-term memory model[13].
Machine learning can perform two different types of tasks: Supervised and unsupervised. Supervised algorithms aim to reach a previously defined targeted outcome and are used for classification and prediction tasks. Labeled input data is presented to the algorithm and the model is trained with direct feedbacks to predict corresponding outputs. The spectrum of supervised approaches includes statistical methods such as logistic regression, linear regression, decision trees as well as support vector machines and random forest. Unsupervised algorithms do not have a predefined target and are used for clustering and dimensionality reduction. Unsupervised models are currently used for disease subtype and biomarker discovery studies[14,15]. Supervised learning has been more commonly used in EUS research; therefore, several important nuances will be summarized to better understand the presented literature. To train supervised learning algorithms, the dataset should be pre-annotated for the targeted outcome, which may be a diagnosis, class, or feature. The algorithm aims to optimize its feature detection ability to match the presented inputs to this annotated targeted output, which is defined as “ground truth”. This optimization, or training, task requires large datasets, therefore, learning algorithms are data hungry. However, such datasets are not commonly available, which necessitates data augmentation techniques be used to expand the dataset by inserting slightly changed copies of previously collected data or by creating new synthetic data with computerized approaches.
During training of the algorithm, available data is split into three sets: Training, validation, and test. Training and validation sets are used to develop and fine-tune the model, whereas the test set is used to assess the performance of the final model product. Of note, this validation is different from its conventional use in medicine and seeks to optimize parameters of the model during the training phase. Two of the most common validation approaches in medical AI research are cross-validation and hold-out validation. Cross-validation occurs when the dataset is randomly resampled and split repetitively – the number of repetitions is designated with k- into training and test sets. Each training and test set is then used to develop a new model, and k repetitions yield new k models. In contrast, hold-out validation is a constant single split of a training set and an independent test set to develop one final model which is simpler to perform but brings an increased risk of sampling error. Another important concept in machine learning is overfitting, which is defined as a falsely superior performance of the model caused by learning irrelevant features of the dataset or ‘noise’ as well as the intended signals. Therefore, a separate test set is important to accurately assess the model’s performance.
There are several nuances in the performance assessment of a machine learning model. Sensitivity (recall), specificity, positive predictive value (precision), negative predictive value and area under the rule operator characteristic (AUC) curve are commonly used for assessing the performance of classification. The area under the precision-recall curve (AUPRC) is used instead of the AUC when observations are not equally distributed for two groups. The Dice coefficient (F1 score) is the harmonic mean of precision and recall. It is commonly used to assess the labeling performance of an image recognition model. In a model where a ground truth area X is labeled by an image recognition model as area Y, Dice coefficient equals the overlap of X and Y areas divided by the total of X and Y areas, multiplied by two. Another similar metric is the Jaccard index, or intersection over union (IoU), defined as the ratio of overlap and union of two areas: the algorithm labeled area and the ground truth area. Both Jaccard index and Dice coefficient’s values range from 0 to 1 signifying 0% to 100% accuracy of labeling with 1 being the highest level of accuracy for both.
While AI has been utilized to investigate numerous gastrointestinal diseases, the study of pancreatic diseases using AI and EUS is limited[5]. In this review, we provide a targeted overview of AI with a summary of the current literature on the use of AI in EUS for the diagnosis of pancreatic diseases.
A nonsystematic search of the current literature was performed for 2015 and 2021 in the MEDLINE, PubMed, Google Scholar, Scopus, Web of Science and Embase databases with the following terms: Machine learning, deep learning, artificial intelligence, EUS, endosonography, endoscopic ultrasound, pancreas, pancreatic disease, pancreatitis, and pancreatic cancer. Review articles were manually screened for any additional studies of interest. Congress abstracts, reviews, correspondences, editorials, and book chapters were excluded. Two authors reviewed all the studies after the initial search and confirmed the appropriateness of each study for inclusion. Our literature search yielded fifteen studies with modern machine learning algorithms (Table 1). Of note, five of the fifteen studies were published in 2021 with only two prospective clinical trials from the same group.
| Field | Ref. | Study population used for training (n) | Task | Machine learning method | Performance (in test population if available) |
| Pancreatic Cysts | Kuwahara et al[23], 2019 | Benign IPMN (27); Malignant IPMN (23) | Differentiate benign from malignant IPMN | Convolutional neural network | AUC = 0.98 |
| Springer et al[20], 2019 | Mucinous cystic neoplasms (153); Serous Cystic Neoplasms (148); IPMN (447); Malignant cysts (114) | Guide clinical management by classify into three risk groups: No risk of malignancyLow risk of progression. High-risk of progression or malignant | Not available | First group: 100% specificity, 46% sensitivity. Second group: 54% specificity, 91% sensitivity. Third group: 30% specificity, 99% sensitivity. | |
| Kurita et al[22], 2019 | Mucinous cystic neoplasms (23); Serous Cystic Neoplasms (15); IPMN (30); Other cyst types (17) | Differentiate benign from malignant cyst | Multi-layered perceptron | AUC = 0.96, sensitivity: 95%, specificity: 91.9% | |
| Nguon et al[21], 2021 | Mucinous cystic neoplasms (59); Serous Cystic Neoplasms (49) | Differentiate mucinous cystic neoplasm and serous cystadenoma | Convolutional neural network | AUC = 0.88 | |
| Pancreatic Cancer | Saftouiu et al[27], 2008 | PDAC (32); Normal pancreas (22); Chronic pancreatitis (11); Pancreatic neuroendocrine tumor (3) | Differentiate benign from malignant masses | Multi-layered perceptron | AUC = 0.96 |
| Saftoiu et al[28], 2012 | PDAC (211); Chronic pancreatitis (47) | Differentiate cancer from benign masses | Multi-layered perceptron | AUC = 0.94 | |
| Ozkan et al[30], 2016 | PDAC (202); Normal pancreas (130) | Differentiate cancer from normal pancreas | Multi-layered perceptron | Accuracy: 87.5%, sensitivity: 83.3%, and specificity: 93.3% | |
| Udristou et al[31], 2021 | PDAC (30); Chronic pancreatitis (20); Pancreatic neuroendocrine tumor (15) | Diagnose focal pancreatic mass | Convolutional neural network and long short-term memory | Mean AUC = 0.98 (Includes PDAC, CP and PNET) | |
| Tonozuka et al[32], 2021 | PDAC (76); Chronic pancreatitis (34); Control (29) | Differentiate pancreatic cancer from chronic pancreatitis and normal pancreas | Convolutional neural network and pseudo-colored heatmap | AUC = 0.94 | |
| Autoimmune pancreatitis | Zhu et al[34], 2015 | AIP (81); Chronic pancreatitis (100) | Differentiate AIP from chronic pancreatitis | Support Vector Machine | Accuracy: 89.3%, sensitivity: 84.1%, and specificity: 92.5% |
| Marya et al[36], 2021 | AIP (146); PDAC (292); Chronic pancreatitis (72); Normal pancreas (73) | Differentiate of AIP from PDAC | Convolutional neural network and pseudo-colored heatmap | AUC for AIP from all other = 0.92 | |
| Procedural assistance | Iwasa et al[38], 2021 | Pancreatic mass (100) | Segmentation of pancreatic masses | Convolutional neural network | Intersection over unit = 0.77 |
| Zhang et al[40], 2020 | EUS videos (339) | Recognition of stations, and segmentation of anatomical landmarks | Convolutional neural network | Accuracy for classification of stations (average) = 0.824, Dice coefficient for segmentation of pancreas (average) = 0.715 |
The application of AI was divided into sonographic image recognition, procedural assistance, and training. Endosonographic images contain cues that may not be recognizable by human visual perception. In this context, deep learning algorithms are promising tools to recognize the patterns from these cues. As such, several important diagnostic challenges in pancreatic diseases with EUS have been addressed, including the classification and risk stratification of pancreatic cysts and the diagnosis of autoimmune pancreatitis (AIP) and pancreatic ductal adenocarcinoma (PDAC).
Pancreatic cysts are increasingly detected in patients undergoing abdominal cross-sectional imaging with up to 20% detection rate on MRI[6,16,17]. Since pancreatic cysts carry a risk of malignancy, this risk should be stratified to guide clinical management. However, in most cases, imaging results are not sufficient for the classification of pancreatic cysts, especially for small lesions[18]. Additionally, assessing the risk of malignant progression remains challenging with current imaging modalities, clinical criteria, cyst fluid analysis or their combinations[18,19]. In this context, ML may help classify pancreatic cysts.
Several studies have investigated the utility of EUS ML models in pancreatic cysts, focusing on malignancy risk assessment and classification. Two studies by Kuwahara et al[23] and Nguon et al[21] used still images of EUS examinations with data augmentation, while Springer et al[20] and Kurita et al[22] applied multimodality approaches that included cyst fluid analyses and clinical data[20-23].
The 2019 study by Kuwahara et al[23] assessed the accuracy of ML to predict malignant intraductal papillary mucinous neoplasms (IPMN). This single-center study included 50 IPMN patients who underwent surgical resection. Therefore, all diagnoses were made from histopathological examination of surgical specimens. A total of 3970 still images were collected from 50 EUS examinations, and the CNN was fed over 500000 images using data augmentation. Ten-fold cross-validation was performed for training. For each case, the output of the CNN model was given as a predictive continuous value ranging from 0 to 1 for benign and malignant assigned probabilities, respectively. When the final model’s predictive values were compared with the surgical diagnoses, predictive values for the benign cases were significantly lower than values for the malignant cases (0.104 vs 0.808, respectively). The optimal cutoff for the predictive value was determined using the Youden Index. This cutoff value (0.49) generated an AUC of 98% for the diagnosis of malignancy. The accuracy of the final model (94%) was significantly higher than that of human preoperative diagnosis which incorporated contrast enhanced EUS examination findings of mural nodule size, diameter main pancreatic duct, cyst size, and growth rate (56%). Multivariate analysis showed that the AI predictive value was the only significant factor for diagnosing malignant IPMN. ML outperformed currently used criteria, including serum CA 19-9, presence of mural nodule, and type of IPMN. This study demonstrated the promise of EUS ML algorithms in predicting malignant IPMNs. However, further prospective studies with larger sample sizes that do not rely solely on internal validation are necessary.
Kurita et al[22] used a multimodality approach to differentiate benign from malignant cysts. This single center study used 85 patients with pancreatic cystic lesions and final diagnosis from surgical pathology or combination of cyst fluid analysis, radiology imaging, and clinical follow-up. The input data consisted of sex, cyst fluid protein markers, cytologic diagnosis and EUS imaging features of the cyst. A Multi-layered Perceptron was used as the ML model. The final model achieved 95.7% sensitivity, 91.9% specificity, and 0.97 AUC for classifying lesions as benign or malignant, which was the primary endpoint. The model showed 92.9% accuracy which was significantly higher than carcinoembryonic antigen (CEA) (71.8%) and cytology (85.9%) alone[22]. An external data set was not available to test the algorithm. In addition, it is unclear why the algorithm did not mention inclusion of known high-risk features including enhancing nodule, solid mass, and dilated main pancreatic duct.
Another large multicenter study used a ML based approach called CompCyst to guide the management of pancreatic cystic lesions and relied heavily on molecular analysis of cyst fluid in addition to clinical and radiologic imaging features. The study population consisted of 862 patients recruited from 16 centers who underwent surgical resection with final diagnosis based on histologic analysis. DNA from cyst fluid were extracted and evaluated for four types of molecular abnormalities including mutations, loss of heterozygosity, aneuploidy as well as protein markers CEA and vascular endothelial growth factor-A (VEGF-A). Then the CompCyst test was used to classify cysts into one of the three following groups using a combination of molecular and imaging features. The first group was defined as cysts without any malignant potential which would not need surveillance. VHL and GNAS were used in this step and achieved 100% specificity and 46% sensitivity. The second group was cysts with small risk of malignant progression which would require surveillance. Multiple gene mutations and solid component in imaging was used in this step yielding 91% sensitivity and 54% specificity in the test cohort. The third group included cysts with high likelihood of malignant progression or malignancy which should be resected. VEGF-A protein expression was used in this step with 99% sensitivity and 30% specificity. The system was compared to standard of care and demonstrated significantly higher accuracy (69% vs 56%, respectively)[20]. This study used a separate validation set and a comprehensive model that incorporated clinical and radiologic findings, however, the wide-ranging molecular analysis is not readily available for routine clinical use.
A recent 2021 study focused on differentiating mucinous cystic neoplasms from serous cystadenomas using a total of 109 cases from two centers[21]. Final diagnoses were determined by endosonographers with over 5 years of experience. Additional cyst fluid or histopathologic examinations were available for only 44% of patients. A total of 221 still images were obtained followed by data augmentation, but the final number of input images was not provided in the study. The ResNet framework was used as the CNN model. Three hold-out validations were performed with 10 cases for testing, and the remaining cases used for training. The result of the study showed 82.75% accuracy and 0.88 AUC to correctly classify mucinous cystic neoplasms and serous cystadenomas from the still EUS images. A pseudo-colored decision map [gradient weighted class activation mapping (GradCAM)] was used to visualize the decision-making process. Presentation of the pseudo-colored decision map is an important asset because it highlights and color codes (red for higher impact and blue for lower impact) the areas in the image which affected the algorithm’s final decision; therefore, this allows clinicians to better comprehend the decision-making process by the model. However, this study has several limitations. First, the most commonly encountered cyst, IPMN, was not included in the dataset that decreases the generalizability of the model. Second, ground truth was endosonographers’ expert opinion and only 44% of patients had cyst fluid or histologic confirmation of diagnosis. Despite various limitations, the studies presented demonstrate the feasibility of image recognition ML models to perform classification tasks for pancreatic cysts and guide clinical management.
PDAC is currently the fourth leading cause of cancer-related mortality in Western countries and is predicted to become the second by 2030[24]. Most cases are diagnosed at later stages with 5-year survival rates less than 10%. A promising strategy is earlier diagnosis to combat this disease[25]. For this, EUS with FNA has superseded the cross-sectional imaging modalities such as CT and MRI, especially in the earlier diagnosis of PDAC[26]. However, EUS is operator dependent, and EUS diagnosis of PDAC is more challenging in patients with baseline abnormal pancreatic imaging (e.g., chronic pancreatitis) who also carry a higher risk. Within this context, ML has been used to improve the diagnostic performance of EUS for pancreatic masses. Four studies used histologically confirmed PDAC cases with normal pancreas as control. Additional control groups were used in different studies to reflect clinical scenarios including chronic pancreatitis and neuroendocrine tumors. EUS images served as inputs for the algorithms. Additional EUS diagnostic technology, such as elastography, digital characteristics, contrast-enhancement, and Doppler imaging were also used. Regarding ML methods, Support-Vector-Machines were used in earlier studies to select the best combination of digital imaging features. In later studies the preferred methods were neural networks with different complexity levels depending on the year of the study. Although the models and populations varied, all studies achieved over 80% specificity and 0.94 AUC, demonstrating the feasibility of ML in this area.
In an early 2008 study by Saftoiu et al[27], ML for EUS elastography images was evaluated to discriminate pancreatic tumors from ‘pseudotumoral’ chronic pancreatitis. The prospective study enrolled 68 patients including PDAC, pancreatic neuroendocrine tumor, chronic pancreatitis, and normal pancreas. Final diagnoses were confirmed with additional pathology, imaging findings, and 6-mo follow-up of patients. From each patient, EUS elastography images were converted to vector data. As the sample size was small, 10-fold cross-validation was performed. The vector data was then analyzed with simple three and four layered ANNs. This ML algorithm yielded an AUC of 0.93 to classify malignant tumors from normal and pseudotumoral pancreatitis. This study was followed by a larger prospective blinded study in 2012 with 258 patients enrolled from 13 European centers. The population consisted of 211 PDAC confirmed by pathology diagnosis and 47 chronic pancreatitis patients diagnosed by clinical, imaging and EUS criteria (at least four of the following: hyperechoic foci, hyperechoic strands, lobularity, calcifications, hyperechoic duct wall, dilated main pancreatic duct, irregular main pancreatic duct, dilated side branches, and cysts). EUS elastography images of the regions of interests were converted to vector data and then analyzed with similar ANNs. One hundred training iterations were performed with the model to increase the statistical power of the results. The mean performance of one hundred models to correctly classify PDAC from chronic pancreatitis showed 0.94 (0.91-0.97) AUC with 85.6% sensitivity and 82.9% specificity compared with 0.85 AUC for hue histogram analysis[28]. These two studies present an excellent example for the roadmap of ML research with an initial proof-of-concept study followed by a larger prospective study. Of note, less complex neural networks were used with fewer layers. Multi-layered Perceptron only accepts numeric data as the input unlike newer CNN algorithms that can directly process the image itself. Therefore, the performance of ML in these studies can be improved.
An early study in 2013 used analysis of digital image characteristics as input to the ML model[29]. The study population consisted of 262 PDAC patients diagnosed by cytology with 126 chronic pancreatitis controls diagnosed by standard EUS criteria and over 2-year follow up. Regions of interests were manually selected by blinded endosonographers. Then 105 digital imaging characteristics of these images were extracted with dedicated software. The final combination of 16 characteristics yielded a strong discriminative performance with 94.2% accuracy, 96% sensitivity and 93% specificity.
Another older study evaluated the use of ML to classify PDAC from normal pancreas[30]. This retrospective study in 2016 included 202 PDAC patients and 130 patients with normal pancreas as controls. The regions of interests from EUS images were annotated by endosonographers. Then digital characteristics of the images (wavelet decomposition energy, boundary fractal, gray level cooccurrence matrix, standard statistical) were extracted. Among 112 digital characteristics, 20 were identified as more effective for classification, and therefore served as the input for the ML algorithm. A three-layered Multi-layered Perceptron model was used as the neural network, which is a less-complex approach accepting numerical data such as the digital characteristics of EUS images and does not require extra image processing. As such, because the images themselves are not being used, important information may not be included in the model. The final model yielded 83% sensitivity, 93% specificity and 87% accuracy for differentiating PDAC from normal pancreas. This model also only compared PDAC images to normal pancreatic tissue and not to other commonly encountered differential diagnoses such as chronic pancreatitis, which limits its adoptability to clinical use.
A recent study in 2021 evaluated the performance of ML to classify focal solid lesions. The study population consisted of 30 patients with PDAC, 20 patients with pseudo tumors in chronic pancreatitis, and 15 patients with pancreatic neuroendocrine tumors[31]. The final diagnoses were confirmed with histologic evaluations of fine-needle specimens and clinical follow-ups. From each EUS examination, 5 sets of images were extracted including grayscale images, color Doppler, contrast-enhanced imaging, and elastography. A total of 1300 collected images was increased to 3360 with data augmentation. Regarding the ML method, a CNN algorithm was combined with a Long Short-Term Memory model. Long Short-Term Memory model is a supervised ML model that has additional feedback learning functions and allows the use of sequential pre- and post-contrast appearance from the same EUS images. Cross-validation was performed for each dataset with 80% of images used as training and 20% as test sets. The final combined model’s overall specificity was 96.4%, and sensitivity was 98.6% for classifying the pancreatic masses. For PDAC cases, the algorithm yielded 96.7% specificity, 98.1% sensitivity, 97.6% accuracy, and 0.97 AUC. When compared to previous studies, Udristoiu et al[31] used a more complex, combined ML approach with CNN and Long Short-Term Memory allowing inclusion of temporal data with contrast-enhanced imaging.
Tonozuka et al[32] also evaluated their own ML algorithm for its performance in classifying pancreatic masses. The 139 total patients included 76 with PDAC, 34 with chronic pancreatitis and 29 normal controls. PDAC was diagnosed using histology from EUS-fine needle biopsy or surgery, and chronic pancreatitis was diagnosed using the Rosemont criteria. All patients were followed for over 6 mo. Ten still images of lesions were chosen from each EUS examination, and the input dataset was increased to over 80000 after data augmentation. From 1390 still images, 920 were used for training and cross-validation, while the remaining 470 images were used for testing. A CNN algorithm with seven layers was used. In addition to the CNN model, a pseudo-colored feature mapping was used to highlight the areas in the image with greater impact on the final model, which makes the decision-making process more comprehensible to the endosonographer. In the test dataset, the model yielded 84.1% specificity, 92.4% sensitivity and 0.94 AUC.
AIP is an increasingly recognized entity that may be challenging to diagnose. Accurate diagnosis is particularly important as the differential often includes PDAC with its different prognostic and management implications. Many diagnostic algorithms have been developed that include clinical, serologic, imaging, and histopathologic criteria, but their performance remains limited. While EUS with biopsy is the most effective diagnostic tool, its diagnostic yield also is suboptimal[33]. Image processing may enhance our ability to diagnose AIP by extracting data and learning from the cues in sonographic images. Two studies have studied the utility of ML in differentiating AIP from other diagnoses, including chronic pancreatitis and PDAC. The studies by Zhu et al[34] and Marya et al[35] used different ML approaches, but both achieved over 80% sensitivity and specificity for diagnosing AIP only from EUS images[34,35].
The earlier 2015 retrospective study by Zhu et al[34] studied a ML algorithm to differentiate AIP from chronic pancreatitis using an EUS image dataset of 81 AIP and 100 chronic pancreatitis cases. AIP diagnoses were based on HISORt criteria. Chronic pancreatitis was diagnosed by standard EUS criteria. Experienced endosonographers selected regions of interest in EUS images, and 115 digital parameters were extracted from each image. Then, a supervised Support Vector Machine algorithm was used to select the best combination of these digital parameters for discriminating AIP from chronic pancreatitis. The final combination of digital parameters yielded 90.6% accuracy, 84.1% sensitivity and 94.0% specificity.
A recent study examined the additive performance of ML with EUS to distinguish AIP from PDAC as well as chronic pancreatitis and normal pancreas. The study included 583 patients (146 AIP, 292 PDAC, 72 chronic pancreatitis, and 74 normal) with all available videos and still images of the pancreatic and peripancreatic regions included in the analysis regardless of whether they included regions of interest[36]. A total of 1174461 still images were extracted from the images and videos. Since all portions of EUS videos were included, there was a risk of oversimplification of diagnosis from certain aspects of the examination, such as presence of metastasis, which were removed from the dataset. The classification was performed with two datasets: the first one included still images obtained from both EUS videos and captured images, while the second dataset only included EUS videos. The CNN algorithm was trained for both datasets. Pseudo-colored feature mapping was also used to visualize decision making. For comparison, seven independent EUS experts evaluated each case using videos. In the final analysis, ML showed 87% specificity, 90% sensitivity and 0.9 AUC for distinguishing AIP from PDAC in the image-only dataset. In the video-only dataset, the metrics were 90%, 93% and 0.96 for specificity, sensitivity, and AUC, respectively. The ML model was superior to expert endosonographers, who had 82.4% specificity and 53.8% sensitivity in differentiating AIP from PDAC. ML also had high sensitivity (99%) and specificity (98%) for distinguishing AIP from normal pancreas. It had inferior performance in separating AIP from chronic pancreatitis (94% sensitivity, 71% specificity, 0.89 AUC). The heatmap analysis yielded interesting results, which may help guide endosonographers, showing that visualizing a hyperechoic plane between the parenchyma and duct or vessel was highly predictive of AIP while post acoustic enhancement deep to a dilated pancreatic duct or vessel was consistent with PDAC. Regarding AI technology, these two studies differ with respect to their approach of utilizing ML with EUS data. Zhu et al[34] used an older ML algorithm, support vector machine, which is a supervised algorithm that classifies two numeric data points. As such, EUS images are converted into numerical data by extracting digital parametric features, and then the ML model is trained with these features. On the other hand, Marya et al[35] used a CNN algorithm, ResNet, with 50 layers that can work directly on the EUS images itself.
EUS is the leading modality for assessing and obtaining tissue from the pancreas with approximately 90% specificity and sensitivity for solid masses[36]. However, interobserver reliability remains an issue in EUS as accuracy relies on the endosonographers’ skills and experience and carries the risk of false-negative results. Pancreatic EUS also has a steep learning curve. ML approaches have been developed to potentially augment the diagnostic performance of EUS and biopsy as well as aid in training.
Iwasa et al[38] tested ML to augment contrast enhanced EUS by dividing the sonographic image into regions with similar appearance and then differentiating regions of interest, also called automatic segmentation. For this study, videos from 100 contrast enhanced EUS examinations of solid pancreatic masses with histologic diagnosis were used. Each video was transformed into 900 still images as input for a U-Net CNN algorithm. The borders of the lesions were manually annotated by two endosonographers and served as the ground truth. IoU was used as the performance output of the algorithm with median IoU for all cases being 0.77, which is greater than the acceptable 0.5 threshold value[37]. The EUS videos were also classified into different categories to understand the effect of respiratory movements and visibility of boundaries of the lesions by the endosonographers. IoU significantly improved to 0.91 in cases with the most visible boundaries and decreased to 0.13 for cases with the least visible boundaries[39]. On the other hand, respiratory movements did not change the performance of the algorithm. This proof-of-concept study suggests that ML can provide real-time assistance in the detection of pancreatic lesions. The classification of exams with respect to the ease of detecting the border of lesions is an important aspect of this study because it demonstrated that ML can also be affected by the quality of the EUS examination and the sonographic characteristics of the lesion, reflected in this case by how well the border was visible.
A case report suggested that a ML model may help target areas to biopsy within pancreatic masses that have the highest diagnostic yield by avoiding areas of necrosis. A CNN algorithm was used to label and highlight the more cellular region in a 6.5 cm solid pancreatic mass, which was predicted to have the highest probability of yielding a diagnosis by discriminating it from neighboring necrotic or inflammatory regions. EUS-fine needle aspiration was performed and yielded a positive diagnosis for PDAC. The technical details, training dataset and methods, validation and model characteristics were not presented in the report[39]. This is a novel idea that may provide valuable intra-procedural assistance, however, needs further evaluation.
ML may aid EUS training by guiding the steps of routine diagnostic EUS evaluation of the pancreas. A novel AI system aimed to assist recognition of fundamental stations and identification of pancreatic and vascular anatomical landmarks. This was performed in four steps: Identifying images, filtering suitable images, recognizing pancreas stations, and segmenting anatomical landmarks and monitoring for loss of visualization of the pancreas. Two expert endosonographers decided on the criteria for suitable images and annotated video clips that served as ground truth. A ResNet model was used as the CNN algorithm. A separate set of prospective EUS examinations were used as a test set. Three different endosonographers classified each image for comparison with the AI model. The final model was tested using an external test set and demonstrated an accuracy of 82.4% to identify six anatomical stations (abdominal aorta, pancreatic body, pancreatic tail, confluence, pancreatic head from stomach, or pancreatic head from descending duodenum), and a Dice of 0.715 to label pancreas and vessels. Comparison of the AI model with the three expert endosonographers yielded strong interobserver agreement with kappa values of 0.846, 0.853 and 0.826[40]. The results of this study demonstrated that a ML model may aid in recognizing stations and anatomic landmarks in sonographic images. This has the potential to assist procedural navigation during EUS examination and improve cognitive aspects of EUS skills. However, the impact of such real-time procedural assistance on the endosonographer’s performance was not assessed in this study and warrants further evaluation.
In this review, we summarize the current literature regarding the use of ML in EUS for diagnosing pancreatic diseases. Our review defined two main areas for AI in the field: visual recognition-classification and procedural assistance and training. AI has been more utilized in transabdominal ultrasonography for detecting liver fibrosis and in CT scans for lesion classification, which have been extensively reviewed elsewhere[41-45]. ML appears to have great potential in assisting EUS examination of the pancreas as sonographic imaging contains vital visual information that the human eye cannot distinguish. The diagnostic accuracy of EUS imaging is highly operator dependent and requires both technical and cognitive skills. Acquisition of these skills currently requires dedicated training with proctorship and procedural experience, which remains limited, apart from dedicated advanced endoscopy fellowship programs. These issues in training limit the widespread adoption of EUS, which is the leading tool for diagnosing pancreatic disorders, including PDAC. AI may assist in the development of cognitive skills and augmentation of procedural efficiency in relatively less experienced endosonographers.
Further opportunities should be explored with AI and pancreatic EUS. However, several limitations exist in the field. First, the number of EUS procedures and the prevalence of pancreatic diseases are lower, which makes it more difficult to train data-hungry machine learning algorithms. Second, annotation of EUS data is more challenging compared to other imaging modalities as the number of experts endosonographers is relatively limited. Third, EUS examinations with histopathologic or cytologic diagnosis is harder to obtain for certain pancreatic diseases and have issues with sensitivity, which further limits the number of studies for AI training. However, these limitations may be overcome with multi-center collaborations and prospective data collection, which will hopefully lead to improved image recognition, procedural assistance, and training for pancreatic EUS.
Provenance and peer review: Invited article; Externally peer reviewed.
Peer-review model: Single blind
Specialty type: Gastroenterology and hepatology
Country/Territory of origin: United States
Peer-review report’s scientific quality classification
Grade A (Excellent): A
Grade B (Very good): B
Grade C (Good): C
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Dabbakuti JRKKK, India; Hanada E, Japan; Wang P, China S-Editor: Liu JH L-Editor: A P-Editor: Liu JH
| 1. | Miller KD, Nogueira L, Mariotto AB, Rowland JH, Yabroff KR, Alfano CM, Jemal A, Kramer JL, Siegel RL. Cancer treatment and survivorship statistics, 2019. CA Cancer J Clin. 2019;69:363-385. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2417] [Cited by in RCA: 3071] [Article Influence: 511.8] [Reference Citation Analysis (0)] |
| 2. | Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68:394-424. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 53206] [Cited by in RCA: 55773] [Article Influence: 7967.6] [Reference Citation Analysis (132)] |
| 3. | Rahib L, Wehner MR, Matrisian LM, Nead KT. Estimated Projection of US Cancer Incidence and Death to 2040. JAMA Netw Open. 2021;4:e214708. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 277] [Cited by in RCA: 782] [Article Influence: 195.5] [Reference Citation Analysis (0)] |
| 4. | Xiao AY, Tan ML, Wu LM, Asrani VM, Windsor JA, Yadav D, Petrov MS. Global incidence and mortality of pancreatic diseases: a systematic review, meta-analysis, and meta-regression of population-based cohort studies. Lancet Gastroenterol Hepatol. 2016;1:45-55. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 549] [Cited by in RCA: 474] [Article Influence: 52.7] [Reference Citation Analysis (0)] |
| 5. | Kichler A, Jang S. Chronic Pancreatitis: Epidemiology, Diagnosis, and Management Updates. Drugs. 2020;80:1155-1168. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 14] [Cited by in RCA: 44] [Article Influence: 11.0] [Reference Citation Analysis (0)] |
| 6. | Megibow AJ, Baker ME, Gore RM, Taylor A. The incidental pancreatic cyst. Radiol Clin North Am. 2011;49:349-359. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 42] [Cited by in RCA: 28] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 7. | Zhang B, Zhu F, Li P, Yu S, Zhao Y, Li M. Endoscopic ultrasound elastography in the diagnosis of pancreatic masses: A meta-analysis. Pancreatology. 2018;18:833-840. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 31] [Cited by in RCA: 39] [Article Influence: 5.6] [Reference Citation Analysis (0)] |
| 8. | Russell S, Norvig P. Artificial intelligence: a modern approach. 2002. |
| 9. | Yu KH, Beam AL, Kohane IS. Artificial intelligence in healthcare. Nat Biomed Eng. 2018;2:719-731. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 732] [Cited by in RCA: 1084] [Article Influence: 154.9] [Reference Citation Analysis (0)] |
| 10. | Penny W, Frost D. Neural networks in clinical medicine. Med Decis Making. 1996;16:386-398. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 113] [Cited by in RCA: 93] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 11. | Yamashita R, Nishio M, Do RKG, Togashi K. Convolutional neural networks: an overview and application in radiology. Insights Imaging. 2018;9:611-629. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1202] [Cited by in RCA: 1184] [Article Influence: 169.1] [Reference Citation Analysis (0)] |
| 12. | Zhang, HM. and B. Dong, A Review on Deep Learning in Medical Image Reconstruction. Journal of the Operations Research Society of China. 2020;8:311-340. [DOI] [Full Text] |
| 13. | Van Houdt G, Mosquera C, Nápoles G. A review on the long short-term memory model. Artif Intell Rev. 2020;53:5929-5955. [RCA] [DOI] [Full Text] [Cited by in Crossref: 134] [Cited by in RCA: 201] [Article Influence: 40.2] [Reference Citation Analysis (0)] |
| 14. | Rintala TJ, Federico A, Latonen L, Greco D, Fortino V. A systematic comparison of data- and knowledge-driven approaches to disease subtype discovery. Brief Bioinform. 2021;22. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1] [Cited by in RCA: 2] [Article Influence: 0.5] [Reference Citation Analysis (0)] |
| 15. | Sinkala M, Mulder N, Martin D. Machine Learning and Network Analyses Reveal Disease Subtypes of Pancreatic Cancer and their Molecular Characteristics. Sci Rep. 2020;10:1212. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 65] [Cited by in RCA: 55] [Article Influence: 11.0] [Reference Citation Analysis (0)] |
| 16. | Zerboni G, Signoretti M, Crippa S, Falconi M, Arcidiacono PG, Capurso G. Systematic review and meta-analysis: Prevalence of incidentally detected pancreatic cystic lesions in asymptomatic individuals. Pancreatology. 2019;19:2-9. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 160] [Cited by in RCA: 135] [Article Influence: 22.5] [Reference Citation Analysis (0)] |
| 17. | Zaheer A, Pokharel SS, Wolfgang C, Fishman EK, Horton KM. Incidentally detected cystic lesions of the pancreas on CT: review of literature and management suggestions. Abdom Imaging. 2013;38:331-341. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 50] [Cited by in RCA: 39] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 18. | Pinho DF, Rofsky NM, Pedrosa I. Incidental pancreatic cysts: role of magnetic resonance imaging. Top Magn Reson Imaging. 2014;23:117-128. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 14] [Cited by in RCA: 13] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 19. | Wu J, Wang Y, Li Z, Miao H. Accuracy of Fukuoka and American Gastroenterological Association Guidelines for Predicting Advanced Neoplasia in Pancreatic Cyst Neoplasm: A Meta-Analysis. Ann Surg Oncol. 2019;26:4522-4536. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 13] [Cited by in RCA: 18] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 20. | Springer S, Masica DL, Dal Molin M, Douville C, Thoburn CJ, Afsari B, Li L, Cohen JD, Thompson E, Allen PJ, Klimstra DS, Schattner MA, Schmidt CM, Yip-Schneider M, Simpson RE, Fernandez-Del Castillo C, Mino-Kenudson M, Brugge W, Brand RE, Singhi AD, Scarpa A, Lawlor R, Salvia R, Zamboni G, Hong SM, Hwang DW, Jang JY, Kwon W, Swan N, Geoghegan J, Falconi M, Crippa S, Doglioni C, Paulino J, Schulick RD, Edil BH, Park W, Yachida S, Hijioka S, van Hooft J, He J, Weiss MJ, Burkhart R, Makary M, Canto MI, Goggins MG, Ptak J, Dobbyn L, Schaefer J, Sillman N, Popoli M, Klein AP, Tomasetti C, Karchin R, Papadopoulos N, Kinzler KW, Vogelstein B, Wolfgang CL, Hruban RH, Lennon AM. A multimodality test to guide the management of patients with a pancreatic cyst. Sci Transl Med. 2019;11. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 154] [Cited by in RCA: 134] [Article Influence: 22.3] [Reference Citation Analysis (0)] |
| 21. | Nguon LS, Seo K, Lim JH, Song TJ, Cho SH, Park JS, Park S. Deep Learning-Based Differentiation between Mucinous Cystic Neoplasm and Serous Cystic Neoplasm in the Pancreas Using Endoscopic Ultrasonography. Diagnostics (Basel). 2021;11. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 5] [Cited by in RCA: 24] [Article Influence: 6.0] [Reference Citation Analysis (0)] |
| 22. | Kurita Y, Kuwahara T, Hara K, Mizuno N, Okuno N, Matsumoto S, Obata M, Koda H, Tajika M, Shimizu Y, Nakajima A, Kubota K, Niwa Y. Diagnostic ability of artificial intelligence using deep learning analysis of cyst fluid in differentiating malignant from benign pancreatic cystic lesions. Sci Rep. 2019;9:6893. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 34] [Cited by in RCA: 55] [Article Influence: 9.2] [Reference Citation Analysis (0)] |
| 23. | Kuwahara T, Hara K, Mizuno N, Okuno N, Matsumoto S, Obata M, Kurita Y, Koda H, Toriyama K, Onishi S, Ishihara M, Tanaka T, Tajika M, Niwa Y. Usefulness of Deep Learning Analysis for the Diagnosis of Malignancy in Intraductal Papillary Mucinous Neoplasms of the Pancreas. Clin Transl Gastroenterol. 2019;10:1-8. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 61] [Cited by in RCA: 127] [Article Influence: 25.4] [Reference Citation Analysis (0)] |
| 24. | Rahib L, Smith BD, Aizenberg R, Rosenzweig AB, Fleshman JM, Matrisian LM. Projecting cancer incidence and deaths to 2030: the unexpected burden of thyroid, liver, and pancreas cancers in the United States. Cancer Res. 2014;74:2913-2921. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 5379] [Cited by in RCA: 5128] [Article Influence: 466.2] [Reference Citation Analysis (0)] |
| 25. | Howlader N, Noone AM, Krapcho M, Garshell J, Miller D, Altekruse SF, Kosary Cl, Yu M, Ruhl J, Tatalovich Z, Mariotto A, Lewis DR, Chen HS, Feuer EJ, Cronin KA. SEER cancer statistics review, 1975–2012. National Cancer Institute, 2015. Available from: https://seer.cancer.gov/archive/csr/1975_2012/. |
| 26. | Kitano M, Yoshida T, Itonaga M, Tamura T, Hatamaru K, Yamashita Y. Impact of endoscopic ultrasonography on diagnosis of pancreatic cancer. J Gastroenterol. 2019;54:19-32. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 170] [Cited by in RCA: 214] [Article Influence: 35.7] [Reference Citation Analysis (0)] |
| 27. | Săftoiu A, Vilmann P, Gorunescu F, Gheonea DI, Gorunescu M, Ciurea T, Popescu GL, Iordache A, Hassan H, Iordache S. Neural network analysis of dynamic sequences of EUS elastography used for the differential diagnosis of chronic pancreatitis and pancreatic cancer. Gastrointest Endosc. 2008;68:1086-1094. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 169] [Cited by in RCA: 173] [Article Influence: 10.2] [Reference Citation Analysis (0)] |
| 28. | Săftoiu A, Vilmann P, Gorunescu F, Janssen J, Hocke M, Larsen M, Iglesias-Garcia J, Arcidiacono P, Will U, Giovannini M, Dietrich CF, Havre R, Gheorghe C, McKay C, Gheonea DI, Ciurea T; European EUS Elastography Multicentric Study Group. Efficacy of an artificial neural network-based approach to endoscopic ultrasound elastography in diagnosis of focal pancreatic masses. Clin Gastroenterol Hepatol. 2012;10:84-90.e1. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 149] [Cited by in RCA: 116] [Article Influence: 8.9] [Reference Citation Analysis (0)] |
| 29. | Zhu M, Xu C, Yu J, Wu Y, Li C, Zhang M, Jin Z, Li Z. Differentiation of pancreatic cancer and chronic pancreatitis using computer-aided diagnosis of endoscopic ultrasound (EUS) images: a diagnostic test. PLoS One. 2013;8:e63820. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 84] [Cited by in RCA: 73] [Article Influence: 6.1] [Reference Citation Analysis (2)] |
| 30. | Ozkan M, Cakiroglu M, Kocaman O, Kurt M, Yilmaz B, Can G, Korkmaz U, Dandil E, Eksi Z. Age-based computer-aided diagnosis approach for pancreatic cancer on endoscopic ultrasound images. Endosc Ultrasound. 2016;5:101-107. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 75] [Cited by in RCA: 66] [Article Influence: 7.3] [Reference Citation Analysis (0)] |
| 31. | Udriștoiu AL, Cazacu IM, Gruionu LG, Gruionu G, Iacob AV, Burtea DE, Ungureanu BS, Costache MI, Constantin A, Popescu CF, Udriștoiu Ș, Săftoiu A. Real-time computer-aided diagnosis of focal pancreatic masses from endoscopic ultrasound imaging based on a hybrid convolutional and long short-term memory neural network model. PLoS One. 2021;16:e0251701. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 26] [Cited by in RCA: 36] [Article Influence: 9.0] [Reference Citation Analysis (0)] |
| 32. | Tonozuka R, Itoi T, Nagata N, Kojima H, Sofuni A, Tsuchiya T, Ishii K, Tanaka R, Nagakawa Y, Mukai S. Deep learning analysis for the detection of pancreatic cancer on endosonographic images: a pilot study. J Hepatobiliary Pancreat Sci. 2021;28:95-104. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 36] [Cited by in RCA: 79] [Article Influence: 15.8] [Reference Citation Analysis (0)] |
| 33. | Facciorusso A, Barresi L, Cannizzaro R, Antonini F, Triantafyllou K, Tziatzios G, Muscatiello N, Hart PA, Wani S. Diagnostic yield of endoscopic ultrasound-guided tissue acquisition in autoimmune pancreatitis: a systematic review and meta-analysis. Endosc Int Open. 2021;9:E66-E75. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 33] [Cited by in RCA: 26] [Article Influence: 6.5] [Reference Citation Analysis (0)] |
| 34. | Zhu J, Wang L, Chu Y, Hou X, Xing L, Kong F, Zhou Y, Wang Y, Jin Z, Li Z. A new descriptor for computer-aided diagnosis of EUS imaging to distinguish autoimmune pancreatitis from chronic pancreatitis. Gastrointest Endosc. 2015;82:831-836.e1. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 18] [Cited by in RCA: 13] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 35. | Marya NB, Powers PD, Chari ST, Gleeson FC, Leggett CL, Abu Dayyeh BK, Chandrasekhara V, Iyer PG, Majumder S, Pearson RK, Petersen BT, Rajan E, Sawas T, Storm AC, Vege SS, Chen S, Long Z, Hough DM, Mara K, Levy MJ. Utilisation of artificial intelligence for the development of an EUS-convolutional neural network model trained to enhance the diagnosis of autoimmune pancreatitis. Gut. 2021;70:1335-1344. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 44] [Cited by in RCA: 86] [Article Influence: 21.5] [Reference Citation Analysis (1)] |
| 36. | Facciorusso A, Wani S, Triantafyllou K, Tziatzios G, Cannizzaro R, Muscatiello N, Singh S. Comparative accuracy of needle sizes and designs for EUS tissue sampling of solid pancreatic masses: a network meta-analysis. Gastrointest Endosc. 2019;90:893-903.e7. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 123] [Cited by in RCA: 107] [Article Influence: 17.8] [Reference Citation Analysis (0)] |
| 37. | Rezatofighi H, Tsoi N, Gwak JY, Sadeghian A, Reid I, Savaresel S. Generalized intersection over union: A metric and a loss for bounding box regression. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition. 2019. |
| 38. | Iwasa Y, Iwashita T, Takeuchi Y, Ichikawa H, Mita N, Uemura S, Shimizu M, Kuo YT, Wang HP, Hara T. Automatic Segmentation of Pancreatic Tumors Using Deep Learning on a Video Image of Contrast-Enhanced Endoscopic Ultrasound. J Clin Med. 2021;10. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 6] [Cited by in RCA: 19] [Article Influence: 4.8] [Reference Citation Analysis (0)] |
| 39. | Yu X, Zhang Z, Zhu N, Tang A, Hu S, Wang X, Tian L. Application of a novel artificial intelligence system in guiding the targeted puncture of a pancreatic mass. Endoscopy. 2021;. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1] [Cited by in RCA: 1] [Article Influence: 0.3] [Reference Citation Analysis (0)] |
| 40. | Zhang J, Zhu L, Yao L, Ding X, Chen D, Wu H, Lu Z, Zhou W, Zhang L, An P, Xu B, Tan W, Hu S, Cheng F, Yu H. Deep learning-based pancreas segmentation and station recognition system in EUS: development and validation of a useful training tool (with video). Gastrointest Endosc. 2020;92:874-885.e3. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 84] [Cited by in RCA: 69] [Article Influence: 13.8] [Reference Citation Analysis (0)] |
| 41. | Chen Y, Luo Y, Huang W, Hu D, Zheng RQ, Cong SZ, Meng FK, Yang H, Lin HJ, Sun Y, Wang XY, Wu T, Ren J, Pei SF, Zheng Y, He Y, Hu Y, Yang N, Yan H. Machine-learning-based classification of real-time tissue elastography for hepatic fibrosis in patients with chronic hepatitis B. Comput Biol Med. 2017;89:18-23. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 82] [Cited by in RCA: 60] [Article Influence: 7.5] [Reference Citation Analysis (0)] |
| 42. | Yang CM, Shu J. Cholangiocarcinoma Evaluation via Imaging and Artificial Intelligence. Oncology. 2021;99:72-83. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 13] [Cited by in RCA: 25] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 43. | Gatos I, Tsantis S, Spiliopoulos S, Karnabatidis D, Theotokas I, Zoumpoulis P, Loupas T, Hazle JD, Kagadis GC. A Machine-Learning Algorithm Toward Color Analysis for Chronic Liver Disease Classification, Employing Ultrasound Shear Wave Elastography. Ultrasound Med Biol. 2017;43:1797-1810. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 80] [Cited by in RCA: 73] [Article Influence: 9.1] [Reference Citation Analysis (0)] |
| 44. | Lee JH, Joo I, Kang TW, Paik YH, Sinn DH, Ha SY, Kim K, Choi C, Lee G, Yi J, Bang WC. Deep learning with ultrasonography: automated classification of liver fibrosis using a deep convolutional neural network. Eur Radiol. 2020;30:1264-1273. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 42] [Cited by in RCA: 83] [Article Influence: 13.8] [Reference Citation Analysis (0)] |
| 45. | Kuwahara T, Hara K, Mizuno N, Haba S, Okuno N, Koda H, Miyano A, Fumihara D. Current status of artificial intelligence analysis for endoscopic ultrasonography. Dig Endosc. 2021;33:298-305. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 24] [Cited by in RCA: 36] [Article Influence: 9.0] [Reference Citation Analysis (0)] |