Published online Apr 28, 2022. doi: 10.35712/aig.v3.i2.46
Peer-review started: December 31, 2021
First decision: February 7, 2022
Revised: February 18, 2022
Accepted: April 28, 2022
Article in press: April 28, 2022
Published online: April 28, 2022
Processing time: 119 Days and 9.8 Hours
Non-alcoholic fatty liver disease (NAFLD) and chronic viral hepatitis are among the most significant causes of liver-related mortality worldwide. It is critical to develop reliable methods of predicting progression to fibrosis, cirrhosis, and decompensated liver disease. Current screening methods such as biopsy and transient elastography are limited by invasiveness and observer variation in analysis of data. Artificial intelligence (AI) provides a unique opportunity to more accurately diagnose NAFLD and viral hepatitis, and to identify patients at high risk for disease progression. We conducted a literature review of existing evidence for AI in NAFLD and viral hepatitis. Thirteen articles on AI in NAFLD and 14 on viral hepatitis were included in our analysis. We found that machine learning algorithms were comparable in accuracy to current methods for diagnosis and fibrosis prediction (MELD-Na score, liver biopsy, FIB-4 score, and biomarkers). They also reliably predicted hepatitis C treatment failure and hepatic encephalopathy, for which there are currently no established prediction tools. These studies show that AI could be a helpful adjunct to existing techniques for diag
Core Tip: Non-alcoholic fatty liver disease (NAFLD) exists on a spectrum from simple hepatocyte steatosis to non-alcoholic steatohepatitis (NASH) with ballooning and fibrosis. Given the lack of efficient screening methods and high rate of asymptomatic disease, it is challenging to identify patients with NAFLD in its various stages. Although liver biopsy remains the gold standard for diagnosing NASH, it is an invasive, costly, and painful procedure. Conventional imaging modalities including ultrasound, computed tomography, magnetic resonance imaging and transient elastography are limited by inter- and intra-observer variability depending on the stage of fibrosis. Similarly, despite recent progress in the prevention and treatment of viral hepatitis, predicting sustained virological response and disease progression remains challenging. Artificial intelligence (AI) is an exciting and increasingly pertinent field in medicine as clinicians incorporate augmenting technology into their daily practice. This review summarizes recent literature on the application of AI in NAFLD and viral hepatitis. Specifically, the review will assess the performance of AI as a non-invasive method for the diagnosis and staging of liver fibrosis and steatosis, as well as for the detection and treatment of chronic viral hepatitis. It will also aim to highlight the potential for AI based methods on their ability to develop therapeutic targets.
- Citation: Gunasekharan A, Jiang J, Nickerson A, Jalil S, Mumtaz K. Application of artificial intelligence in non-alcoholic fatty liver disease and viral hepatitis. Artif Intell Gastroenterol 2022; 3(2): 46-53
- URL: https://www.wjgnet.com/2644-3236/full/v3/i2/46.htm
- DOI: https://dx.doi.org/10.35712/aig.v3.i2.46
Non-alcoholic fatty liver disease (NAFLD) exists on a spectrum from simple hepatocyte steatosis to inflammation, ballooning and fibrosis. Given the lack of efficient screening methods and high rate of asymptomatic disease, it is challenging to identify patients with various stages of NAFLD[1,2]. Non-alcoholic steatohepatitis (NASH) patients with significant fibrosis are at increased risk for cirrhosis and progressive liver failure, which has led NASH to become one of the leading causes of liver transplantation in the United States[3]. NASH affects approximately 3% to 6% of the US population, and this number continues to increase. It affects approximately 25% of the population worldwide[4].
Although liver biopsy remains the gold standard for diagnosing NASH, it is an invasive, costly, and painful procedure. Therefore, serial liver biopsies for surveillance are not always feasible. Conventional imaging modalities including ultrasound, computed tomography (CT), magnetic resonance imaging (MRI) and transient elastography are limited by inter- and intra-observer variability depending on the stage of fibrosis[1,2]. Similarly, despite recent progress in the prevention and treatment of viral hepatitis, predicting sustained virological response (SVR) and disease progression remains challenging.
Artificial intelligence (AI) is an exciting and increasingly pertinent field in medicine as clinicians incorporate augmenting technology into their daily practice. AI is the concept of teaching a computer to simulate the cognitive abilities of the human brain. Machine learning (ML) entails allowing the computer to simulate the human brain independently. It can either be supervised (through specific feedback from humans) or unsupervised, in which case there is no guidance provided and the computer is able to independently synthesize and analyze the output[1]. AI is increasingly applied to the diagnosis and prediction of various diseases. Researchers are developing machine learning (ML) algorithms to predict risk and outcomes using multiple demographic, clinical, biochemical, and imaging parameters for diagnosis and prognosis related to liver fibrosis and steatosis, including NAFLD and viral hepatitis[1].
Current methods of assessing liver fibrosis progression and mortality in both NAFLD and viral hepatitis have many limitations. These include the intra- and inter-observer variability in staging fibrosis, the inability to place fibrosis along a continuum, and the lack of identifiable markers for disease progression[1,2]. These limitations and the ability of ML models to overcome them will be discussed further in this review. This review will also highlight how ML models have the potential to present opportunities for drug discovery and prediction of therapeutic and toxic effects of drugs. Machine learning models based on AI provide promising features that could not only enhance screening for NAFLD, but also help with fibrosis staging in patients with NASH and viral hepatitis.
This review summarizes recent literature on the application of AI in NAFLD and viral hepatitis. The main objective is to assess the performance of AI as a non-invasive method for the diagnosis and staging of liver fibrosis and steatosis, as well as the detection and treatment of chronic viral hepatitis.
A review of current literature in the areas of AI in NAFLD and viral hepatitis was conducted using two separate searches on PubMed. First, we used the search terms “non-alcoholic fatty liver disease”, “NAFLD”, and “deep learning” in combination with “artificial intelligence”, “histology”, “omics” and “radiology.” The second search was conducted using the search terms “viral hepatitis” in combination with “hepatitis A”, “hepatitis B”, “hepatitis C”, “hepatitis E”, “machine learning”, “artificial intelligence”, “histology” and “radiology”.
Most articles on NASH and NAFLD published between 2018 and 2021 were included in this review. Articles were excluded if they did not offer comparisons between AI modalities and existing methods for screening or prediction (MELD score, elastography, etc.). Twenty-seven articles were included in our review, 13 on NAFLD and 14 on chronic viral hepatitis. For studies on viral hepatitis, described machine learning algorithms fell into one of three categories: Predicting prevalence, screening for complications (including fibrosis, hepatocellular carcinoma, decompensated cirrhosis, and death), and predicting response to treatment.
It is estimated that half of patients infected with hepatitis C worldwide are unaware of their diagnosis and only 17% have undergone liver fibrosis staging[5]. This rate is even lower for hepatitis B, for which only 10.5% of infected patients are aware of their status. In March 2020, the USPSTF recommended hepatitis C screening for all adults over 18; however, there are currently no population-based screening recommendations for hepatitis A and B. Primary care offices do not routinely test for hepatitis B. Machine learning has been used both to determine regional prevalence of chronic hepatitis and to identify undiagnosed cases.
Zheng et al[6] compared two algorithms (Elman neural network and autoregressive integrated moving average, or ARIMA) designed to predict incidence of hepatitis B in Guangxi, China. ARIMA is a type of model that can capture the randomness of data and is often used for infectious disease prediction. Predictions were compared to the reported cases of hepatitis B cases from the Health Commission of Guangxi, China. The neural network was the more predictive model, with a root-mean-square error (RMSE) of 0.89 and mean absolute error (MAE) of 0.70, while the ARIMA had an RSME of 0.94 and an MAE of 0.81.
A 2020 study by Doyle et al[7] aimed to predict chronic hepatitis C (HCV) positive status by using patient claims data to develop four algorithms, all with a predictive accuracy of over 95%. Algorithms included logistic regression, gradient boosted trees, a stacked ensemble, and random forests. The stacked ensemble performed the best, with a precision of 97% at recall levels > 50%. Key predictors of HCV infection included nonsteroidal anti-inflammatory drug use, opioids, healthcare utilization, patient age and osteoarthritis or glomerulonephritis treatment. We were unable to find any study to date using AI to screen for NAFLD/NASH.
Existing histologic models not only rely on scoring of fibrosis by a pathologist but are also unable to place fibrosis along a continuum. Artificial intelligence enables the placement of fibrosis along a continuum, identifies risk factors for progression of fibrosis, allows enhanced scoring of fibrosis stages, leading to better selection of patients for clinical trials This also allows for identification of therapeutic targets[2].
Lu et al[8] developed a light gradient-boosting machine model to predict liver fibrosis and cirrhosis in treatment-naive chronic hepatitis B patients at four centers in China. The model, named Fibro Box, outperformed transient elastography, APRI, and FIB-4, with area under the curve (AUC) 0.88 in external validation sets for significant fibrosis and 0.87 for cirrhosis. Input variables included fibroscan results, platelets, alanine aminotransferase (ALT), Prothrombin time (PT), and splenic vein diameter.
A 2013 study by Zheng et al[9], used an artificial neural network (ANN) to predict 3-month mortality of individuals with acute-on-chronic liver failure due to hepatitis B (HBV-ACLF). Patient characteristics included in this model were age, PT, serum sodium, total bilirubin, E antigen positivity status and hemoglobin. The ANN predicted mortality more accurately than MELD-based scoring systems, with area under the curve receiver operating characteristic (AUCROC) 0.765 in the validation cohort compared to 0.599 for MELD.
Similarly, Huo et al[10] developed ANNs to predict 28- and 90-d mortality in HBV-ACLF. Data were retrospectively reviewed from 684 patients admitted for ALF at 8 hospitals in various Chinese provinces with 423 cases in the training cohort and 261 in the validation cohort. In the training cohorts, the neural network had a significantly higher accuracy than MELD, MELD-Na, CLIF-ACLF, and Child-Pugh score, with AUC 0.948 and 0.913 for 28- and 90-d mortality, respectively. In the validation cohort, the model performed significantly better than MELD and insignificantly better than other scoring systems, with AUC 0.748 and 0.754 for 28- and 90-d mortality. Significant mortality predictors included age, presence of HE, sodium, PT, gamma-glutamyl transpeptidase (GGT), e antigen, alkaline phosphatase, and bilirubin.
In another study, Wang et al[11] used deep learning radiomics of elastography (DLRE) to assess stages of liver fibrosis in patients with chronic hepatitis B. DLRE was compared to 2D shear wave elastography and biomarkers (AST: Platelet ratio, fibrosis index), with liver biopsy as the reference standard. 1990 images from 398 patients were used to develop the models. AUCROCs for DLRE were 0.97 for cirrhosis, 0.98 for advanced fibrosis, and 0.85 for significant fibrosis; this performed better than other methods except for elastography in severe fibrosis.
Like viral hepatitis, there are several studies establishing the role of AI in assessing fibrosis in NAFLD/NASH. In one study by Forlano et al[2], liver biopsy specimens were annotated by two expert pathologists using the clinical research network (CRN) score as a measurable scale of degree of steatosis, inflammation, ballooning and fibrosis. The machine learning model was built using 100 patients with NAFLD in the derivation group and 146 patients in the validation group. There was good concordance when the machine learning model was compared to the scoring of the expert histopathologist on the liver biopsy specimens; the interclass correlation coefficients were 0.97 (95%CI, 0.95-0.99; P value < 0.001) for steatosis, 0.96 (95%CI, 0.9-0.98; P value < 0.001) for inflammation, 0.94 (95%CI, 0.87-0.98; P value < 0.001 for ballooning, and 0.92 for fibrosis (95%CI, 0.88-0.96; P value < 0.001). A subgroup analysis showed that quantitative analysis performed better than the CRN score in differentiating between the various stages of NAFLD. Another CNN model developed by Qu et al[12], showed that a convolutional neural network (CNN) model had an area under the curve (AUC) of 63% for all four subsets of the NAFLD scoring, while the AUC’s were 90.48% for steatosis, 81.06% for ballooning, 70.18% for inflammation and 83.85% for fibrosis. These studies underscore the utility of ML models in illustrating the heterogeneity of liver pathology in NAFLD[9,26].
In another study by Taylor-Weiner et al[13], a CNN model was developed that allowed for assessment of fibrosis along a continuum, which is not possible with pathologist scoring alone. The CRN and Ishak scores were applied to each pixel within a given image, allowing for evaluation of heterogeneity in fibrosis as well. In addition, the CNN served as a prediction model allowing for identification of features associated with disease progression. The model’s predictions correlated significantly with the pathologist scoring in all three studies, the STELLAR-3, STELLAR-4, and ATLAS, whose participants were used to build and validate the ML model - steatosis, ρ = 0.60; P value < 0.001; lobular inflammation, ρ = 0.35; P value < 0.001; and HB, ρ = 0.41; P value < 0.001. The model’s level of agreement with pathologist scoring was within the range of agreement between individual pathologists. The weighted Cohen’s kappa was 0.801 for NASH CRN and 0.817 for the Ishak classifications.
Another study by Gawrieh et al[14] built a ML model using support vector machines (SVM) to better characterize architectural patterns in fibrosis. This ML model was built to differentiate between six different patterns of fibrosis and had a strong correlation with the pathologist’s semi-quantitative scores for fibrosis, with a coefficient of determination of automated CPA ranging between 0.60 to 0.86 when compared with the pathologist score. The model was built using a trichrome-stained liver biopsy specimen which was marked with 987 annotations for different fibrosis types. As noted in the study, the model’s AUROCs were 78.6% for detection of periportal fibrosis, 83.3% for pericellular fibrosis, 86.4% for portal fibrosis, and > 90% for detection of normal fibrosis, bridging fibrosis and presence of nodules/cirrhosis.
There is an increasing number of studies focusing on metabolomics that allow for non-invasive identification of targets associated with development and progression of NAFLD. These biomarkers may differentiate between patients with and without cirrhosis, and between a healthy liver and NAFLD or NASH[3,15,16]. Several direct and indirect blood-based biomarkers currently exist to assess fibrosis. These have been incorporated to form scoring systems such as NAFLD fibrosis score (NFS), Fibrosis-4 (FIB-4), AST to platelet ratio index (APRI), BARD Score, FibroSURE and Enhanced liver fibrosis score[3]. ML allows for analysis of many multi-omics and clinical variables to screen for NASH and NAFLD and to build models for disease progression.
An eXtreme Gradient Boosting Model (XG Boost) was developed using the NIDDK database by Docherty et al[16], which contains a large real-world patient population. This model used confirmed NASH and non-NASH patients within this subset. The unique feature of this study is that it used several demographic variables and clinical biomarkers run through recursive feature elimination in combination with confirmed histologic cases to build an efficient model with a high specificity. When a greater number of markers were used in predicting patients with NASH, the AUROC was 0.82, sensitivity 81%, and precision 81%.
In a study of adults of European ancestry by Atabaski-Pasdar et al[15], patients with type 2 diabetes and others with high-risk features for the development of NASH were assessed for liver fat content using MRI. Several multi-omics and clinical data, including laboratory markers, were entered into the least absolute shrinkage and selection operator to select the most relevant features, which then underwent random forest analysis for the development of the algorithm. The model developed using this method produced a cross-validated AUROC of 0.84 (95%CI 0.82, 0.86; P value < 0.001) and outperformed existing prediction tools for NAFLD. However, unlike other studies, the model was built in comparison to MRI fat content, which is not reflective of the continuum of NAFLD, and thus cannot be used to monitor disease progression.
Another study based in China by Ma et al[17] identified BMI, triglycerides, GGT, the serum ALT and uric acid as the most common features contributing to NAFLD when a Bayesian network model was used. The model had an accuracy of 83%, specificity of 0.878, sensitivity of 0.675, and F-measure score of 0.655. The F-measure score is an indicator of whether there can be a balance between precision and recall of these variables, and it was higher than for logistic regression models in machine learning.
Like markers discussed previously, many studies have combined machine learning with imaging modalities to more effectively assess liver fat content and to better define fibrosis scores. This would allow for more accurate monitoring of patients for disease progression and their selection for clinical trials.
Current modalities for estimation of liver fat content include conventional ultrasound (US), which is limited by variable accuracy, operator dependency, and its qualitative nature. The measurement of proton density fat fraction (PDFF) by MRI is proving to be an effective method for quantification of hepatic steatosis, but it is expensive and there is variability in results due to dependence on calibration. In a study by Han et al[18], one-dimensional CNN was applied to ultrasound radiofrequency signals for the diagnosis of NAFLD and quantitation of hepatic fat content with an AUC of 0.98 (95%CI: 0.94, 1.00). In diagnosing NAFLD, the model had an accuracy of 96%, sensitivity of 97%, and specificity of 94%, PPV of 97% and NPV of 94%. The ML model also correlated with MRI-PDFF with a Pearson correlation coefficient of 0.85 (P value < 0.001). The same method was applied to animal models in a study by Nguyen et al[19] and it showed that CNN outperformed quantitative ultrasound in differentiating between NAFLD and normal liver. Further support for ML comes from a recent study by Das et al[20] on pediatric patients which used an ensemble model comprising SVM, Neural Net and XG Boost that had an AUC of 0.92 (95%CI, 0.91-0.94) when tested in an external validation cohort.
Nonenhanced CT also remains superior to histopathologic quantification of liver fat content like MRI-PDFF, but it is also more commonly performed in clinical practice for other reasons when compared to MRI. It currently uses a manual region-of-interest (ROI) for estimation of liver fat content. A study by Graffy et al[21] developed a deep-learning based automated liver segmentation tool and applied it to estimate liver fat content using three-dimensional CNN, without having to depend on manual ROI. The pearson correlation coefficient was 0.93. This allows for large population level estimation of liver fat content to determine the prevalence of NAFLD. It would also determine normal liver fat content based on a large sample. Used in combination with other non-invasive modalities such as serum biomarkers, it could help identify patients who will need closer monitoring for NAFLD progression to cirrhosis. In a similar study by Hou et al[22], the automated liver attenuation ROI-based measurement model had a pearson coefficient of 0.94 when compared with manual ROI.
In addition to differentiating healthy liver from NAFLD, ML models have also been used to reduce variability in detecting fibrosis, specifically F2 fibrosis, which is a limiting feature of shear wave elastography. A study by Brattain et al[23] combined the use of shear wave elastography with CNN to better assess F2 fibrosis. This approach not only assessed image quality, but also selected ROI, unlike the previous studies. This ML model detected F2 fibrosis with AUC of 0.89 compared to AUC of 0.74 when image quality and ROI were not incorporated into a ML model. This demonstrates the importance of ML models once again in selecting patients for clinical trials, and in assessing response to treatment.
The rate of SVR for hepatitis C with modern direct acting antiviral (DAA) regimens is estimated to be over 90%; however, variability remains in treatment length and efficacy. Patients with prior DAA exposure, cirrhosis, and other risk factors may require a longer treatment course[18,24]. Machine learning has been applied to predicting treatment response and duration based on patient-specific factors.
Haga et al[24] applied nine machine learning algorithms to identify the optimized combination of HCV genotypic variants that predict SVR after DAA therapy. HCV genomes were sequenced from the serum of 173 patients (including 64 without SVR). The support vector machine algorithm was found to be the most predictive, with a validation accuracy of 0.95. Feldman et al[25] used data from 60 million beneficiaries of a managed care plan (including 3943 cases of hepatitis C who received sofos
The only approved treatment for NAFLD is weight reduction. We were unable to find AI based algorithms and predictive models for NAFLD due to lack of pharmacologic management options.
Among the algorithms described, more complex models performed better, with machine learning consistently outperforming more basic logistic regression models. The highest-performing models incorporated both demographic and radiologic/serologic variables. AI models also predicted complications more accurately than biomarkers and scoring systems like MELD and FIB-4. These models could be used to predict the incidence and prevalence of viral hepatitis in regions without robust, widespread screening programs. Additionally, they could be helpful in the initiation of treatment and predicting response to antivirals for individual patients, for which no gold standard currently exists.
Limitations of the current AI models are notably due to the lack of large scale, randomized controlled trials. Further research is necessary to demonstrate the utility of AI. With further advancements, ML models could potentially be incorporated into all aspects of a patient’s care, from screening the general population for NAFLD or NASH, to monitoring disease progression and treatment response in clinical trials by enhancing classification of steatosis, ballooning, inflammation, and fibrosis. In this regard, more population-based studies are needed to study the applications of ML models in screening. Additionally, large scale, randomized controlled trials are needed to study serologic and histologic markers for disease progression. Further studies are also warranted to explore the potential of ML algorithms to provide target-specific medications, yielding efficacious pharmacotherapy in a disease such as NASH where good treatment options are lacking at this time. Though AI is promising in terms of its potential to develop therapeutic targets, we were unable to find any studies to date describing the use of AI in drug discovery.
Future directions also include using AI to actively improve outcomes with viral hepatitis by increasing adherence to DAAs or identifying individuals at risk for contracting viral hepatitis. Machine learning models could also help identify barriers to accessing treatment.
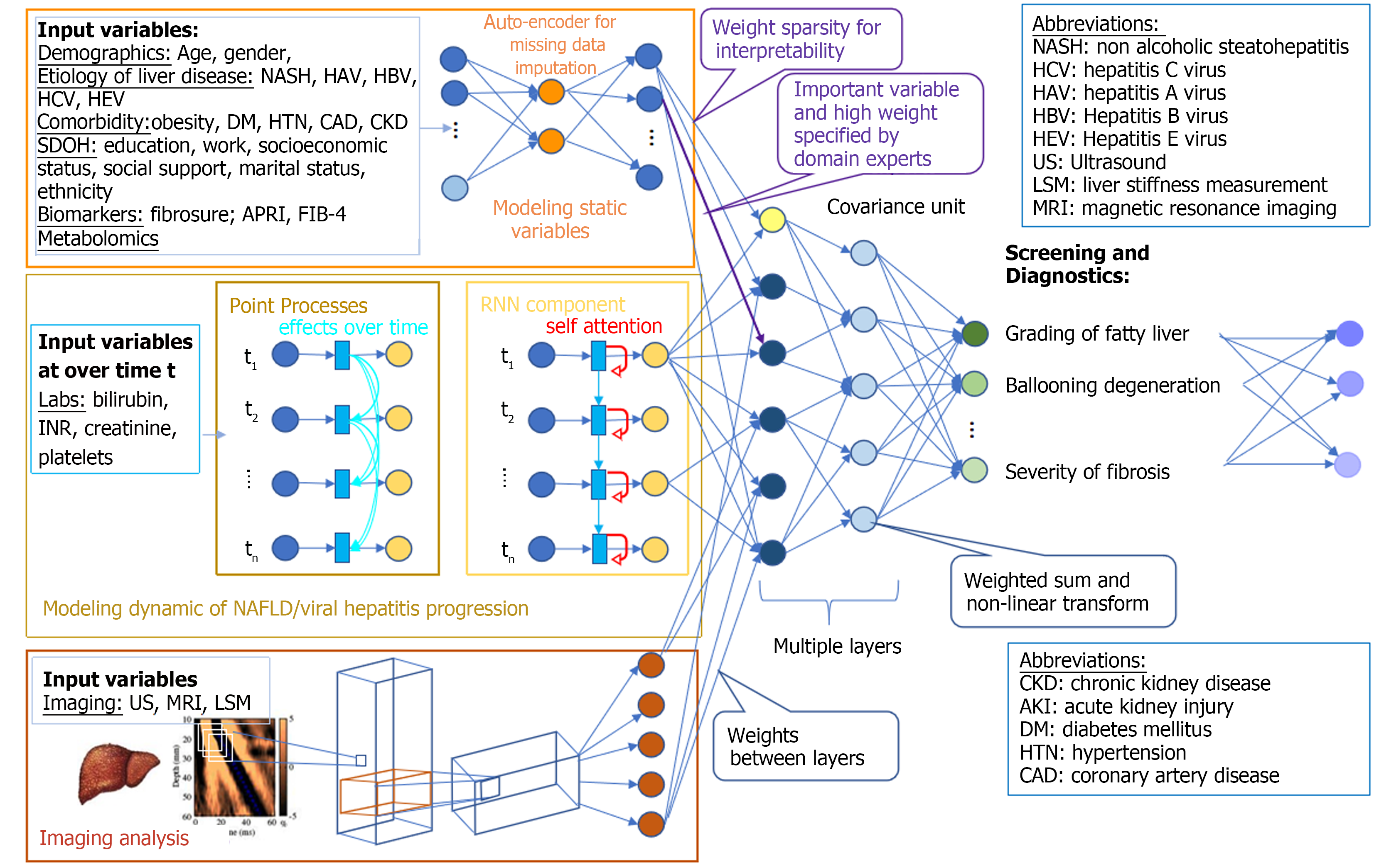
Machine learning models focus on various aspects of liver disease, including demographics, biochemical labs, histologic assessment and patterns, identification of non-invasive biomarkers, and liver imaging techniques (Figure 1). Overall, the studies outlined above are promising in their reliance on non-invasive methods as opposed to conventional liver biopsy to study the stages of fibrosis, as well as their ability to place fibrosis along a continuum and identify markers for disease progression. This could reduce healthcare costs by allowing better selection of patients in whom a liver biopsy is performed. It would also benefit patients by decreasing the number of them who undergo this invasive procedure. AI can also improve efficiency of pathologist and sonographer scoring of samples when added to existing methods. This will allow for a better understanding of the pathophysiology of diseases like NAFLD, which would not only allow for appropriate screening for disease progression, but also improve the ability to develop therapeutic targets.
Provenance and peer review: Invited article; Externally peer reviewed.
Peer-review model: Single blind
Specialty type: Gastroenterology and hepatology
Country/Territory of origin: United States
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): 0
Grade C (Good): C
Grade D (Fair): 0
Grade E (Poor): E
P-Reviewer: Cannella R, Italy; Mandal P, India S-Editor: Liu JH L-Editor: A P-Editor: Cai YX
| 1. | Aggarwal P, Alkhouri N. Artificial Intelligence in Nonalcoholic Fatty Liver Disease: A New Frontier in Diagnosis and Treatment. Clin Liver Dis (Hoboken). 2021;17:392-397. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 5] [Cited by in RCA: 5] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 2. | Forlano R, Mullish BH, Giannakeas N, Maurice JB, Angkathunyakul N, Lloyd J, Tzallas AT, Tsipouras M, Yee M, Thursz MR, Goldin RD, Manousou P. High-Throughput, Machine Learning-Based Quantification of Steatosis, Inflammation, Ballooning, and Fibrosis in Biopsies From Patients With Nonalcoholic Fatty Liver Disease. Clin Gastroenterol Hepatol. 2020;18:2081-2090.e9. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 54] [Cited by in RCA: 94] [Article Influence: 18.8] [Reference Citation Analysis (0)] |
| 3. | Perakakis N, Polyzos SA, Yazdani A, Sala-Vila A, Kountouras J, Anastasilakis AD, Mantzoros CS. Non-invasive diagnosis of non-alcoholic steatohepatitis and fibrosis with the use of omics and supervised learning: A proof of concept study. Metabolism. 2019;101:154005. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 96] [Cited by in RCA: 99] [Article Influence: 16.5] [Reference Citation Analysis (0)] |
| 4. | Leow WQ, Bedossa P, Liu F, Wei L, Lim KH, Wan WK, Ren Y, Chang JP, Tan CK, Wee A, Goh GB. An Improved qFibrosis Algorithm for Precise Screening and Enrollment into Non-Alcoholic Steatohepatitis (NASH) Clinical Trials. Diagnostics (Basel). 2020;10. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 6] [Cited by in RCA: 13] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 5. | Yehia BR, Schranz AJ, Umscheid CA, Lo Re V 3rd. The treatment cascade for chronic hepatitis C virus infection in the United States: a systematic review and meta-analysis. PLoS One. 2014;9:e101554. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 360] [Cited by in RCA: 354] [Article Influence: 32.2] [Reference Citation Analysis (0)] |
| 6. | Zheng Y, Zhang L, Zhu X, Guo G. A comparative study of two methods to predict the incidence of hepatitis B in Guangxi, China. PLoS One. 2020;15:e0234660. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 8] [Cited by in RCA: 10] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 7. | Doyle OM, Leavitt N, Rigg JA. Finding undiagnosed patients with hepatitis C infection: an application of artificial intelligence to patient claims data. Sci Rep. 2020;10:10521. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 20] [Cited by in RCA: 20] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 8. | Lu XJ, Yang XJ, Sun JY, Zhang X, Yuan ZX, Li XH. FibroBox: a novel noninvasive tool for predicting significant liver fibrosis and cirrhosis in HBV infected patients. Biomark Res. 2020;8:48. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 5] [Cited by in RCA: 8] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 9. | Zheng MH, Shi KQ, Lin XF, Xiao DD, Chen LL, Liu WY, Fan YC, Chen YP. A model to predict 3-month mortality risk of acute-on-chronic hepatitis B liver failure using artificial neural network. J Viral Hepat. 2013;20:248-255. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 44] [Cited by in RCA: 31] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 10. | Huo Y, Terry JG, Wang J, Nair S, Lasko TA, Freedman BI, Carr JJ, Landman BA. Fully automatic liver attenuation estimation combing CNN segmentation and morphological operations. Med Phys. 2019;46:3508-3519. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 15] [Cited by in RCA: 30] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 11. | Wang K, Lu X, Zhou H, Gao Y, Zheng J, Tong M, Wu C, Liu C, Huang L, Jiang T, Meng F, Lu Y, Ai H, Xie XY, Yin LP, Liang P, Tian J, Zheng R. Deep learning Radiomics of shear wave elastography significantly improved diagnostic performance for assessing liver fibrosis in chronic hepatitis B: a prospective multicentre study. Gut. 2019;68:729-741. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 226] [Cited by in RCA: 352] [Article Influence: 58.7] [Reference Citation Analysis (1)] |
| 12. | Qu H, Minacapelli CD, Tait C, Gupta K, Bhurwal A, Catalano C, Dafalla R, Metaxas D, Rustgi VK. Training of computational algorithms to predict NAFLD activity score and fibrosis stage from liver histopathology slides. Comput Methods Programs Biomed. 2021;207:106153. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 11] [Cited by in RCA: 18] [Article Influence: 4.5] [Reference Citation Analysis (0)] |
| 13. | Taylor-Weiner A, Pokkalla H, Han L, Jia C, Huss R, Chung C, Elliott H, Glass B, Pethia K, Carrasco-Zevallos O, Shukla C, Khettry U, Najarian R, Taliano R, Subramanian GM, Myers RP, Wapinski I, Khosla A, Resnick M, Montalto MC, Anstee QM, Wong VW, Trauner M, Lawitz EJ, Harrison SA, Okanoue T, Romero-Gomez M, Goodman Z, Loomba R, Beck AH, Younossi ZM. A Machine Learning Approach Enables Quantitative Measurement of Liver Histology and Disease Monitoring in NASH. Hepatology. 2021;74:133-147. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 48] [Cited by in RCA: 125] [Article Influence: 31.3] [Reference Citation Analysis (0)] |
| 14. | Gawrieh S, Sethunath D, Cummings OW, Kleiner DE, Vuppalanchi R, Chalasani N, Tuceryan M. Automated quantification and architectural pattern detection of hepatic fibrosis in NAFLD. Ann Diagn Pathol. 2020;47:151518. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 20] [Cited by in RCA: 44] [Article Influence: 8.8] [Reference Citation Analysis (0)] |
| 15. | Atabaki-Pasdar N, Ohlsson M, Viñuela A, Frau F, Pomares-Millan H, Haid M, Jones AG, Thomas EL, Koivula RW, Kurbasic A, Mutie PM, Fitipaldi H, Fernandez J, Dawed AY, Giordano GN, Forgie IM, McDonald TJ, Rutters F, Cederberg H, Chabanova E, Dale M, Masi F, Thomas CE, Allin KH, Hansen TH, Heggie A, Hong MG, Elders PJM, Kennedy G, Kokkola T, Pedersen HK, Mahajan A, McEvoy D, Pattou F, Raverdy V, Häussler RS, Sharma S, Thomsen HS, Vangipurapu J, Vestergaard H, 't Hart LM, Adamski J, Musholt PB, Brage S, Brunak S, Dermitzakis E, Frost G, Hansen T, Laakso M, Pedersen O, Ridderstråle M, Ruetten H, Hattersley AT, Walker M, Beulens JWJ, Mari A, Schwenk JM, Gupta R, McCarthy MI, Pearson ER, Bell JD, Pavo I, Franks PW. Predicting and elucidating the etiology of fatty liver disease: A machine learning modeling and validation study in the IMI DIRECT cohorts. PLoS Med. 2020;17:e1003149. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 37] [Cited by in RCA: 59] [Article Influence: 11.8] [Reference Citation Analysis (0)] |
| 16. | Docherty M, Regnier SA, Capkun G, Balp MM, Ye Q, Janssens N, Tietz A, Löffler J, Cai J, Pedrosa MC, Schattenberg JM. Development of a novel machine learning model to predict presence of nonalcoholic steatohepatitis. J Am Med Inform Assoc. 2021;28:1235-1241. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 10] [Cited by in RCA: 28] [Article Influence: 7.0] [Reference Citation Analysis (0)] |
| 17. | Ma H, Xu CF, Shen Z, Yu CH, Li YM. Application of Machine Learning Techniques for Clinical Predictive Modeling: A Cross-Sectional Study on Nonalcoholic Fatty Liver Disease in China. Biomed Res Int. 2018;2018:4304376. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 92] [Cited by in RCA: 70] [Article Influence: 10.0] [Reference Citation Analysis (0)] |
| 18. | Han A, Byra M, Heba E, Andre MP, Erdman JW Jr, Loomba R, Sirlin CB, O'Brien WD Jr. Noninvasive Diagnosis of Nonalcoholic Fatty Liver Disease and Quantification of Liver Fat with Radiofrequency Ultrasound Data Using One-dimensional Convolutional Neural Networks. Radiology. 2020;295:342-350. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 45] [Cited by in RCA: 84] [Article Influence: 16.8] [Reference Citation Analysis (0)] |
| 19. | Nguyen TN, Podkowa AS, Park TH, Miller RJ, Do MN, Oelze ML. Use of a convolutional neural network and quantitative ultrasound for diagnosis of fatty liver. Ultrasound Med Biol. 2021;47:556-568. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 6] [Cited by in RCA: 6] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 20. | Das A, Connell M, Khetarpal S. Digital image analysis of ultrasound images using machine learning to diagnose pediatric nonalcoholic fatty liver disease. Clin Imaging. 2021;77:62-68. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3] [Cited by in RCA: 4] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 21. | Graffy PM, Sandfort V, Summers RM, Pickhardt PJ. Automated Liver Fat Quantification at Nonenhanced Abdominal CT for Population-based Steatosis Assessment. Radiology. 2019;293:334-342. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 53] [Cited by in RCA: 101] [Article Influence: 16.8] [Reference Citation Analysis (0)] |
| 22. | Hou Y, Zhang Q, Gao F, Mao D, Li J, Gong Z, Luo X, Chen G, Li Y, Yang Z, Sun K, Wang X. Artificial neural network-based models used for predicting 28- and 90-day mortality of patients with hepatitis B-associated acute-on-chronic liver failure. BMC Gastroenterol. 2020;20:75. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 3] [Cited by in RCA: 19] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 23. | Brattain LJ, Telfer BA, Dhyani M, Grajo JR, Samir AE. Objective Liver Fibrosis Estimation from Shear Wave Elastography. Annu Int Conf IEEE Eng Med Biol Soc. 2018;2018:1-5. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 10] [Cited by in RCA: 16] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 24. | Haga H, Sato H, Koseki A, Saito T, Okumoto K, Hoshikawa K, Katsumi T, Mizuno K, Nishina T, Ueno Y. A machine learning-based treatment prediction model using whole genome variants of hepatitis C virus. PLoS One. 2020;15:e0242028. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 10] [Cited by in RCA: 16] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 25. | Feldman TC, Dienstag JL, Mandl KD, Tseng YJ. Machine-learning-based predictions of direct-acting antiviral therapy duration for patients with hepatitis C. Int J Med Inform. 2021;154:104562. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1] [Cited by in RCA: 5] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 26. | Wei W, Wu X, Zhou J, Sun Y, Kong Y, Yang X. Noninvasive Evaluation of Liver Fibrosis Reverse Using Artificial Neural Network Model for Chronic Hepatitis B Patients. Comput Math Methods Med. 2019;2019:7239780. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 6] [Cited by in RCA: 10] [Article Influence: 1.7] [Reference Citation Analysis (0)] |