Published online Sep 28, 2020. doi: 10.35712/aig.v1.i3.51
Peer-review started: June 29, 2020
First decision: July 28, 2020
Revised: September 25, 2020
Accepted: September 27, 2020
Article in press: September 27, 2020
Published online: September 28, 2020
Processing time: 91 Days and 9.4 Hours
Artificial intelligence (AI) is gaining incredible momentum as a companion diagnostic in a number of fields in oncology. In the present mini-review, we summarize the main uses and findings of AI applied to the analysis of digital histopathological images of slides from colorectal cancer (CRC) patients. Machine learning tools have been developed to automatically and objectively recognize specific CRC subtypes, such as those with microsatellite instability and high lymphocyte infiltration that would optimally respond to specific therapies. Also, AI-based classification in distinct prognostic groups with no studies of the basic biological features of the tumor have been attempted in a methodological approach that we called “biology-agnostic”.
Core Tip: Artificial intelligence (AI) is gaining incredible momentum as a companion diagnostic in a number of fields in oncology. In the present mini-review, we summarize the main uses and findings of AI applied to the analysis of digital histopathological images of slides from colorectal cancer patients.
- Citation: Formica V, Morelli C, Riondino S, Renzi N, Nitti D, Roselli M. Artificial intelligence for the study of colorectal cancer tissue slides. Artif Intell Gastroenterol 2020; 1(3): 51-59
- URL: https://www.wjgnet.com/2644-3236/full/v1/i3/51.htm
- DOI: https://dx.doi.org/10.35712/aig.v1.i3.51
Artificial intelligence (AI) refers to any form of machine activity that attempts to mimic human intelligence. The main abilities of the human intelligence objective of AI research are performing complex tasks and achieving goals through processes that are typical of the human cognitive functions, such as “learning” [which in this case is termed “machine learning” (ML)] and “problem solving”[1-4].
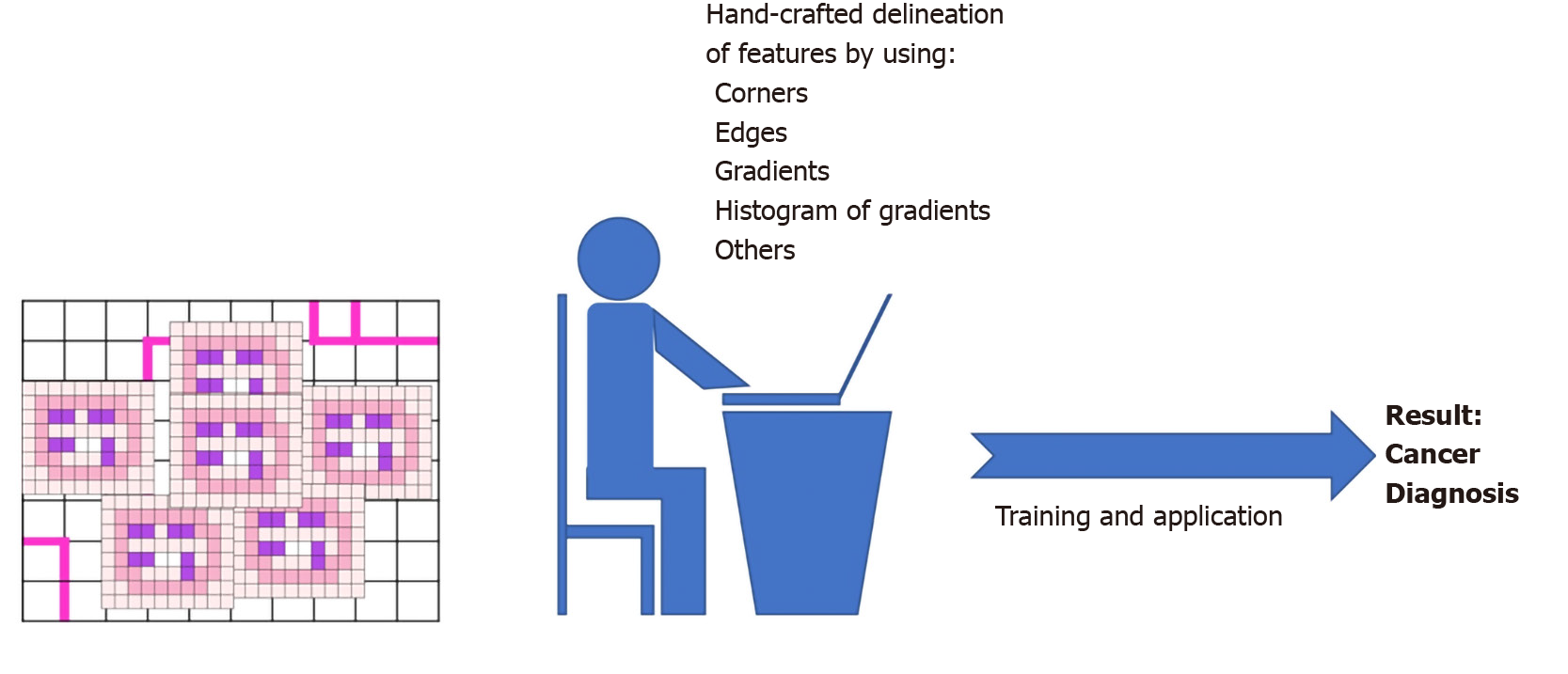
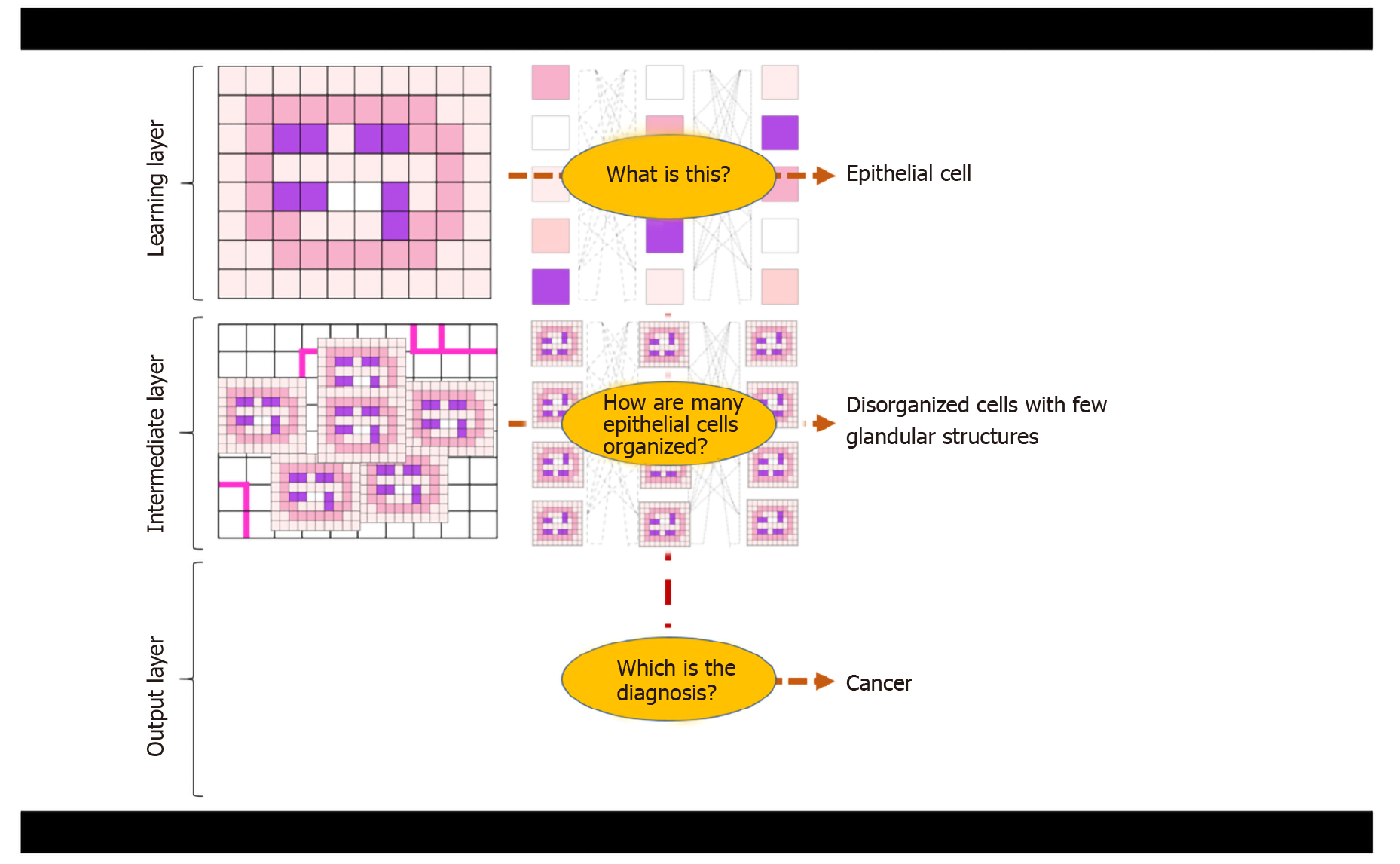
“Deep” learning (DL) is a basic method of ML. It is based on specific computer algorithms that recall neural networking and that try to model data-rich input for the identification of general and meaningful features/patterns or for the prediction of specific outcomes[5]. The term “deep” refers to the organization in layers of the artificial neural network, with deep layers of analysis being crucial to infer more complex and higher-level characteristics up to the final layer of the label/output that is pursued (Figure 1 and Figure 2).
ML has been successfully applied in the so-called digital pathology[6-8], where histological images are deeply analyzed in every single pixel for color and light intensity by computer algorithms, to deduce a final diagnosis or prognostic/predictive feature.
The use of AI in cancer histopathology has been possible since the advent of histologic images digitalization[6]. Widely available image scanners are currently used in clinical practice, also for the acquisition of initial hematoxylin and eosin (HE) stained slides. Mukhopadhyay et al[9] performed a comparison between digital pathology and classical microscopy in a large multi-center cohort of patients and confirmed the exceptional utility of the digital support. Early studies were based on the analysis and segmetation of specific slide regions, such as the tumor center, margins, stroma or others, mainly in order to recognize specific objects and structures[10]. More recently, with the ability of the computational analysis of higher digital dimension, automated analysis of whole HE-stained tumor tissue slides has been possible and inference of patient outcome and prognosis has been attempted[11,12]. In the present mini-review, we summarize the latest findings for the application of AI tools for the digital pathology of colorectal cancer (CRC). In general, two research fields have been identified: AI instruments for identification of specific biologic features and AI instruments for prediction of patient outcome independently of the cancer biology.
One typical approach of AI in pathology is to use pre-defined key image features as building block for AI-algorithm development. These are usually termed “hand-crafted” features and are engineered based on biological insights. Typical examples of “hand-crafted” features are shape and orientation of the cell nuclei[13], that can be used to recognize cancer cells or tumor infiltrating lymphocytes or other cell types.
Shape and orientation of cancer cells are among the most commonly assessed variables to predict patient outcome[14]. A predictive, ML-based algorithm for lymph node metastasis (LNM) on whole slide images in early CRC with pathological submucosal invasion (pT1) has proven useful when using data from cancer cell morphology[16]. After delineating cancer cell regions by using immunohistochemistry for cytokeratins, Takamatsu et al[15] analyzed tissue slides from over 300 CRC patients diagnosed as pT1, looking for predictors of LNM. They used the popular ImageJ software, released by the United States’ National Institutes of Health, and JPEG images. Digital parameters extracted from the slides essentially referred to shape, circularity, orientation and organization of cancer cells (e.g., Feret’s diameter), and these were used to feed a supervised ML algorithm based on random forest classifier. AI prediction was proved to be superior to human conventional assessment, with a discriminatory power of area under the curve (commonly known as AUC) 94% vs 83%, respectively.
Sailem et al[16] proposed a data-rich integrated platform that utilized a ML approach to conjugate high-throughput gene perturbation analysis with morphological features of CRC and, in particular, single cell morphology and cell population organization. After identification of TGFβ and WNT signaling genes and olfactory receptors genes as key altered genes in CRC, they validated their association with single cell and cell population morphology and grade of differentiation in CRC. Those specific gene alterations were associated to abnormal organization of HCT116 CRC cell cultures and also to specific CRC molecular subtypes (the well-known consensus molecular subtypes – CSM[17]) in over 550 patients from The Cancer Genome Atlas (referred to as TCGA) database.
In recent years, the development of immunotherapy has revolutionized the care of cancer[18], and great attention has been placed upon assessment of the immune response surrounding the tumor tissue[19]. Most reports are focused on specific immune cells and chiefly on tumor-induced cytotoxic T cells. Indeed, a high T-cell infiltration appears to be associated with a decreased risk of tumor dissemination and improved survival in most solid tumors. Väyrynen et al[20] have recently proposed an AI-based assessment of CRC slides, simultaneously evaluating different cell populations involved in immune response against the tumor. Scanned HE-stained images of CRC cases from two United States’ prospective cohorts [the Nurses’ Health Study (referred to as NHS), and the Health Professionals Follow-up Study (referred to as HPFS)] were analyzed with an open source software (QuPath v0.1.2), enabling the recognition of cancer epithelial cells and four immune cell types with distinct morphological features: lymphocytes, plasma cells, eosinophils, and neutrophils. Moreover, QuPath v0.1.2 was able to distinguish between two tissue regions: the intraepithelial region and stromal region[21]. Densities of immune cells in the intraepithelial and stromal regions, together with a function (G-cross function) measuring the proximity of immune cells to cancer cells (expressed as AUC within a distance of 20 μm) were calculated. Findings from NHS and HPFS cohorts were validated in a set of 570 CRC from TGCA with available digitalized pathological images in the data portal. Automated quantification of immune cells was compared to manual count by pathologists.
A high correlation between measurement of immune cell densities performed by automated AI classifiers and manual counts performed by experienced pathologist was found (Spearman’s rho 0.71-0.96). Moreover AI-determined stromal density of lymphocytes and eosinophils were independently correlated with survival in a multivariable Cox regression analysis, with higher densities associated with longer cancer-specific survival (P for trend across quartiles of density < 0.001 for both cell types). The multivariate analysis included very important molecular factors which were available for the post-hoc analysis of NHS and HPFS populations and partly associated with immune response, such as microsatellite instability (MSI), CpG island methylator phenotype, KRAS, BRAF, and PIK3CA mutations, and LINE-1 methylation level. High G cross function for Tumor: Lymphocyte and Tumor: Eosinophil proximity was also associated with longer cancer-specific survival (Ptrend 0.002 and < 0.001, respectively). High stromal eosinophil density was associated with better cancer-specific survival also in the TGCA cohort (Ptrend < 0.001).
AI can also improve CRC cancer staging and, consequently, patient prognosis. Reichling and co-workers[22], using a LASSO algorithm, DGMate (DiGital tuMor pArameTErs), combined the analysis of tumor stroma and tumor cell intrinsic variables, in association with immune cell infiltrate of tumor samples from stage III CRC patients of the PETACC8 study cohort, and were able to detect digital parameters within the tumor cells related to patients’ outcomes[23]. Similarly, clinically relevant information were gained in the study of Yoo et al[24], in which ML-based analysis was applied to study the quantitative parameters of immune infiltrate within the tumor immune microenvironment. By quantifying intraepithelial tumor-infiltrating lymphocytes, stromal tumor-infiltrating lymphocytes, and the tumor-stroma ratio from CD3 and CD8 immunohistochemical stained whole-slide images, the authors classified five CRC subgroups with distinctive biological features and different prognostic behaviors. Indeed, the CD3+ and CD8+ T-cell infiltration is considered an important independent prognostic factor predicting CRC patient survival[25]. An automated image analysis–based workflow quantifying the tumor-infiltrating immune cells and tumor budding, at the invasive front, combined with a ML approach, demonstrated that the spatial association of lymphocytes and tumor buddings could provide a high prognostic significance in stage II CRC patients[26].
MSI-high (MSI-H) tumors are a peculiar molecular subgroup of CRC characterized by deficient expression of mismatch repair proteins (dMMR phenotype)[27]. MSI-H is associated with the inherited Lynch syndrome and found to be a marker of increased tumor mutational burden, neoantigen generation and exceptional sensitivity to immune checkpoint inhibitors[28]. It accounts for 4%-5% of metastatic CRC and its early recognition is now crucial to avoid useless treatment with conventional chemotherapy and to tailor adequate and effective immunotherapy. Kather et al[29] have recently built an AI-classifier of conventionally HE-stained slides of gastric and CRC for the detection of MSI-H. The model used Resnet18, a residual learning convolutional neural network (CNN), which was trained and validated, both internally and externally, in a very large cohort of gastrointestinal cancer. In particular, Resnet18 was initially trained to automatically identify tumor areas within the normal tissue background in common histological slides. In a second step, it was trained to define the degree of MSI using TCGA bank, and specifically 315 and 360 formalin-fixed paraffin embedded gastric and CRC specimens, respectively, and 378 snap-frozen CRC samples. TGCA of stomach cancer was 80% from non-Asian patients, and authors tested the possible influence of ethnicity on the model performance by validating the AI classifier in an external cohort of 185 Japanese patients.
The discriminatory power of the AI tool for the MSI-H status identification was impressively high (AUC around 80% in almost all experiments) across all the different sets: gastric vs CRC tissues, Asian vs non-Asian patients, and formalin-fixed paraffin embedded vs snap-frozen samples. Moreover, it was found that minimal required number of histological “tiles” on which to train and interrogate the algorithm (the ML process starts by fragmenting the entire histologic digital image in multiple elementary color-normalized slide tiles) was as small as those encompassed in a common core needle biopsy (performance plateaued at approximately 100 “slide tiles” of 256 μm edge length). The logical next step in this field will be to test the predictive value of AI-based MSI-H diagnosis for immunotherapy efficacy.
In another study by the same research group, a DL model for MSI definition was trained and validated in a cohort of nearly 9000 patients from Germany, Netherlands, United Kingdom and United States by using standard HE-stained slides and immunohistochemistry for mismatch repair proteins or conventional genetic test of microsatellite regions as ground truth for MSI status. In the validation step, the DL tool demonstrated an impressively high ability to correctly classify MSI-H patients with a discriminatory power > 95% (AUROC 0.95-096)[30].
In CRC, the so-called desmoplastic reaction, i.e. a pronounced stromal tissue growth within the tumor mass, has been associated with reduced prognosis[31]. Desmoplastic reaction is driven by differentiated cells responsible for the deposition of extracellular fibrotic material, the cancer-associated fibroblasts (CAFs). Kather et al[32] have used DL for automatic quantification of stromal tissue proportion in CRC tissues and set up a “deep stroma score”. They first trained the DL instrument with over 100000 digital image patches of HE-stained tumors from 86 CRC patients where stromal and non-stromal areas were hand-delineated. The DL tool so obtained was then tested in an independent set of over 7000 images from 25 patients, where it demonstrated an accuracy of > 94%.
The tool, that was able to recognize tissue-specific features, was finally used to produce the “deep stroma score” which was calculated for a cohort of 500 colorectal patients from the TCGA repository and correlated with survival. The stroma score significantly and independently associated with overall survival with a hazard ratio of 1.99, P = 0.003. In the same patient group, manual stroma quantification by experienced pathologists or stromal assessment by means of gene expression profiles attributable to CAFs had an inferior performance in terms of survival prediction across the different tumor stages (from I to IV). The DL stroma score yielded similar prognostic efficacy in an independent validation cohort of over 400 patients from Germany: hazard ratio for overall survival 2.29, P = 0.0004.
DL is a novel type of ML that uses a more “blind” approach to analyze the input data with no pre-definition of hand-crafted features to train the model. An extreme way to apply DL is the direct correlation of the digital input data to the final outcome, for example risk of disease relapse vs definitive care after surgical removal of the tumor, with no specific “biological insights” fed into the machine. Skrede et al[33] have applied an advanced DL method to analyze whole-slide image, using CNN. CNN makes use of mathematical convolutions, i.e. resulting mathematical functions obtained by combining more primary functions reversed and shifted in time. In Skrede et al[33]’s experiment, no clinical or biological information (such as grade of histological differentiation, neurovascular invasion, depth of tumor infiltration within the colon wall, or others) was fed into the model, and the final objective was to identify “biology-agnostic” digital image profiles (purely based on patterns of pixel color or intensity of the digital images) predictive of CRC specific survival in surgically-resected patients (stages II and III). Four large cohorts of approximately 1000 patients each were used (training, tuning, test and validation cohorts). The CNN-based automatic prognostic classifier, that authors called DoMore-v1-CRC, was initially trained for outcome prediction in a cohort of patients constituted of two subgroups with “pre-labeled” distinct outcomes: good (survival > 2 years) vs poor (relapse within 6 mo) outcome. DoMore-v1-CRC was then tuned, tested and validated in cohorts with non-prelabeled outcome to define its prognostic ability. The DoMore-v1-CRC had an output of three possible classes, good vs uncertain vs poor prognosis. The DoMore-v1-CRC–based classification had an independent prognostic value in a multivariable survival analysis that included and adjusted for well-known and broadly-used clinical and biological factors, such as pN stage, pT stage, lymphatic invasion, and venous vascular invasion (hazard ratio for DoMore-v1-CRC-defined poor vs good prognosis 3.04, P < 0.0001). Accuracy in predicting cancer-specific survival was around 70% in all cohorts. The prognostic power remained high for both stage II and stage III tumors, and for groups of slides prepared in different laboratories, thus confirming its robustness against inter-laboratory variability in tissue preparation and staining. Moreover, the prognostic yield remained high when two different image scanners were compared (NanoZoomer XR vs Aperio AT2 scanner). Interestingly, even though derived from a biology-agnostic approach, DoMore-v1-CRC significantly correlated with clinical and biological markers, such as age, pN stage, pT stage, histological grade, tumor sidedness, BRAF mutation, and microsatellite instability.
Even if still at the early stages, AI in digital pathology is increasingly gaining momentum, for a number of reasons: (1) worldwide transferability; (2) high-dimension of analyzable data; and (3) automated procedural approaches. Far from being conceived as substitutive of human intelligence, it can be seen as a useful companion diagnostic in oncology and as pre-screening/pre-selection tool. Both biology-driven and biology-agnostic algorithms have been pursued in the implementation of AI in the CRC pathology field (Table 1).
| Target | Description | Ref. |
| Cancer cell shape and organization to predict N+ | Digital assessment of cancer cells using Feret’s diameters allows to predict lymph node metastasis in pT1 colon cancer | [11] |
| Assessment of anti-cancer immune response | Simultaneous assessment of all immune cell subpopulations and demonstration that eosinophils, other than T cells, may play a role in CRC immune response | [12,14,15,17] |
| Identification of Microsatellite instable tumors | Rapid and large size screening of histopathologic features in conventional HE-stained slides increasing the probability of being a MSI-H tumor, without the need of specific immunohistochemical or molecular testing | [18] |
| Quantification of stroma within the tumor | Algorithms for tissue –specific recognition, even when sparse within the tumor mass have been development. These algorithms have allowed the validation of “deep stroma score” which is significantly associated with survival in CRC | [20] |
| Biology-agnostic prediction of survival | Development of tissue “digital” profiles without specific underlying biologic background or significance that are predictive of distinct survivals, bad vs good outcome | [21] |
The main challenges for AI-supported digital pathology are currently to demonstrate that can diffusely be applied and significantly help in the clinical decision-making especially as predictor of efficacy of specific therapeutic options. Such a demonstration can only come with an enhancement of the AI application itself to different cancer patient settings and different clinical scenarios and problem-solving tasks. Future directions rely on the prospective validation of AI-based tools in randomized phase III trials, which delineates the so-called “IA level” of evidence.
Manuscript source: Invited manuscript
Specialty type: Oncology
Country/Territory of origin: Italy
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): 0
Grade C (Good): C, C
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Cheng X, Lalmuanawma S S-Editor: Wang JL L-Editor: Filipodia P-Editor: Ma YJ
| 1. | Bi WL, Hosny A, Schabath MB, Giger ML, Birkbak NJ, Mehrtash A, Allison T, Arnaout O, Abbosh C, Dunn IF, Mak RH, Tamimi RM, Tempany CM, Swanton C, Hoffmann U, Schwartz LH, Gillies RJ, Huang RY, Aerts HJWL. Artificial intelligence in cancer imaging: Clinical challenges and applications. CA Cancer J Clin. 2019;69:127-157. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 848] [Cited by in RCA: 820] [Article Influence: 136.7] [Reference Citation Analysis (3)] |
| 2. | Dasgupta P. Science, technology and artificial intelligence. BJU Int. 2018;122:913. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2] [Cited by in RCA: 2] [Article Influence: 0.3] [Reference Citation Analysis (0)] |
| 3. | He J, Baxter SL, Xu J, Xu J, Zhou X, Zhang K. The practical implementation of artificial intelligence technologies in medicine. Nat Med. 2019;25:30-36. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 597] [Cited by in RCA: 952] [Article Influence: 158.7] [Reference Citation Analysis (0)] |
| 4. | Le Berre C, Sandborn WJ, Aridhi S, Devignes MD, Fournier L, Smaïl-Tabbone M, Danese S, Peyrin-Biroulet L. Application of Artificial Intelligence to Gastroenterology and Hepatology. Gastroenterology. 2020;158:76-94.e2. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 230] [Cited by in RCA: 326] [Article Influence: 65.2] [Reference Citation Analysis (1)] |
| 5. | Sherbet GV, Woo WL, Dlay S. Application of Artificial Intelligence-based Technology in Cancer Management: A Commentary on the Deployment of Artificial Neural Networks. Anticancer Res. 2018;38:6607-6613. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 21] [Cited by in RCA: 20] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 6. | Bera K, Schalper KA, Rimm DL, Velcheti V, Madabhushi A. Artificial intelligence in digital pathology - new tools for diagnosis and precision oncology. Nat Rev Clin Oncol. 2019;16:703-715. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 858] [Cited by in RCA: 844] [Article Influence: 140.7] [Reference Citation Analysis (0)] |
| 7. | Niazi MKK, Parwani AV, Gurcan MN. Digital pathology and artificial intelligence. Lancet Oncol. 2019;20:e253-e261. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 507] [Cited by in RCA: 631] [Article Influence: 105.2] [Reference Citation Analysis (0)] |
| 8. | Ruffle JK, Farmer AD, Aziz Q. Artificial Intelligence-Assisted Gastroenterology- Promises and Pitfalls. Am J Gastroenterol. 2019;114:422-428. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 71] [Cited by in RCA: 91] [Article Influence: 15.2] [Reference Citation Analysis (0)] |
| 9. | Mukhopadhyay S, Feldman MD, Abels E, Ashfaq R, Beltaifa S, Cacciabeve NG, Cathro HP, Cheng L, Cooper K, Dickey GE, Gill RM, Heaton RP, Kerstens R, Lindberg GM, Malhotra RK, Mandell JW, Manlucu ED, Mills AM, Mills SE, Moskaluk CA, Nelis M, Patil DT, Przybycin CG, Reynolds JP, Rubin BP, Saboorian MH, Salicru M, Samols MA, Sturgis CD, Turner KO, Wick MR, Yoon JY, Zhao P, Taylor CR. Whole Slide Imaging Versus Microscopy for Primary Diagnosis in Surgical Pathology: A Multicenter Blinded Randomized Noninferiority Study of 1992 Cases (Pivotal Study). Am J Surg Pathol. 2018;42:39-52. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 213] [Cited by in RCA: 257] [Article Influence: 32.1] [Reference Citation Analysis (0)] |
| 10. | Al-Kofahi Y, Lassoued W, Lee W, Roysam B. Improved automatic detection and segmentation of cell nuclei in histopathology images. IEEE Trans Biomed Eng. 2010;57:841-852. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 475] [Cited by in RCA: 327] [Article Influence: 20.4] [Reference Citation Analysis (0)] |
| 11. | Lu C, Romo-Bucheli D, Wang X, Janowczyk A, Ganesan S, Gilmore H, Rimm D, Madabhushi A. Nuclear shape and orientation features from H&E images predict survival in early-stage estrogen receptor-positive breast cancers. Lab Invest. 2018;98:1438-1448. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 61] [Cited by in RCA: 84] [Article Influence: 12.0] [Reference Citation Analysis (0)] |
| 12. | Corredor G, Wang X, Zhou Y, Lu C, Fu P, Syrigos K, Rimm DL, Yang M, Romero E, Schalper KA, Velcheti V, Madabhushi A. Spatial Architecture and Arrangement of Tumor-Infiltrating Lymphocytes for Predicting Likelihood of Recurrence in Early-Stage Non-Small Cell Lung Cancer. Clin Cancer Res. 2019;25:1526-1534. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 144] [Cited by in RCA: 170] [Article Influence: 28.3] [Reference Citation Analysis (0)] |
| 13. | Sirinukunwattana K, Ahmed Raza SE, Yee-Wah Tsang, Snead DR, Cree IA, Rajpoot NM. Locality Sensitive Deep Learning for Detection and Classification of Nuclei in Routine Colon Cancer Histology Images. IEEE Trans Med Imaging. 2016;35:1196-1206. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 784] [Cited by in RCA: 511] [Article Influence: 56.8] [Reference Citation Analysis (0)] |
| 14. | Gisselsson D, Björk J, Höglund M, Mertens F, Dal Cin P, Akerman M, Mandahl N. Abnormal nuclear shape in solid tumors reflects mitotic instability. Am J Pathol. 2001;158:199-206. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 152] [Cited by in RCA: 169] [Article Influence: 7.0] [Reference Citation Analysis (0)] |
| 15. | Takamatsu M, Yamamoto N, Kawachi H, Chino A, Saito S, Ueno M, Ishikawa Y, Takazawa Y, Takeuchi K. Prediction of early colorectal cancer metastasis by machine learning using digital slide images. Comput Methods Programs Biomed. 2019;178:155-161. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 35] [Cited by in RCA: 47] [Article Influence: 7.8] [Reference Citation Analysis (0)] |
| 16. | Sailem HZ, Rittscher J, Pelkmans L. KCML: a machine-learning framework for inference of multi-scale gene functions from genetic perturbation screens. Mol Syst Biol. 2020;16:e9083. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 8] [Cited by in RCA: 7] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 17. | Guinney J, Dienstmann R, Wang X, de Reyniès A, Schlicker A, Soneson C, Marisa L, Roepman P, Nyamundanda G, Angelino P, Bot BM, Morris JS, Simon IM, Gerster S, Fessler E, De Sousa E Melo F, Missiaglia E, Ramay H, Barras D, Homicsko K, Maru D, Manyam GC, Broom B, Boige V, Perez-Villamil B, Laderas T, Salazar R, Gray JW, Hanahan D, Tabernero J, Bernards R, Friend SH, Laurent-Puig P, Medema JP, Sadanandam A, Wessels L, Delorenzi M, Kopetz S, Vermeulen L, Tejpar S. The consensus molecular subtypes of colorectal cancer. Nat Med. 2015;21:1350-1356. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 3408] [Cited by in RCA: 3558] [Article Influence: 355.8] [Reference Citation Analysis (0)] |
| 18. | Waldman AD, Fritz JM, Lenardo MJ. A guide to cancer immunotherapy: from T cell basic science to clinical practice. Nat Rev Immunol. 2020;. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2409] [Cited by in RCA: 2667] [Article Influence: 533.4] [Reference Citation Analysis (0)] |
| 19. | Byrne A, Savas P, Sant S, Li R, Virassamy B, Luen SJ, Beavis PA, Mackay LK, Neeson PJ, Loi S. Tissue-resident memory T cells in breast cancer control and immunotherapy responses. Nat Rev Clin Oncol. 2020;17:341-348. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 80] [Cited by in RCA: 202] [Article Influence: 40.4] [Reference Citation Analysis (0)] |
| 20. | Väyrynen JP, Lau MC, Haruki K, Väyrynen SA, Dias Costa A, Borowsky J, Zhao M, Fujiyoshi K, Arima K, Twombly TS, Kishikawa J, Gu S, Aminmozaffari S, Shi S, Baba Y, Akimoto N, Ugai T, Da Silva A, Song M, Wu K, Chan AT, Nishihara R, Fuchs CS, Meyerhardt JA, Giannakis M, Ogino S, Nowak JA. Prognostic Significance of Immune Cell Populations Identified by Machine Learning in Colorectal Cancer Using Routine Hematoxylin and Eosin-Stained Sections. Clin Cancer Res. 2020;26:4326-4338. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 33] [Cited by in RCA: 49] [Article Influence: 9.8] [Reference Citation Analysis (0)] |
| 21. | Bankhead P, Loughrey MB, Fernández JA, Dombrowski Y, McArt DG, Dunne PD, McQuaid S, Gray RT, Murray LJ, Coleman HG, James JA, Salto-Tellez M, Hamilton PW. QuPath: Open source software for digital pathology image analysis. Sci Rep. 2017;7:16878. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 5567] [Cited by in RCA: 5010] [Article Influence: 626.3] [Reference Citation Analysis (0)] |
| 22. | Taieb J, Tabernero J, Mini E, Subtil F, Folprecht G, Van Laethem JL, Thaler J, Bridgewater J, Petersen LN, Blons H, Collette L, Van Cutsem E, Rougier P, Salazar R, Bedenne L, Emile JF, Laurent-Puig P, Lepage C; PETACC-8 Study Investigators. Oxaliplatin, fluorouracil, and leucovorin with or without cetuximab in patients with resected stage III colon cancer (PETACC-8): an open-label, randomised phase 3 trial. Lancet Oncol. 2014;15:862-873. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 188] [Cited by in RCA: 211] [Article Influence: 19.2] [Reference Citation Analysis (0)] |
| 23. | Reichling C, Taieb J, Derangere V, Klopfenstein Q, Le Malicot K, Gornet JM, Becheur H, Fein F, Cojocarasu O, Kaminsky MC, Lagasse JP, Luet D, Nguyen S, Etienne PL, Gasmi M, Vanoli A, Perrier H, Puig PL, Emile JF, Lepage C, Ghiringhelli F. Artificial intelligence-guided tissue analysis combined with immune infiltrate assessment predicts stage III colon cancer outcomes in PETACC08 study. Gut. 2020;69:681-690. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 55] [Cited by in RCA: 84] [Article Influence: 16.8] [Reference Citation Analysis (0)] |
| 24. | Yoo SY, Park HE, Kim JH, Wen X, Jeong S, Cho NY, Gwon HG, Kim K, Lee HS, Jeong SY, Park KJ, Han SW, Kim TY, Bae JM, Kang GH. Whole-Slide Image Analysis Reveals Quantitative Landscape of Tumor-Immune Microenvironment in Colorectal Cancers. Clin Cancer Res. 2020;26:870-881. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 17] [Cited by in RCA: 44] [Article Influence: 7.3] [Reference Citation Analysis (0)] |
| 25. | Galon J, Mlecnik B, Bindea G, Angell HK, Berger A, Lagorce C, Lugli A, Zlobec I, Hartmann A, Bifulco C, Nagtegaal ID, Palmqvist R, Masucci GV, Botti G, Tatangelo F, Delrio P, Maio M, Laghi L, Grizzi F, Asslaber M, D'Arrigo C, Vidal-Vanaclocha F, Zavadova E, Chouchane L, Ohashi PS, Hafezi-Bakhtiari S, Wouters BG, Roehrl M, Nguyen L, Kawakami Y, Hazama S, Okuno K, Ogino S, Gibbs P, Waring P, Sato N, Torigoe T, Itoh K, Patel PS, Shukla SN, Wang Y, Kopetz S, Sinicrope FA, Scripcariu V, Ascierto PA, Marincola FM, Fox BA, Pagès F. Towards the introduction of the 'Immunoscore' in the classification of malignant tumours. J Pathol. 2014;232:199-209. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1121] [Cited by in RCA: 1076] [Article Influence: 97.8] [Reference Citation Analysis (0)] |
| 26. | Nearchou IP, Lillard K, Gavriel CG, Ueno H, Harrison DJ, Caie PD. Automated Analysis of Lymphocytic Infiltration, Tumor Budding, and Their Spatial Relationship Improves Prognostic Accuracy in Colorectal Cancer. Cancer Immunol Res. 2019;7:609-620. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 47] [Cited by in RCA: 63] [Article Influence: 10.5] [Reference Citation Analysis (0)] |
| 27. | Innocenti F, Ou FS, Qu X, Zemla TJ, Niedzwiecki D, Tam R, Mahajan S, Goldberg RM, Bertagnolli MM, Blanke CD, Sanoff H, Atkins J, Polite B, Venook AP, Lenz HJ, Kabbarah O. Mutational Analysis of Patients With Colorectal Cancer in CALGB/SWOG 80405 Identifies New Roles of Microsatellite Instability and Tumor Mutational Burden for Patient Outcome. J Clin Oncol. 2019;37:1217-1227. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 156] [Cited by in RCA: 240] [Article Influence: 40.0] [Reference Citation Analysis (0)] |
| 28. | Overman MJ, Lonardi S, Wong KYM, Lenz HJ, Gelsomino F, Aglietta M, Morse MA, Van Cutsem E, McDermott R, Hill A, Sawyer MB, Hendlisz A, Neyns B, Svrcek M, Moss RA, Ledeine JM, Cao ZA, Kamble S, Kopetz S, André T. Durable Clinical Benefit With Nivolumab Plus Ipilimumab in DNA Mismatch Repair-Deficient/Microsatellite Instability-High Metastatic Colorectal Cancer. J Clin Oncol. 2018;36:773-779. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1013] [Cited by in RCA: 1475] [Article Influence: 210.7] [Reference Citation Analysis (0)] |
| 29. | Kather JN, Pearson AT, Halama N, Jäger D, Krause J, Loosen SH, Marx A, Boor P, Tacke F, Neumann UP, Grabsch HI, Yoshikawa T, Brenner H, Chang-Claude J, Hoffmeister M, Trautwein C, Luedde T. Deep learning can predict microsatellite instability directly from histology in gastrointestinal cancer. Nat Med. 2019;25:1054-1056. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 992] [Cited by in RCA: 761] [Article Influence: 126.8] [Reference Citation Analysis (0)] |
| 30. | Echle A, Grabsch HI, Quirke P, van den Brandt PA, West NP, Hutchins GGA, Heij LR, Tan X, Richman SD, Krause J, Alwers E, Jenniskens J, Offermans K, Gray R, Brenner H, Chang-Claude J, Trautwein C, Pearson AT, Boor P, Luedde T, Gaisa NT, Hoffmeister M, Kather JN. Clinical-Grade Detection of Microsatellite Instability in Colorectal Tumors by Deep Learning. Gastroenterology. 2020;. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 210] [Cited by in RCA: 220] [Article Influence: 44.0] [Reference Citation Analysis (0)] |
| 31. | Danielsen HE, Hveem TS, Domingo E, Pradhan M, Kleppe A, Syvertsen RA, Kostolomov I, Nesheim JA, Askautrud HA, Nesbakken A, Lothe RA, Svindland A, Shepherd N, Novelli M, Johnstone E, Tomlinson I, Kerr R, Kerr DJ. Prognostic markers for colorectal cancer: estimating ploidy and stroma. Ann Oncol. 2018;29:616-623. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 41] [Cited by in RCA: 53] [Article Influence: 7.6] [Reference Citation Analysis (0)] |
| 32. | Kather JN, Krisam J, Charoentong P, Luedde T, Herpel E, Weis CA, Gaiser T, Marx A, Valous NA, Ferber D, Jansen L, Reyes-Aldasoro CC, Zörnig I, Jäger D, Brenner H, Chang-Claude J, Hoffmeister M, Halama N. Predicting survival from colorectal cancer histology slides using deep learning: A retrospective multicenter study. PLoS Med. 2019;16:e1002730. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 555] [Cited by in RCA: 492] [Article Influence: 82.0] [Reference Citation Analysis (0)] |
| 33. | Skrede OJ, De Raedt S, Kleppe A, Hveem TS, Liestøl K, Maddison J, Askautrud HA, Pradhan M, Nesheim JA, Albregtsen F, Farstad IN, Domingo E, Church DN, Nesbakken A, Shepherd NA, Tomlinson I, Kerr R, Novelli M, Kerr DJ, Danielsen HE. Deep learning for prediction of colorectal cancer outcome: a discovery and validation study. Lancet. 2020;395:350-360. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 332] [Cited by in RCA: 365] [Article Influence: 73.0] [Reference Citation Analysis (0)] |